CellBio
Vol.2 No.3(2013), Article ID:37014,14 pages DOI:10.4236/cellbio.2013.23017
Characterization of the Intracellular Distribution of Adenine Nucleotide Translocase (ANT) in Drosophila Indirect Flight Muscles
1Department of Biology, University of Vermont, Burlington, USA
2Berg Biosystems, Framingham, USA
3Department of Cancer Biology, University of Massachusetts Medical School, Worcester, USA
4Department of Molecular Physiology and Biophysics, University of Vermont, Burlington, USA
Email: *jvigorea@uvm.edu
Copyright © 2013 Vivek K. Vishnudas et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received July 25, 2013; revised August 22, 2013; accepted August 29, 2013
Keywords: Drosophila; Insect Flight Muscle; Adenine Nucleotide Translocase; Phosphagen System
ABSTRACT
Background: The high power output necessary for insect flight has driven the evolution of muscles with large myofibrils (primary energy consumers) and abundant mitochondria (primary energy suppliers). The intricate functional interrelationship between these two organelles remains largely unknown despite its fundamental importance in understanding insect flight bioenergetics. Unlike vertebrate muscle that relies on a phosphagen (creatine phosphate/creatine kinase) system to regulate high energy phosphate flux, insect flight muscle has been reported to lack mitochondrial arginine kinase (analogous to creatine kinase), a key enzyme that enables intracellular energy transport. Creatine kinase is known to interact with mitochondrial adenine nucleotide translocase (ANT) in the transfer of ADP and ATP into and out of the mitochondria. Results: Here, we use quantitative immunogold transmission electron microscopy to show that in Drosophila melanogaster indirect flight muscles (IFM), ANT is present in the mitochondria as well as throughout the myofibril. To confirm this unexpected result, we created a transgenic line that expresses a chimeric GFP-ANT protein and used an anti-GFP antibody to determine the intracellular distribution of the fusion protein in the IFM. Similar to results obtained with anti-ANT, the fusion GFP-ANT protein is detected in myofibrils and mitochondria. We confirmed the absence of arginine kinase from IFM mitochondria and show that its sarcomeric (i.e., intramyofibrillar) distribution is similar to that of ANT. Conclusions: These results raise the possibility that direct channeling of nucleotides between mitochondria and myofibrils is assisted by an ANT protein thereby circumventing the need for a phosphagen shuttle in the IFM. The myofibrillar ANT may represent a unique adaptation in the muscles that require efficient exchange of nucleotides between mitochondria and myofibrils.
1. Background
Insect flight is widely regarded as one of the most metabolically demanding forms of animal locomotion [1]. Respiratory rates that increase 50 - 100 fold from rest to flight have been reported for some insects [2]. In particular, insects that rely on high wing beat frequencies to achieve the power needed to sustain flight are known to have very high mass-specific metabolic rates in comparison to other animals. ATP turnover rates during flight are among the highest ever recorded [3]. The primary consumer of metabolic energy in muscle is myosin, the molecular motor that generates the forces for contraction. The Drosophila indirect flight muscle (IFM) myosin is especially adapted for very fast muscle speed, including a very low affinity for MgATP [4]. Thus a very high concentration of MgATP is required in the IFM to satisfy the energetic demand of flight and to compensate for myosin’s weak MgATP affinity. High MgATP concentration is ensured by the large and abundant mitochondria that occupy up to 37% of the cellular space in the flight muscle [5,6].
While the role of the mitochondria as the main energy supplier in insect muscle is well described, the mechanism by which the intracellular energy flow is established and maintained is less clear. The adenine nucleotide translocase (ANT) is a highly abundant integral protein of the inner mitochondrial membrane that plays an essential and well established role in cellular bioenergetics [7] as well as in apoptosis [8] and cell signaling [9]. ANT catalyzes the exchange of ADP for ATP across the inner mitochondrial membrane thereby coupling oxidative phosphorylation to cell metabolism. In vertebrates and some invertebrates, ANT fulfills this important energetic function through its interaction with mitochondrial creatine kinase (Mi-CK), an enzyme that transfers high energy phosphates to creatine, thus forming the important metabolic currency, creatine phosphate (CP) [10]. Mi-CK works in conjunction with a soluble CK isoform, MM-CK, as part of the “phosphagen shuttle” responsible for transporting CP from mitochondria through the cytosol to major sites of ATP catalysis such as the myofibril and sarcoplasmic reticulum. This system creates metabolic capacitance by means of spatial and temporal buffering [6,11].
In Drosophila IFM, as in other insect muscles, the phosphagen system consists of arginine phosphate (AP) and arginine kinase (AK) that is assumed to operate in a manner analogous to the CK-CP system in vertebrates [12]. However, one study has questioned the existence of a mitochondrial AK isoenzyme raising the possibility that the AK-AP system may function in a manner fundamentally different from the better characterized CK-CP system [13]. Unlike vertebrate skeletal muscle, sarcoplasmic reticulum is very scarce in Drosophila IFM given its asynchronous mode of contraction. Thus, very little energy is expended on calcium cycling allowing the metabolic energy to be channeled preferentially to the myofibril, the site of contractile activity. How metabolic energy utilization by the myofibril is matched effectively to its production by the mitochondria remains largely unexplored.
Drosophila ANT is encoded by two tandem duplicated genes, stress sensitive B (sesB) and ANT2 [14]. Functional ANT activity in Drosophila adults is encoded by sesB [15]. Mutations in sesB cause paralysis and loss of synaptic transmission in the visual system [16] and lead to failure of synaptic vesicle cycling at the neuromuscular junction during periods of high demand [17]. Like the synapse, the IFM is likely to rely heavily on ANT for metabolic capacitance. Here, we have generated transgenic Drosophila strains that express a Green fluorescent protein-sesB (GFP-sesB) fusion protein in order to gain insight into the function of ANT in the IFM. We present new evidence of a myofibril-associated ANT that may be involved in facilitating the transport of ATP and ADP into and out of the myofibril, respectively. We also confirm earlier results that Drosophila IFM lack a mitochondrial AK isozyme. The study provides new molecular details of a potentially novel pathway of metabolic energy exchange involving tight functional coupling between myofibrils and mitochondria in the IFM.
2. Results
2.1. ANT Localizes to the Mitochondria and the Myofibrils of Drosophila IFM
A rabbit polyclonal antibody generated against bovine cardiac ANT was used to examine the distribution of Drosophila ANT in the IFM. Drosophila and mammalian ANT share ~80% amino acid sequence identity [18, 19]. The antibody has been reported to cross react with mouse ANT [20] and we confirmed, by western blot analysis, the specificity of the antibody for Drosophila sesB-encoded ANT (not shown, but see Figure 3). Studies on adult fly frozen sections revealed strong immunostaining of IFM compared to the jump muscle and other thoracic muscles (data not shown). We then performed electron microscopy on sections of the IFM to examine the intracellular distribution of ANT (Figure 1(A)). Gold particles were detected in the mitochondria and in the myofibril in about equal distribution, but were largely absent in the cytosol (Figure 1(D)). In contrast, a b ATP synthase antibody (Figures 1(B) and (E)) specifically labels the mitochondria and a flightin antibody (Figures 1(C) and (F)) labels the myofibrillar A band, as expected. Two negative controls, pre-immune rabbit serum and no primary antibody (pAG10 only) showed no gold distribution (data not shown).
2.2. Expression of GFP-sesB Fusion Protein in Drosophila Schneider Cells and Transgenic IFM
To confirm the myofibrillar localization of ANT in the IFM, we made transformation vectors encoding ANT with an N-terminal GFP tag. Zhang et al. reported that two adult transcripts of 1.6 KB and 1.2 KB generated from sesB result from alternative 3’ ends [18]. We conducted RT-PCR of total mRNA from late pupae and found two similar size transcripts. DNA sequence analysis of the PCR products confirmed that the differences were only in the 3' UTRs (results not shown). To ensure that the GFP tag does not alter the mitochondrial localization of ANT, we transfected Drosophila Schneider cells with pCaSpeRhs GFP-sesB 1.6 KB and pCaSpeRhs GFP-sesB 1.2 KB constructs. Figure 2 shows confocal images of tissue culture cells labeled with anti-GFP rabbit polyclonal antibodies and with Mitotracker. Both constructs express fusion proteins that are localized to the mitochondria, confirming that the addition of an N-terminal GFP tag does not affect protein targeting.
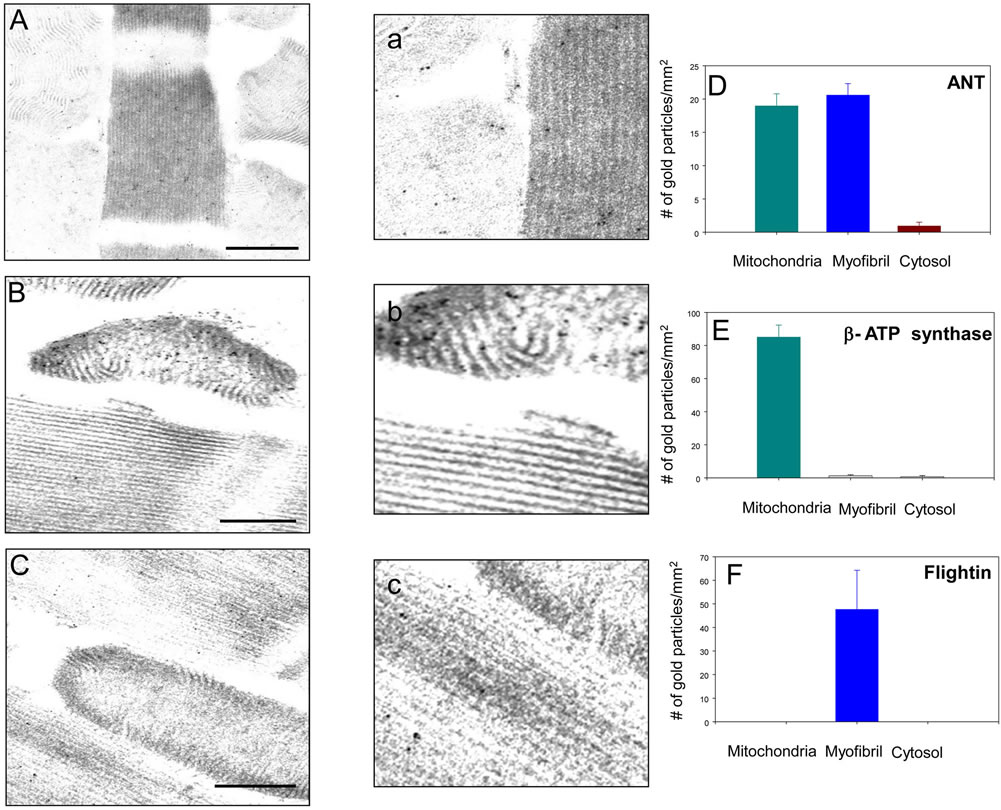
Figure 1. ANT is present in the mitochondria and myofibrils of Drosophila IFM. ((A), (a), and (D)) Intracellular distribution of ANT in the IFM determined with an anti-bovine cardiac ANT antibody (Bar = 200 nm). Quantification of immunogold particles revealed the dual localization of ANT (n = 12); ((B), (b), and (E)) Immunogold labeling of IFM with an anti-β ATP synthase antibody (Bar = 200 nm). Quantification of gold particle distribution shows a strictly mitochondrial localization (n = 12); ((C), (c), and (F)) Immunogold labeling with an anti-Drosophila flightin antibody (Bar = 200 nm). Quantification of gold particle distribution shows a strictly myofibrillar localization (n = 12). Panels a, b, and c are magnifications of a myofibril-mitochondria interface of panels (A), (B), and (C), respectively.
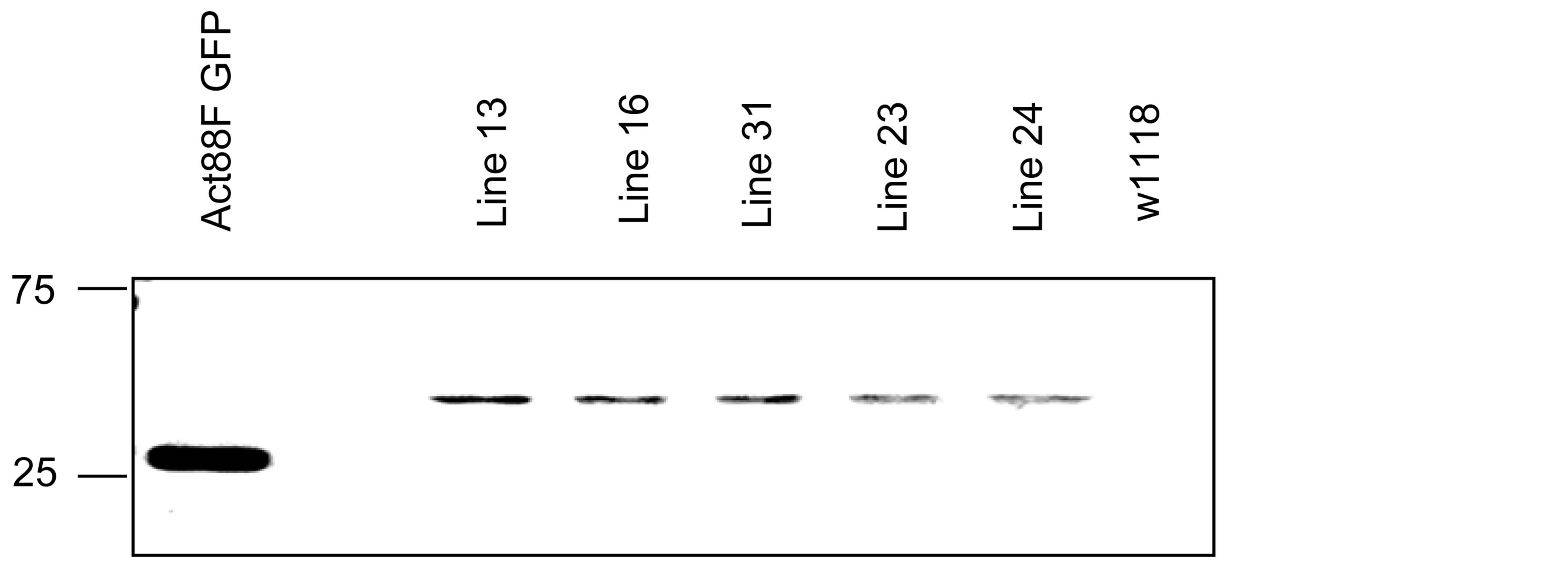
We confirmed the specificity of the anti-bovine cardiac ANT antisera for endogenous Drosophila sesB-encoded protein and GFP chimeric protein by western blot analysis of proteins extracted from the IFM of 5 transgenic lines and 2 control lines. The transgene is inserted in chromosome 1 (lines 13, 16, and 31), chromosome 2 (line 23), or chromosome 3 (line 24). The expression of the fusion protein is demonstrated by western blot with anti-GFP rabbit antibody (Figure 3(a)). Figure 3(b) is a western blot of 12% SDS-PAGE gel containing total thoracic proteins from transgenic and control lines. The anti-bovine cardiac ANT antibody cross reacts with an endogenous ~30 kD protein expressed in all transgenic and control lines, and with a ~57 kD protein expressed only in transgenic lines. The latter matches the expected size of the GFP-sesB fusion protein. In addition, the results show that the fusion protein expression is highest in line 31, where it is expressed at levels comparable to or higher than the endogenous ANT protein. Figure 3(c) is a Sypro stained 12% SDS-PAGE gel showing the expression of the fusion proteins in the five transgenic lines. Note that a band of ~57 kD appears most prominently in the heat shocked induced lane of line 31 suggesting that this line expresses the highest levels of fusion protein, in agreement with the western blot results. Line 31 was therefore selected for further study.
We characterized the localization of GFP-sesB fusion protein in line 31 by immunogold transmission electron
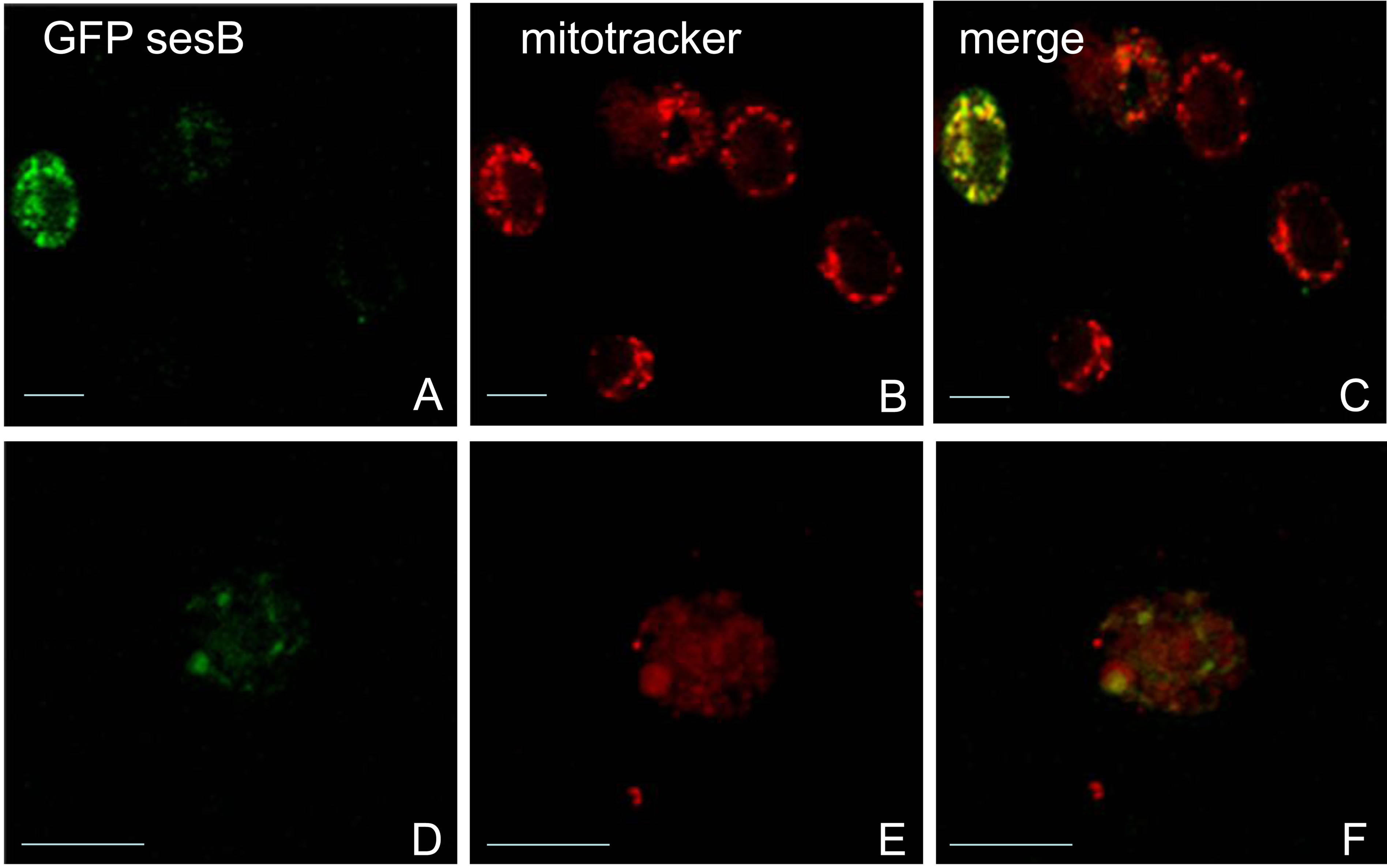
Figure 2. Confocal images of Schneider cells transfected with ANT expression vectors. ((A), (B), (C)) cells transfected with pCaSpeR hs GFP-sesB 1.6 KB ((D), (E), (F)) cells transfected with pCaSpeR hs GFP-sesB 1.2 KB. Protein expression was induced by heat shock. Cells were treated with anti-GFP antisera ((A) and (D)) or mitotracker ((B) and (E)). (C) and (F) are merged images from the two channels. Both constructs target GFP-ANT fusion protein to the mitochondria. Bar = 10 μm.
microscopy with an anti-GFP antibody. As controls, we used an Act88F-GFP strain, a line that expresses GFP in the IFM driven by the Act88F promoter [21], and the w1118 parental strain. As seen in Figure 4(A), GFP labeling is found in all three sub-compartments of the muscle cell, highest in the myofibril and about equal distribution in the cytosol and mitochondria. To determine if this pattern results from a random distribution, we calculated the relative cellular area covered by each sub-compartment. In the images examined, myofibrils occupy ~70% of the area, while mitochondria and cytosol occupy ~17% and 12%, respectively (n = 8). The particle counts approximate the percent area covered by each sub-compartment (Figure 4(D)) suggesting that GFP distribution is random. In contrast, GFP-sesB shows a non-random distribution with approximately equal counts in myofibrils and mitochondria and low counts in the cytosol (Figures 4(B) and (E)). This distribution resembles the one seen with the anti-bovine cardiac ANT antibody (Figure 1(A)). Very few gold particles were detected in the control line w1118 (Figures 4(C) and (F)), and additional controls using pre-immune rabbit serum and pAG10 on line 31 did not show any gold distribution in the IFM (data not shown).
Several studies have shown that slow (oxidative) skeletal and cardiac muscle ANT are functionally coupled to Mi-CK [22,23]. Microfractionation studies of Drosophila IFM have shown that AK is found primarily in the cytosolic fraction and not associated with the mitochondria [13]. We performed immunoelectron microscopy with a rabbit antiDrosophila AK polyclonal antibody to extend the study of Wyss et al. [13] and to determine if the myofibrillar localization of ANT is associ-
 (a)
(a)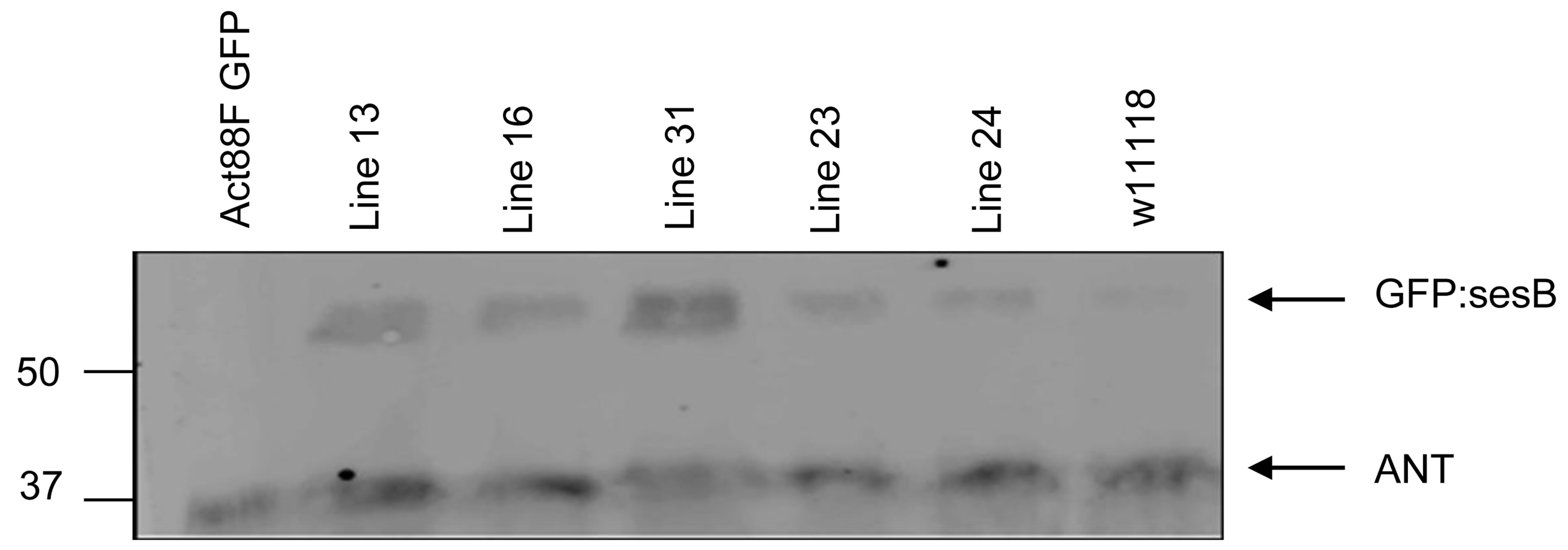 (b)
(b)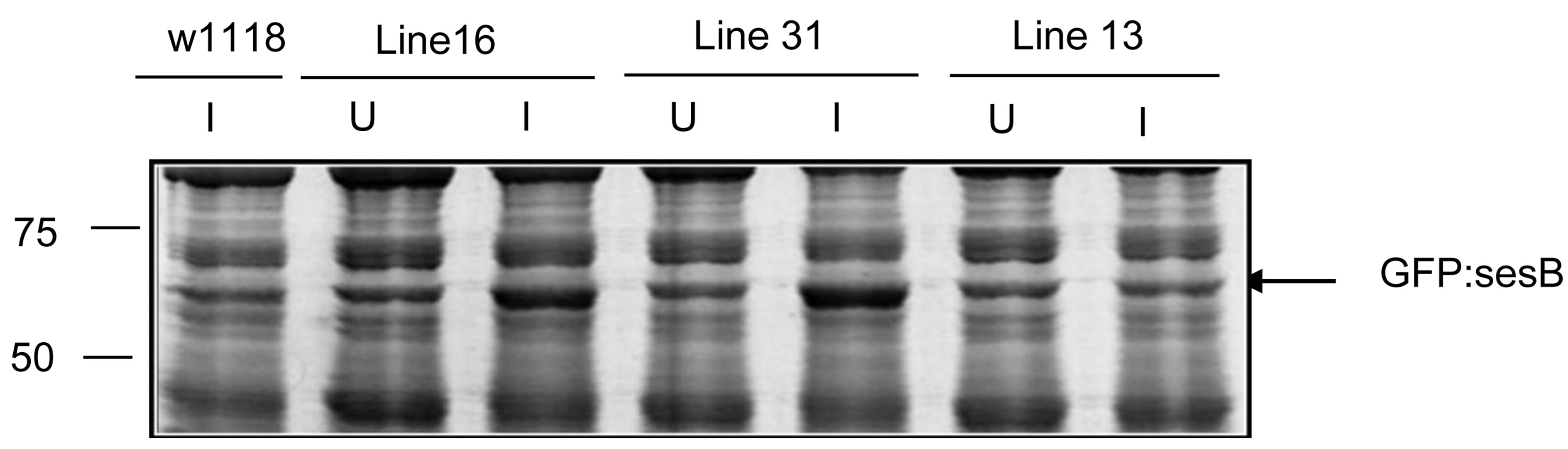 (c)
(c)
Figure 3. Expression of GFP-sesB fusion protein in transgenic lines. (a) Western blot showing expression levels of heat shocked induced GFP-sesB fusion protein in whole thorax protein extracts from transgenic and control lines using an anti-GFP antibody. The antibody detects a 30 kD band in the Act88F-GFP transgenic line corresponding to GFP. An ~57 kD band detected in lines 13, 16, 31, 23, and 24 correspond to the GFP-sesB fusion protein. No bands are detected in the non-transgenic w1118 sample; (b) Western blot showing expression levels of heat shocked induced GFP-sesB fusion protein analyzed in whole thorax protein extracts from transgenic and control lines using an antibovine cardiac ANT antibody. The antibody cross reacts with both endogenous ANT (30 kD band in all lanes) and GFP-sesB fusion protein (57 kD band in lines 13, 16, 31, 23, and 24; (c) Sypro stained gel analysis of thorax proteins from uninduced (U) and heat shock induced (I) expression of GFP-ANT transgene. Note prominent band in induced lane of lines 16 and 31 (arrow).
ated with AK. As seen in Figure 5, most of the AK localizes to the myofibril. Unlike CK in vertebrate slow skeletal and cardiac muscle, and consistent with the results of Wyss et al. [13], Drosophila AK is largely excluded from the mitochondria. We further looked at the sarcomeric distribution of ANT and AK. Figure 6 shows that ANT and AK are primarily distributed throughout the A band. There is no difference in the gold particle distribution for the two treatments (p = 0.827, Kruskal Wallis test) suggesting that ANT and AK may co-localize in the sarcomere. In contrast, the distribution of GFP differs significantly from that of ANT (p < 0.001), AK (p < 0.001), and GFP-sesB (p < 0.05).
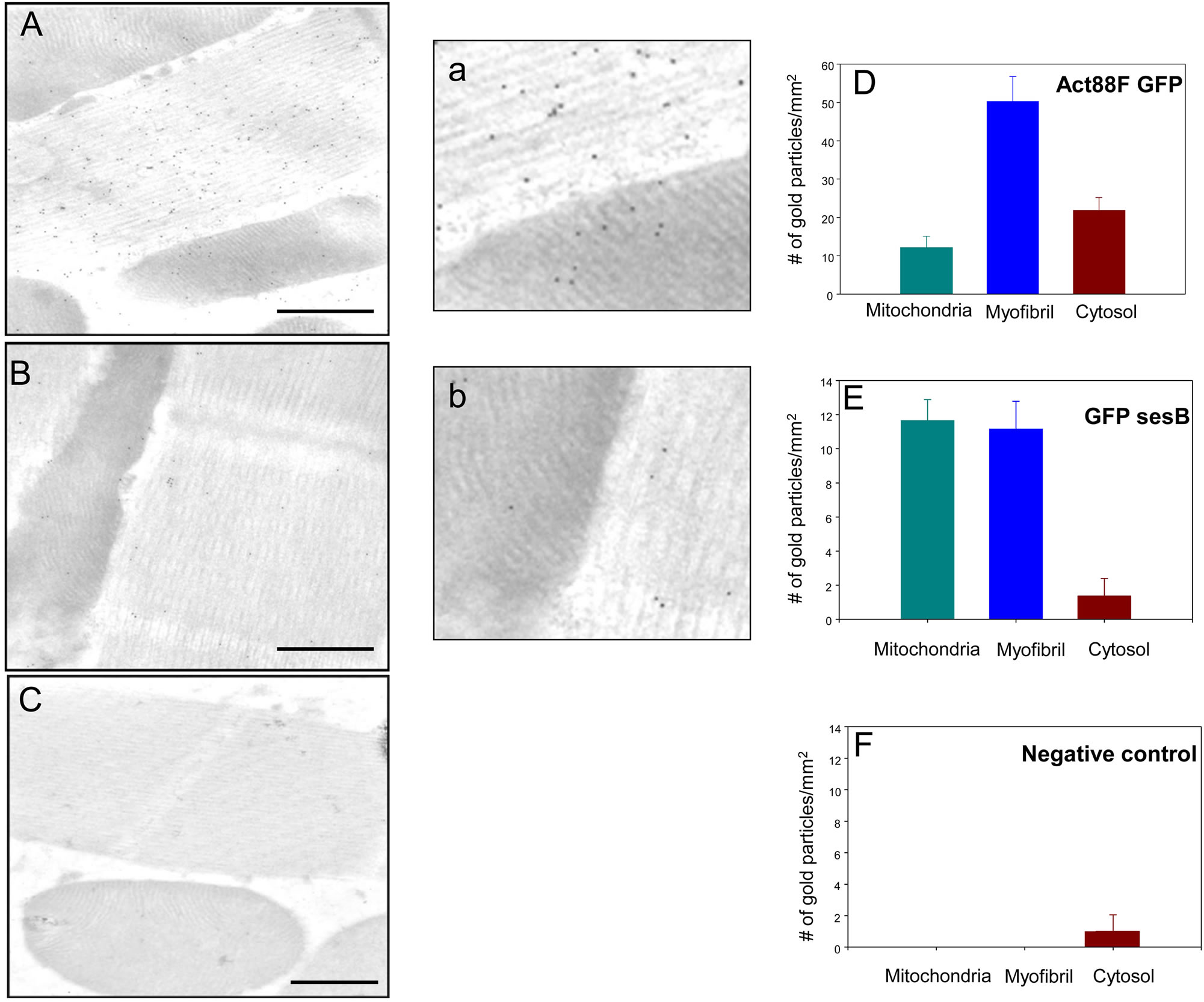
Figure 4. GFP-sesB is present in mitochondria and myofibrils of Drosophila IFM. Distribution of GFP signals in ((A), (a)) Act88F-GFP transgenic control line, ((B), (b)) transgenic Line 31, and (C) w1118 non-transgenic control line. All sections were probed with an anti-GFP antibody (Bar = 200 nm). (D) Quantification of gold particle distribution shows GFP is distributed randomly in transgenic control line (n = 12). (E) Quantification of gold particles shows GFP-sesB is preferentially targeted to mitochondria and myofibrils in transgenic line 31 (n = 12). This distribution pattern parallels that of ANT in wild-type flies (compare to Figures 1(A) and (D)). (F) No GFP signal is detected in the w1118 control line, confirming the specificity of the antibody. Panels a and b are magnifications of a myofibril-mitochondria interface of panels A and B, respectively.
3. Discussion
ANT is a resident protein of the inner mitochondrial membrane and research over the past several years have shown that ANT interacts with a variety of proteins in fulfilling multiple roles in energy metabolism, apoptosis, and cellular signaling [24-27]. As a component of the multiprotein permeability transition pore complex (PTPC), ANT interacts with proteins of the outer mitochondrial membrane, most notably the voltage dependent anion channel (VDAC), as well as with multiple proteins involved in metabolism and cell signaling [28]. These ANT-mediated protein interactions promote functional interplay between the mitochondria and other organelles [28]. Physical contacts have been described between the mitochondria, the endoplasmic reticulum [29,30], the sarcoplasmic reticulum [31], and adherens complexes [32], but it remains to be established if ANT forms part of the molecular connection that links mitochondria with its partners. A function for ANT outside the mitochondria has not been described. However, other mitochondrial proteins have been shown to fulfill roles outside this organelle including cytochrome c [33], ATP synthase β [34], and VDAC [35].
Here we confirmed the results of Zhang et al. that
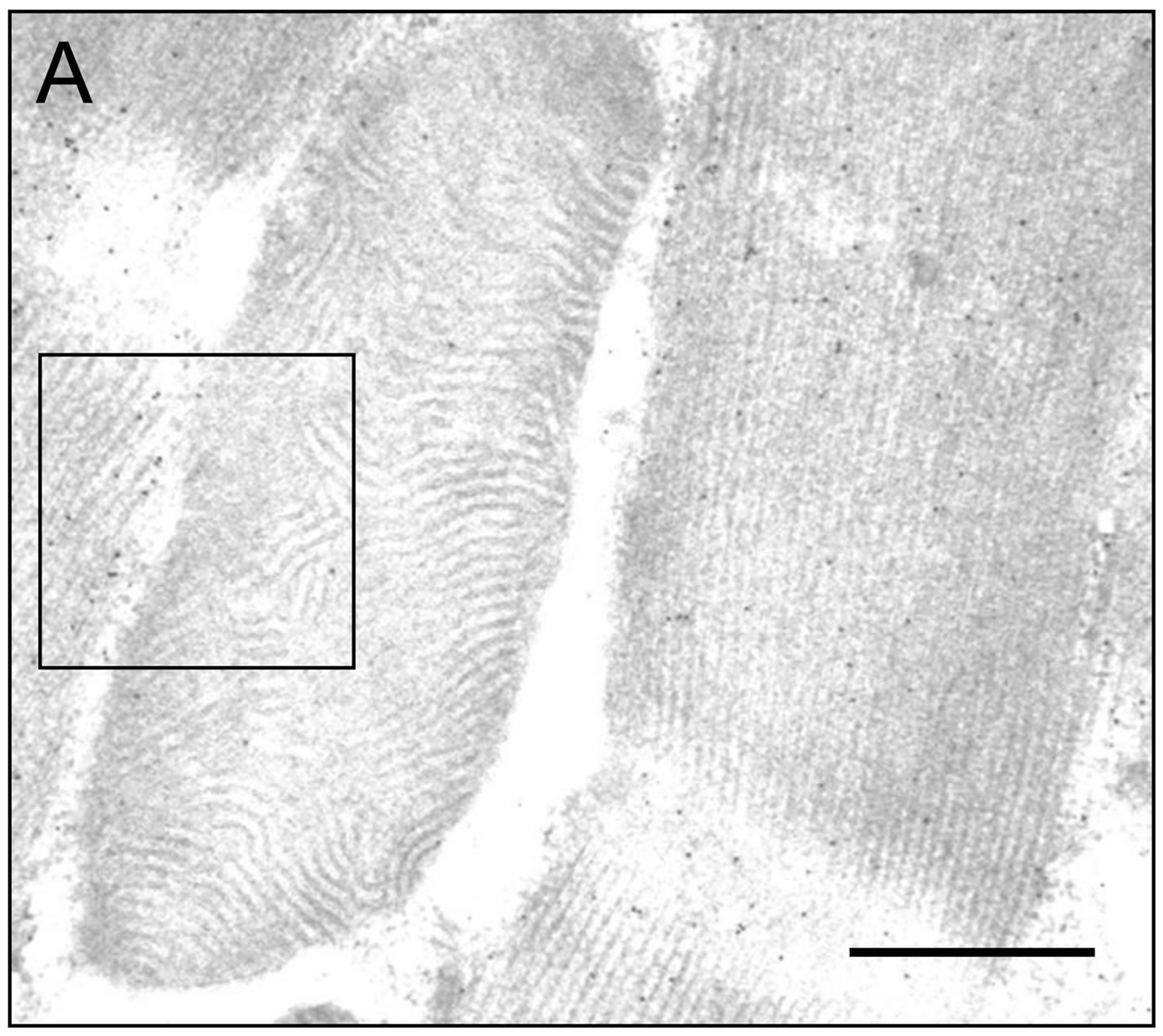
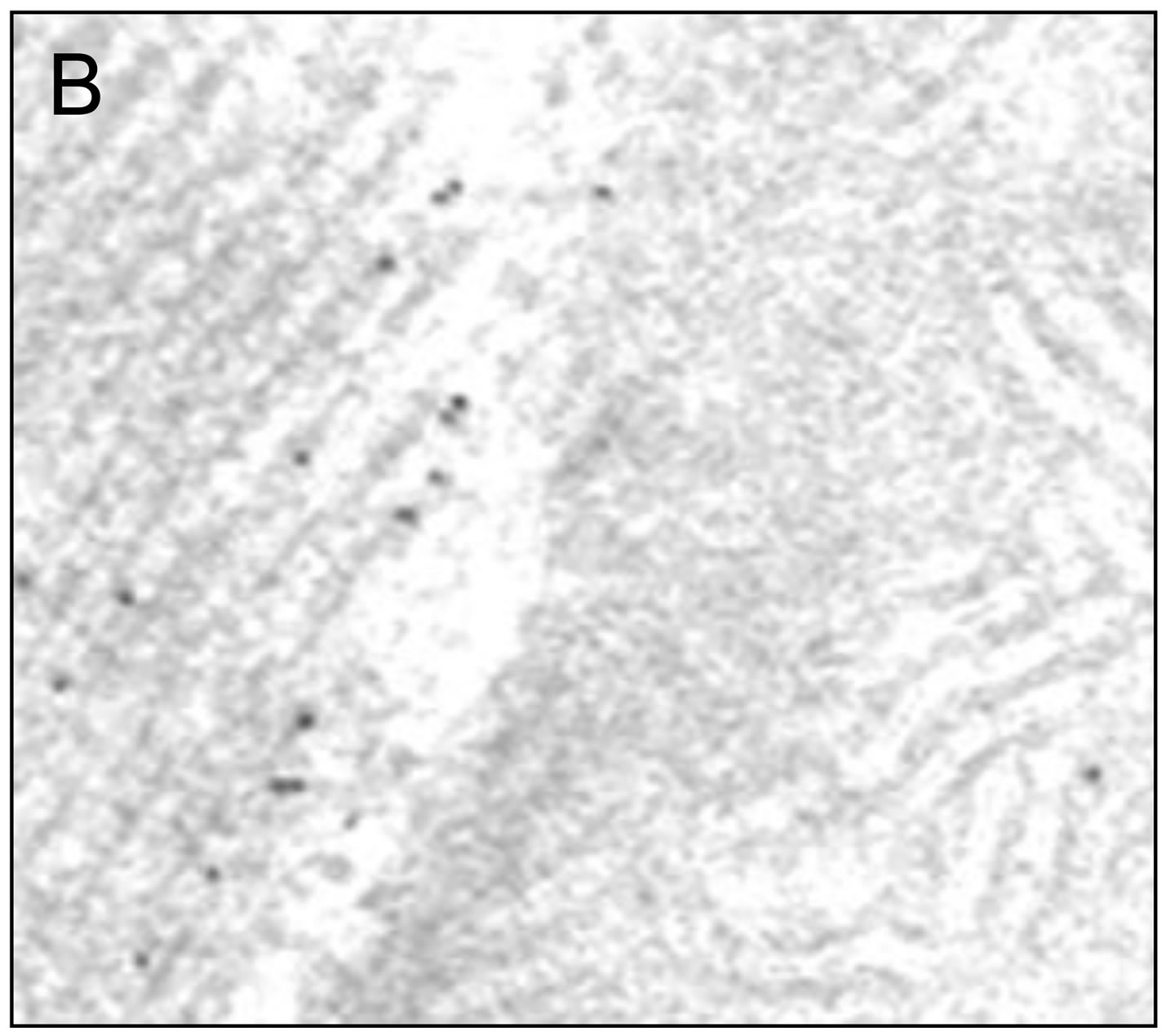
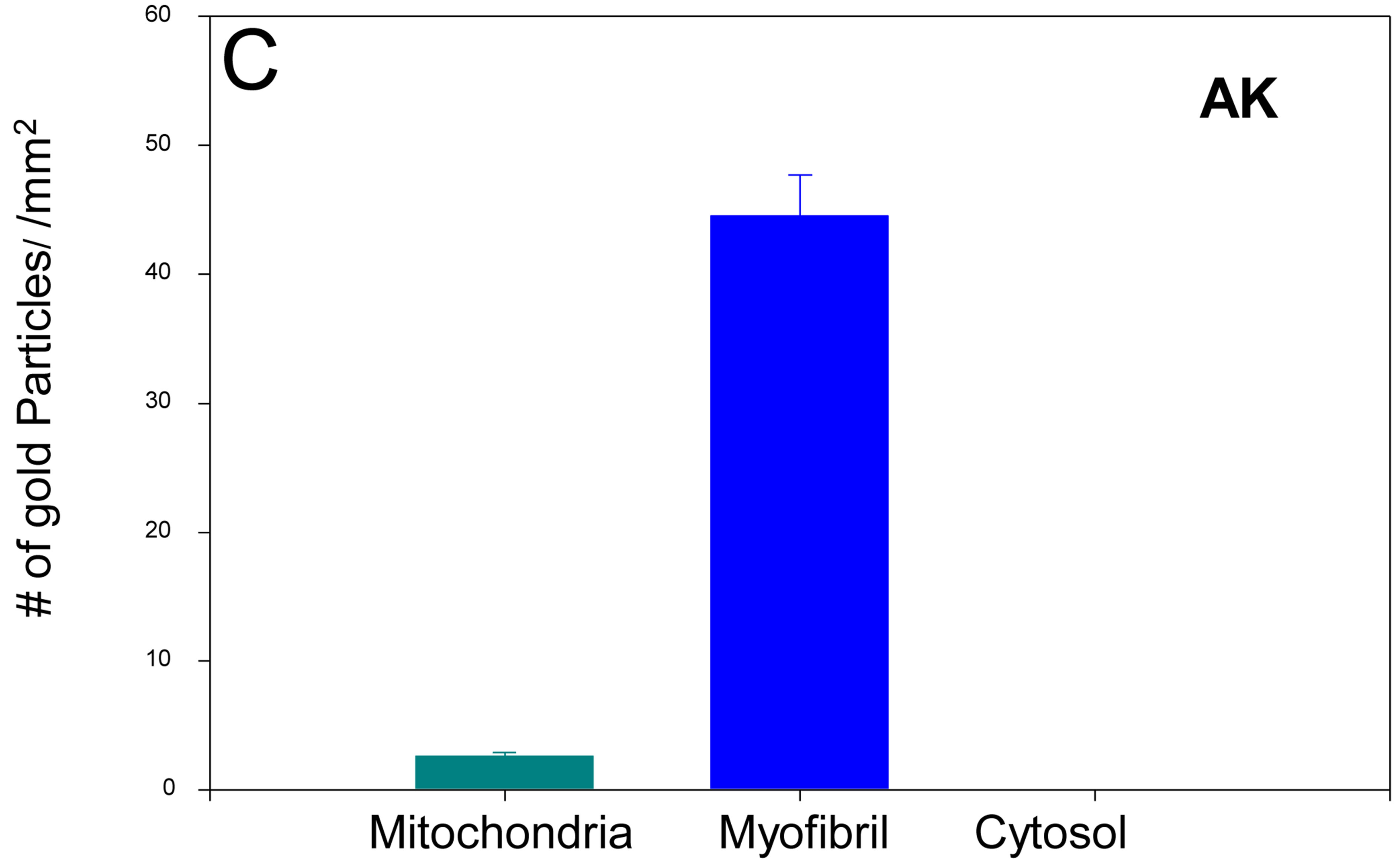
Figure 5. IFM Arginine kinase is predominantly localized to the myofibrils. (A) IFM section treated with an antiDrosophila arginine kinase antibody (Bar = 200 nm); (B) Boxed area in (A) shown in greater magnification. Note the majority of gold particles localize to the myofibril; (C) Quantification of gold particle distribution demonstrates that arginine kinase is predominantly associated with the myofibril (n = 6).
Drosophila sesB encodes two transcripts of ~1.2 KB and 1.6 KB that differ in their 3’ UTR [14]. The functional role of the different 3’ UTRs is not known but one possibility is that cis elements within the UTR determine the intracellular distribution of ANT. sesB transcripts, like those of other nuclear genome encoded mitochondrial proteins, rely on 3’ UTR cis elements for their targeting
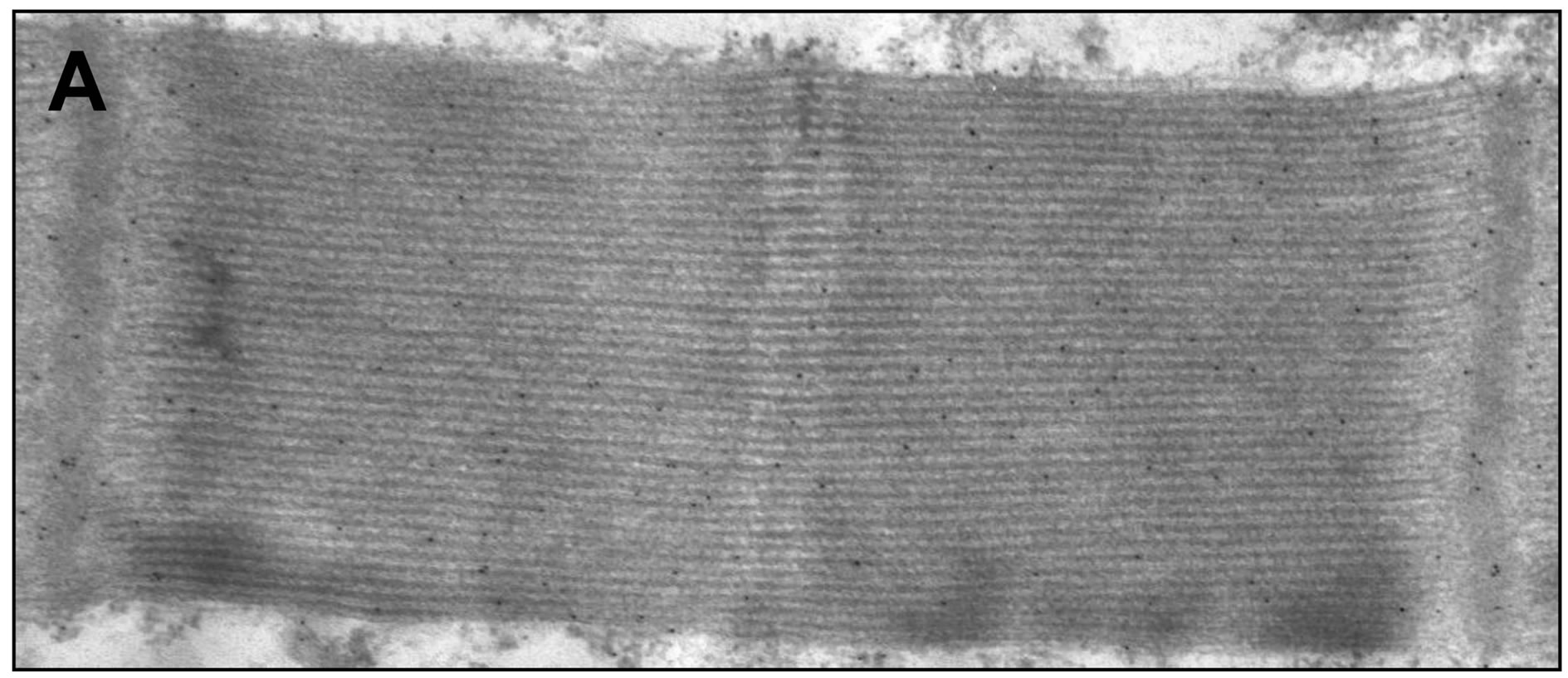
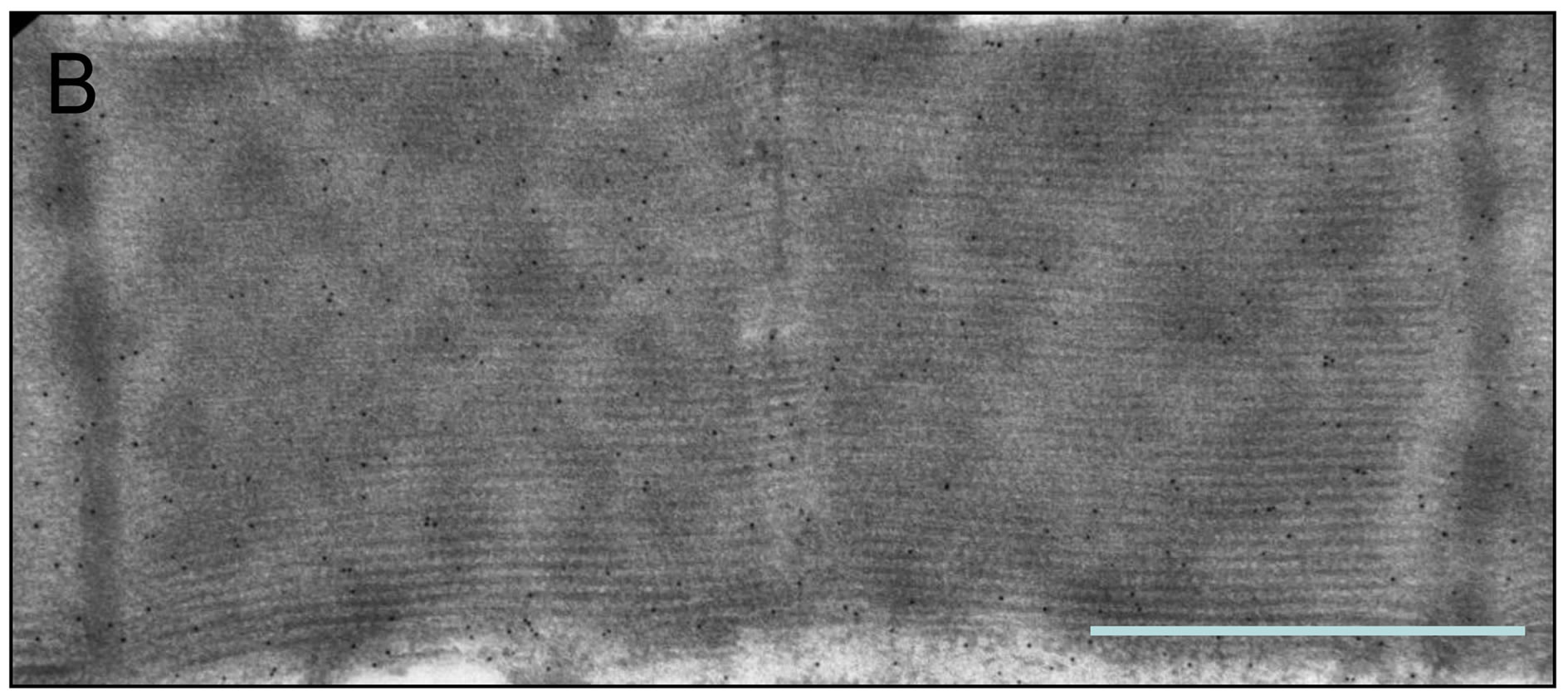
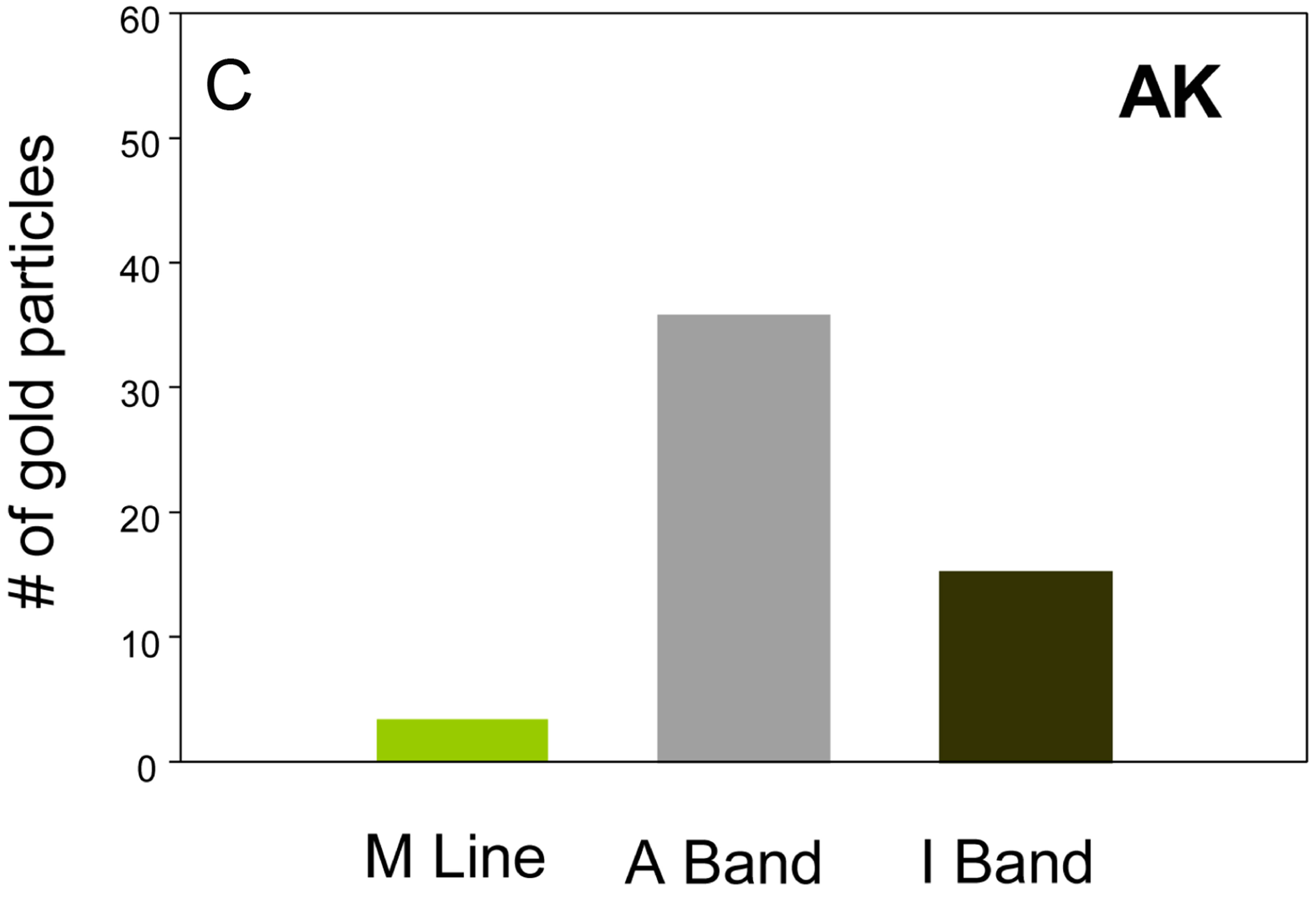
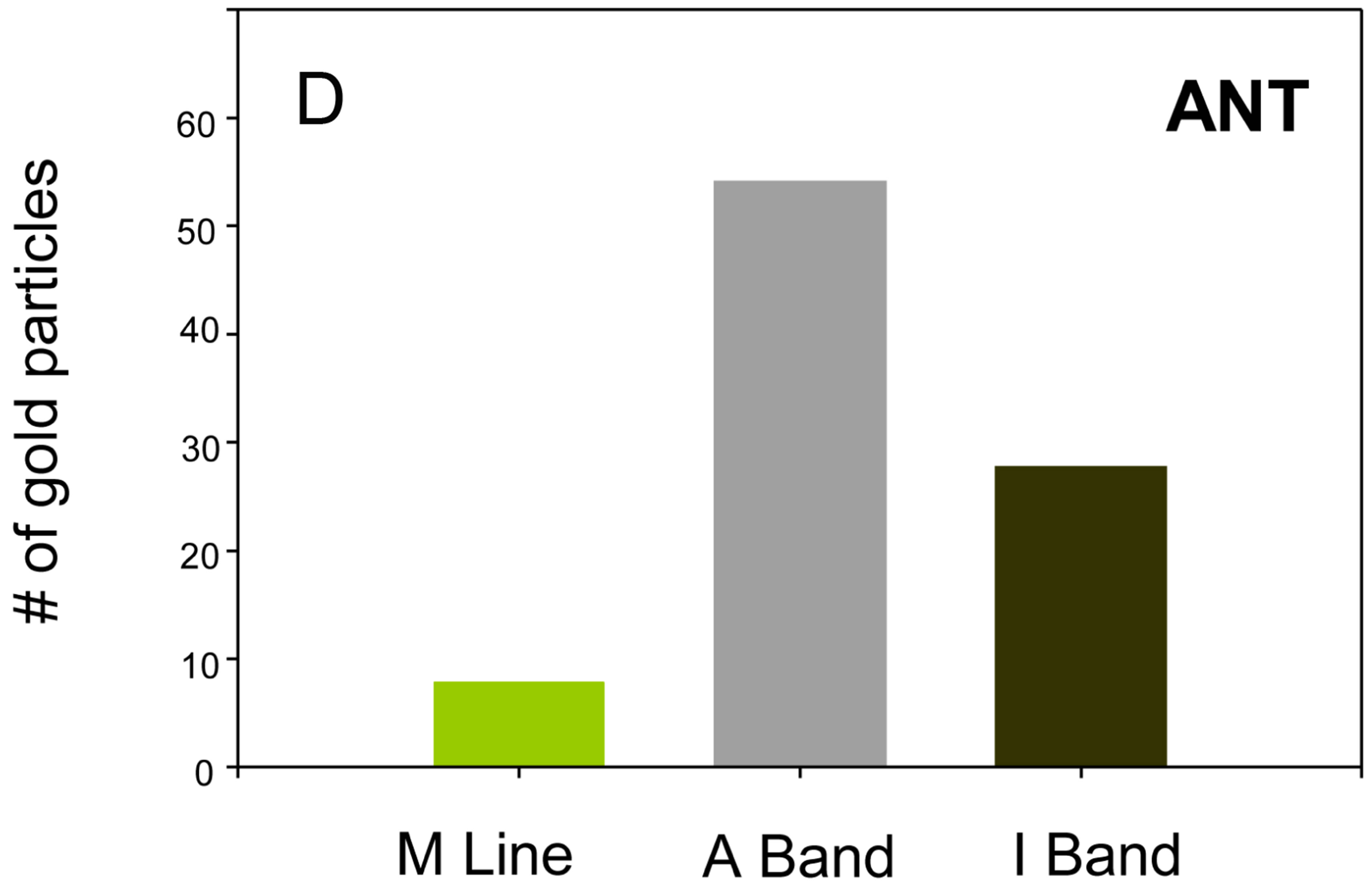
Figure 6. Sarcomeric distribution of AK ((A) and (C)) and ANT ((B) and (D)). The two proteins have similar sarcomeric distributions. Bar = 1 μm.
to polysomes associated with the outer mitochondrial membrane [36]. The first 150 nucleotides of the 3’ UTRs are common to both sesB transcripts; the 1.6 KB transcript has an additional 440 nucleotides downstream of the shared region [14]. The shared region contains a sequence element (uguaaaua) that has been identified as a mitochondrial localization motif [37]. As shown here in Schneider S2 cultured cells, both transcripts target GFPsesB to the mitochondria. Preliminary secondary structure analysis revealed that the mitochondria localization motif in the 1.6 KB transcript may form base pairs with other internal sequences and therefore would not be accessible to binding proteins involved in mitochondrial targeting. Sequence motifs within the additional 440 nucleotide of the 1.6 KB transcript 3’ UTR may override the mitochondrial localization signal. Of particular note is the cis element “acaccc” motif that has been identified as a binding site for MLP1 (the human homologue of Drosophila Muscleblind protein) [38]. This motif is required for the specific targeting of integrin alpha3 protein to distinct cytoplasmic loci [38]. Muscleblind in Drosophila is involved in alternative RNA splicing and is also required for terminal differentiation of muscle [39]. Further experiments will be needed to determine if sequences within the 3’UTR of the 1.6 KB transcript are responsible for an extra-mitochondrial ANT.
The functional coupling between ANT and CK in vertebrate muscles has been the subject of many studies (e.g., [10,22]. By comparison, little is known about the putative interaction between ANT and AK in non-vertebrate muscle [12]. Available evidence suggest that insect flight muscle lacks a mitochondrial AK [12,13]. However, a mitochondrial AK has been identified in the cardiac muscle of the horseshoe crab and the blue crab [40]. While a direct interaction between AK and ANT has not been demonstrated, the localization of mitochondrial AK to the intermembrane space and the outer region of the inner membrane in the crab would be conducive for such interaction to occur.
The AK/AP phosphagen system is likely to play similar physiological roles as the CK/CP phosphagen system, namely: 1) a spatio-temporal “buffering” role for adenine nucleotides, fulfilled by the cytoplasmic enzyme isoform, and 2) a ‘”shuttle” role for energy transfer from sites of energy production (mitochondria) to sites of energy consumption (e.g., myofibril) [23,41]. The shuttle function relies on the presence of a mitochondrial isoform at the production end and a myofibril isoform at the utilization end. Fast skeletal muscle express the cytosolic CK isoform almost exclusively, while slow skeletal muscle and heart ventricular muscle express higher proportions of mitochondrial and myofibrillar CK isoforms [23]. The functional organization of the AK/AP system in Drosophila IFM is not known, but given the highly oxidative nature of this muscle and the physical proximity of mitochondria to myofibrils, one would expect the system would more likely be analogous to that of vertebrate slow and cardiac muscle than to the one in skeletal muscle. However, we have confirmed the results of Wyss et al. [13] that Drosophila IFM lacks a mitochondrial AK, consistent with results of others who have found insect flight muscle lack mitochondrial AK [42]. It remains possible that a mitochondrial AK exists (as originally reported, [43]) that is not recognized by the antibodies used in this study and by Wyss et al. While only one AK gene has been identified in the Drosophila genome, this gene is predicted to encode six different polypeptides (flybase.org). Based on microfractionation studies of IFM fibers, Wyss et al. concluded that most of the AK in Drosophila IFM is cytosolic [13]. In contrast, Lang et al. identified AK associated with the Z line of washed myofibrils [44]. Our immunoelectron microscopy results show that most of the AK is associated with the sarcomeric A band, with no detectable distribution in the cytosol. These seemingly contradictory results can be explained if the association of AK with the myofibril is weak and fails to endure the fractionation and isolation procedures used in prior investigations. Coincidently, glycerination of Lethocerus IFM fibers has been shown to cause migration of zeelins from the A band to the Z band [45]. We speculate that the myofibril isolation procedure used by Lang et al. may have, as in the case for zeelins, lead to a redistribution of AK from the A band, as detected in this study, to the Z band given that our immunolocalization studies were conducted on IFM fixed within the intact thorax immediately following bisection. The redistribution is not unexpected assuming the interaction between AK and the myofibril is purely electrostatic, as has been shown for vertebrate CK and the sarcomeric M line [46].
The absence of a mitochondrial AK isoform would indicate that the organization of the AK/AP system in insect IFM is fundamentally different from the analogous CK/CP system in vertebrate muscle. Unlike vertebrate skeletal muscle, insect flight muscle is regarded to be functionally independent of phosphagens as evidenced by the low levels of AP in resting muscle and by low AK activity [47,48]. Nevertheless, the AK reaction plays a small role in providing metabolic capacitance during sustained flight [11,49,50]. Additionally, myofibrillar AK may be involved in phosphate (Pi) removal as increasing levels of Pi decreases work output at low ATP concentrations [4].
There is an interesting parallel between the situation in IFM and that of mice that are deficient for CK. In gastrocnemius (fast skeletal) muscle of these mice, mitochondria are more abundant than in the muscle of normal mice and are relocated toward the A band, in an apparent attempt to minimize the diffusion distance of nucleotides as a way to overcome the absence of mitochondrial and cytosolic CK [51]. Cellular remodeling presumably to enhance direct nucleotide channeling between mitochondria and organelles also has been observed in the cardiac cells of CK deficient mice [23]. We suspect that the close apposition of mitochondria and myofibrils in the IFM compensates for the lack of a cytosolic buffering phosphagen system and a mitochondrial AK, as seen in the CK deficient mice.
The IFM phosphagen system evolved in conjunction with other features of the IFM that underlie this muscle’s ability for fast and sustained contractions and high power output. Among these are a myosin motor with weak affinity for ATP [4], abundant mitochondria in close apposition to the myofibrils [6], and broad myofibrils with an increased number of force producing thick filaments [5]. These modifications impose certain challenges that the cellular energetic system must overcome in order for the muscle to operate effectively. Principal among these is the large diffusion distance for metabolites in the cell and more specifically, within the confines of the myofibril. Fibers from asynchronous flight muscle have been reported to have a larger cross-sectional area than fibers from synchronous flight muscle [5]. A similar relationship is seen for myofibrils, those from asynchronous muscle have approximately twice the diameter and four times the cross-sectional area as those from synchronous muscle [5]. In crustacean tail muscle, limitations in phosphagen diffusion increase with increases in fiber size [52] and this raises the possibility that asynchronous muscles may face similar limitations. Previous studies have shown that ADP is the most diffusion constrained of all nucleotides [53]. It has been estimated that direct diffusion (i.e., in the absence of cytoplasmic CK) of ADP and ATP would compromise metabolic capacity for diffusion lengths greater than 2 μm [54]. This would be significant in the Drosophila IFM whose myofibrils have a diameter of nearly 2 μm. Taking these factors together we consider that diffusion of ATP and ADP alone provides a rudimentary, if any, metabolic capacitance in the IFM and therefore propose the existence of active mechanisms in these fast and powerful muscles.
The weak affinity of IFM myosin for MgATP implies that a very high concentration of this nucleotide must be present in the IFM, in the vicinity of the myosin motors. Based on skinned fiber mechanics experiments, a MgATP concentration of ~15 mM is required to attain maximal frequency of maximum work [4]. Given the limited sarcoplasmic reticulum in the IFM, and therefore limited requirement to feed SERCA pumps, most of the ATP can be channeled to the myofibril. We have no information on how ATP/ADP and AP/arginine are distributed throughout the IFM fiber but given the complexity of the cytoplasm and the high organelle occupancy, the distribution is unlikely to be homogeneous. The existence of compartmentalized pools and direct channeling of nucleotides has been demonstrated in mouse heart [55]. More recently, the existence of structural barriers that restrict the diffusion of nucleotides and metabolites in heart muscle fibers has been supported by three dimensional finite element models [56]. The large surface area and relatively large radius of IFM myofibrils could impose a significant diffusion constraint on ATP, ADP and arginine phosphate.
A myofibril-associated ANT could assist in creating and maintaining differences in ATP and ADP concentrations across compartmentalized microdomains. While mitochondria are abundant and in close proximity to the myofibril, their ATP contribution may be spatially restricted to the periphery and other means of delivering ATP deep within the confines of the myofibril may be required. In addition to weak MgATP affinity, other kinetic properties of IFM myosin appear to dictate the need for a highly efficient mechanism for nucleotide transport within the myofibril. Unlike skeletal muscle, the ratelimiting step of IFM myosin is phosphate release, i.e., the rate of ADP release is very fast [4]. ADP is characterized by an inherently slow diffusion rate that in cardiac muscle is further restricted by intracellular structures and biochemical processes [57]. Thus the need to prevent pools of high ADP concentration to build up in the vicinity of the myosin motors may be as important as maintaining high ATP concentrations. While myosin motor driven fluctuations may assist in diffusing ADP, the presence of ANT may facilitate directed transfer to the mitochondria that in turn stimulates mitochondrial oxidative phosphorylation. Our observation that ANT and AK have similar cellular and myofibrillar distribution patterns raises the possibility that these proteins might directly and more efficiently buffer ADP and provide sufficient ATP required for sustaining muscle contractions at high frequencies. Further experiments will be needed to establish if and how ANT and AK, operating separately or together, contribute to nucleotide and phosphagen diffusion in the IFM. A direct transfer of nucleotides between ANT and AK would be analogous to that proposed for ANT and CK [22].
4. Conclusions
In this study we used two complementary approaches, one direct and one indirect, to examine the intracellular distribution of ANT in Drosophila IFM. We created a transgenic line expressing a GFP-sesB fusion protein and showed, using anti-GFP specific antibodies and quantitative immunoelectron microscopy, that the fusion protein is equally distributed between mitochondria and myofibrils. The distribution of the fusion protein is similar to that of the endogenous ANT observed using anti-ANT specific antibodies. The novel finding of a myofibril localized ANT is consistent with the results of a prior study that found ANT to be present among the proteins of an IFM myofibrillar preparation [58]. Furthermore, we showed that AK is primarily localized to the sarcomeric A band and is excluded from the mitochondria.
In summary, the quest for bigger myofibrils, necessary to achieve the high levels of power output required for flight, may have propelled the evolution of energy buffering systems within the confines of the myofibril. Our results add to the growing body of evidence that phosphagen-based energy systems are highly adaptable and remarkably versatile in meeting the cellular energetic demands of specific muscle types. These findings provide a new perspective on the versatility of cellular energy transfer pathways as their adaptation to meet the energetic demands of an ultrafast, strictly oxidative and high power muscle. More studies will be needed to fully understand how energy production and utilization are functionally integrated in insect flight muscle.
5. Methods
5.1. Fly Stocks
Drosophila melanogaster w1118, an otherwise wild type strain except for a white eye mutation, was obtained from the Bloomington Stock Center and was used as host for generation of transgenic lines and as a control line. w/w; T(2;3) apxa/ Cyo;TM3 Sb e strain was used for mapping the insertion in the transgenic lines. Act88F-GFP line [21], obtained from John Sparrow, was used as control line for GFP expression while Oregon R was used as the wild-type strain.
5.2. RT-PCR and Cloning
In order to generate an N-terminal GFP tag, an eGFP clone (Clonetech), was modified by Polymerase Chain Reaction (PCR) to remove the stop codon. The forward primer 5’ attaccatggtgagcaagggc 3’ with a NcoI restriction site (shown in bold) and a reverse primer 5’ attatagcggccgccttgctt 3’ with a Not I was used to amplify the coding region of the eGFP clone. The PCR product was cut with NcoI and Not I and cloned into an eGFP vector that was cut with the same combination of enzymes.
Total mRNA was isolated from wild type late stage pupae using Trizol (Invitrogen) and quantified using a Nanodrop spectrophotometer (Thermo Scientific). RTPCR of sesB mRNA was performed using anchored oligo dTs [59]. The oligo dT primers 5’ actagt-t(16)-t 3’, 5’ actagt-t(16)-a3’, 5’ actagt-t(16)-g3’, 5’ actcgt-t(16)-c3’ were used for first strand synthesis. These primers have a single nucleotide 3’ anchor (bold italics) preceded by oligo dT and the Spe I sequence. First strand synthesis was carried out using superscript II RNase H− Reverse transcriptase (Invitrogen) and each of the anchored oligo dTs. PCR was carried out using a forward primer that was specific to the sesB sequence at its 3’ end and had a Not I restriction site at the 5’ end. Using this combination of the forward primer and the anchored oligo dT, the sesB cDNAs that includes its 3’ UTR was amplified and cloned into the modified GFP vector, immediately down stream of the GFP coding sequence using the Not I and Spe I restriction site so that GFP and sesB cDNAs are in a continuous reading frame. The GFP-sesB cDNA with Sma I and Spe I ends was subcloned into a modified pCaSpeR-hs, P-element transformation vector digested with Hpa I and Xba I. The pCaSpeR-hs vector was modified by BamHI digestion and the cut vector was religated so as to eliminate the hsp70 3’ UTR. All ligations were performed using the clonables ligation/transformation kit (Novagen, Madison, WI).
5.3. Generation of Transgenic Lines and Heat Shock Treatment of Flies
The pCaSpeR-hs GFP-sesB vector was amplified by transformation into chemically competent Top10 cells (Invitrogen) and purified using the Qiagen endotoxin free Maxi prep kit (Qiagen, Valencia, CA, USA). The DNA was checked for purity and approximately 2 mg of DNA was shipped to Microinjection Services Inc (Sudbury, MA) for transformation. w1118 strain transformants were identified by orange eye color in the G1 generation. Homozygous strains were generated by single pair mating of siblings with the darkest eye color until no white eye progeny were produced. Each transgene was mapped to its resident chromosome using w/w; T(2;3) apxa/Cyo; TM3 Sb e strain by standard crossing techniques. Five independent lines were generated.
To induce expression of the transgene, early larvae from homozygous transgenic lines were heat shocked by placing vials in a 37˚C water bath for 15 minutes followed by a five-minute incubation at room temperature. This was repeated at least 4 times every day until late pupae stages.
5.4. Preparation of Samples for Gel Electrophoresis and Western Blot Analysis
To prepare samples for gel electrophoresis, flies were placed in acetone overnight at −20˚C. The next morning the acetone was removed and the flies spun for 1 hour in a speed vac after which the thorax was separated from the head, abdomen, wings and legs. The thorax was homogenized in Laemmli sample buffer with 8 mol·L−1 urea and protease inhibitors [60]. Electrophoresis on 12% polyacrylamide SDS denaturing gels were performed as described previously [60] using the discontinuous buffer system [61]. The gels were stained overnight in sypro stain (BIORAD) and viewed using an UV light box and photographed with a digital camera (Kodak Digital Science EDAS 120). For western blots, samples were separated on a 12% SDS gel and transferred onto nitrocellulose as described previously [60]. Membranes were blocked in blocking buffer (LICOR Biosciences) for one hour after which they were incubated with one of the following primary antibodies diluted in 1:1 of blocking buffer and TBST; 1) Anti-bovine cardiac ANT antibodies (Rabbit polyclonal, 1:2000), provided by Gerard Brandolin; 2) Anti-GFP polyclonal antibodies from Molecular Probes (Rabbit polyclonal, 1:2000); 3) Anti-Drosophila bATP synthase antibodies (Rabbit polyclonal, 1:2000) provided by Rafael Garesse. Blots were incubated in primary antibody for one hour at room temperature followed by three washes for 15 minutes each in TBST (Tris Buffer Saline 0.1% Tween). The blots were incubated in secondary anti-rabbit or anti-mouse Alexafluor 698 (Molecular Probes, Eugene, OR, USA) for one hour and then were washed in TBST and scanned on an Odyssey fluorescent scanner (LI-COR Bioscience, Lincoln, NE, USA). The images were converted from color to gray scale in Adobe Photoshop.
5.5. Insect Cell Culture, Transient Transfections and Heat Shock Treatment of Cells
Drosophila Schneider (S2) cells were cultured in 60 × 15 mm style Falcon culture plates (Becton Dickinson, Franklin Lakes, NJ, USA) in Shield and Sank–M3 Medium until 70% to 80% confluence was attained. S2 cells were grown in a 25˚C incubator. For transfection, 10 µg of pCaSpeR hs GFP-sesB1.6 K or pCaSpeR hs GFP-1.2 K were used. Transfections were performed using the Cellfectin reagent (Invitrogen,). Heat shock was administered by leaving cultures in a water bath at 37˚C for 30 minutes after which cells were moved back into 25˚C and allowed to recover for two hours before being processed for confocal microscopy.
5.6. Confocal Microscopy of Drosophila Schneider Cells
Cells were spun down at 4000 RPM for 5 minutes, permeabilized with 0.2% triton X-100 in 0.01 M phosphate buffer PH 7.4 for 5 minutes at room temperature, and fixed with 3.7% formaldehyde in 0.01 M phosphate buffer pH 7.4 for 15 minutes at room temperature. They were washed in 0.01 M PBS three times to remove formaldehyde and triton X-100. Coverslips overlayed with Cell Tak (BD Biosciences) in 3 M NaHCO3 were prepared, cells were applied to the cover slip and incubated at room temperature for 3 minutes. The mitochondria were stained with 25 nM Mitotracker Red CMX Ros (M7512, Molecular Probes). Subsequently, cells were stained with a primary anti-GFP polyclonal antibody (Molecular Probes, 1:250 dilution) for one hour at room temperature followed by secondary goat antirabbit IgG (H + L), Alexafluor 488. All light sensitive reactions were performed in a light proof chamber. Cells were finally washed in PBS for one minute to remove Mitotracker and antibodies. A drop of mounting medium (Vecta shield, Vector Laboratories, Inc, Burlingame, CA 94010) was placed on the coverslip and it was inverted on to a slide. Slides were viewed using the BioRad 1024 confocal laser scanning microscope. Lasersharp 2000 version 5 software (originally from Bio-Rad Laboratories, Hercules, CA, now part of Carl Zeiss Micro imaging, Thornwood, NY) was used for image analysis and processing.
5.7. Immunogold Transmission Electron Microscopy and Image Analysis
Drosophila half thoraces were fixed for one hour at 4˚C in 0.1% glutaraldehyde and 3.0% paraformaldehyde in 0.01M Phosphate Buffered Saline (PBS), pH 7.4. After rinsing in PBS 3 × 5 min each, free aldehyde groups were quenched by washing the tissue 2 × 30 min at 4˚C in 0.05 M ammonium chloride in PBS, followed by storage overnight at 4˚C in PBS. The next day, the tissue was dehydrated in ethanol at progressively lower temperatures and embedded in Lowicryl K4M at −35˚C. Polymerization of the blocks was achieved by exposure to ultraviolet light for 24 - 48 hrs at −35˚C, followed by 48 - 72 hrs at room temperature under UV light. Semi thin sections (1 µm) were cut with a glass knive on a Reichert ultracut microtome, stained with methylene blue–azure II, and evaluated for areas of interest. Ultrathin sections (60 - 80 nm) were cut with a diamond knife, retrieved onto Carbon-Formvar coated 150 mesh nickel grids. Immunohistochemistry experiments were performed as follows: The grids were blocked with 0.5% ovalbumin solution for 20 minutes at room temperature. The following primary antibodies were used. Anti-ANT polyclonal (1:500), anti-Drosophila βATP synthase polyclonal (1:250), antiDrosophila flightin polyclonal (1:30) [62], anti-Drosophila arginine kinase polyclonal (1:200) (provided by Theo Wallimann), and anti-GFP polyclonal (1:10 dilution). As a negative control, rabbit pre-immune serum was used since all primary antibodies were raised in rabbit. Sections were incubated in primary antibodies overnight at 4˚C and were washed 3 times for 5 minutes each in 1% BSA in PBS. Protein A conjugated to 10 nm gold particles (pAg10) (diluted in PBS, Optical Density of 0.06 @ 525 nm) was used for detection of primary antibodies. Sections were incubated in pAg10 diluted in PBS for 60 minutes at room temperature followed by washes in PBS and rinses in distilled water. Sections were then contrasted with uranyl acetate (3% in deionized H2O) and lead citrate, and examined with a JEOL 1210 TEM (JEOL USA, Inc., Peabody, Ma) operating at 60 kV. The negatives were scanned with an Epson scanner and the quantitation of gold spots using MetaMorph software (version 6.3 r7) was done as follows. The program was calibrated using standard files that take into account the magnification and size of the image. Random boxes were drawn on the image using the draw tool. Integrated morphometric analysis (IMA) function was used to calculate the area of each box (in µm2) and the number of gold dots per area. Individual smaller boxes were drawn within the first box such that each covered mitochondria, myofibril, or cytosol. The areas of these smaller boxes and the number of gold dots per area were calculated using the IMA function. All the gold particles counted were normalized to the area and plotted using Sigma plot. Two separate flies were done for each of the treatments (five antibodies and control) and each fly was probed in duplicate, for a total of four grids per treatment. Three frames were randomly photographed from each grid totaling 12 frames per treatment.
5.8. Statistical Analysis
All analyses were performed in MATLAB using nonparametric tests. To compare distributions of gold particles in the different groups, we converted raw data (counts) into percentage data. We then performed Kruskal Wallis tests on the percent data for all groups. Further tests using the RANKSUM function (equivalent to MannWhitney U test) were done to compare the distribution of the GFP control to the experimental groups.
6. Authors’ Contributions
VKV generated the transgenic lines and carried out the immuno EM studies and protein expression studies. He participated in the design of the study and the analysis of the data, and drafted the manuscript. SSG assisted in the characterization of the transgenic lines. PL provided overall technical assistance and participated in the immuno EM studies. DWM participated in the design and coordination of the study and helped draft the manuscript. JOV conceived of the study and had overall responsibility for its design and coordination. He assisted in data analysis and interpretation and drafting of the manuscript. All authors read and approved the final manuscript.
7. Acknowledgements
We thank Cedric Wesley for advice on the tissue culture and supplying S2 cells, and Teresa Ruiz and Michele Von Turkovich for assistance with electron microscopy. We are grateful to members of the Vigoreaux and Maughan labs for providing useful comments. We are indebted to the following individuals for supplying antibodies: Gerard Brandolin (ANT antibodies), Rafael Garesse (ATP synthase antibody) and David Sullivan (arginine kinase antibodies). Supported by NSF grants MCB-0315865, IOS-0718417 and MCB-1050834 to JOV. VKV was supported in part by a predoctoral fellowship of the Vermont Genetics Network through NIH Grant number 1 P20 RR16462 from the BRIN program of the National Center for Research Resources.
REFERENCES
- J. F. Harrison and S P. Roberts, “Flight Respiration and Energetics,” Annual Review of Physiology, Vol. 62, 2000, pp. 179-205. doi:10.1146/annurev.physiol.62.1.179
- A. E. Kammer and B. Heinrich, “Insect Flight Metabolism,” In: J. E. Treherne, M. J. Berridge and V. B. Wigglesworth, Eds., Advances in Insect Physiology, Vol. 13, Academic Press, London, 1978, pp. 133-228.
- T. M. Casey, C. P. Ellington and J. M. Gabriel, “Allometric Scaling of Muscle Performance and Metabolism: Insects,” Advances in Bioscience and Biotechnology, Vol. 84, 1992, pp. 152-162.
- D. M. Swank, V. K. Vishnudas and D. W. Maughan, “An Exceptionally Fast Actomyosin Reaction Powers Insect Flight Muscle,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 103, No. 46, 2006, pp. 17543-17547. doi:10.1073/pnas.0604972103
- R. K. Josephson, J. G. Malamud and D. R. Stokes, “Asynchronous Muscle: A Primer,” The Journal of Experimental Biology, Vol. 203, Pt. 18, 2000, pp. 2713- 2722.
- V. Vishnudas and J. O. Vigoreaux, “Sustained High Power Performance: Possible Strategies for Integrating Energy Supply and Demand in Flight Muscles,” In: J. O. Vigoreaux, Ed., Nature’s Versatile Engine: Insect Flight Muscle Inside and out, Springer/Landes Bioscience, New York, 2006, pp. 188-196. doi:10.1007/0-387-31213-7_15
- E. Pebay-Peyroula and G. Brandolin, “Nucleotide Exchange in Mitochondria: Insight at a Molecular Level,” Current Opinion in Structural Biology, Vol. 14, No. 4, 2004, pp. 420-425. doi:10.1016/j.sbi.2004.06.009
- A. Dorner and H. P. Schultheiss, “Adenine Nucleotide Translocase in the Focus of Cardiovascular Diseases,” Trends in Cardiovascular Medicine, Vol. 17, No. 8, 2007, pp. 284-290. doi:10.1016/j.tcm.2007.10.001
- J. D. Sharer, J. F. Shern, H. Van Valkenburgh and D. C. Wallace and R. A. Kahn, “ARL2 and BART Enter Mitochondria and Bind the Adenine Nucleotide Transporter,” Molecular Biology of the Cell, Vol. 13, No. 1, 2002, pp. 71-83. doi:10.1091/mbc.01-05-0245
- K. Guerrero, B. Wuyam, P. Mezin, I. Vivodtzev, M. Vendelin, J. C. Borel, R. Hacini, O. Chavanon, S. Imbeaud, V. Saks and C. Pison, “Functional Coupling of Adenine Nucleotide Translocase and Mitochondrial Creatine Kinase Is Enhanced after Exercise Training in Lung Transplant Skeletal Muscle,” American Journal of Physiology—Regulatory, Integrative and Comparative Physiology, Vol. 289, No. 4, 2005, pp. R1144-R1154. doi:10.1152/ajpregu.00229.2005
- H. L. Sweeney, “The Importance of the Creatine Kinase Reaction: The Concept of Metabolic Capacitance,” Medicine & Science in Sports & Exercise, Vol. 26, No. 1, 1994, pp. 30-36. doi:10.1249/00005768-199401000-00007
- W. R. Ellington, “Evolution and Physiological Roles of Phosphagen Systems,” Annual Review of Physiology, Vol. 63, 2001, pp. 289-325.
- M. Wyss, D. M. Maughan and T. Wallimann, “ReEvaluation of the Structure and Physiological Function of Guanidino Kinases in Fruitfly (Drosophila), Sea Urchin (Psammechinus miliaris) and Man,” Biochemical Journal, Vol. 309, Pt. 1, 1995, pp. 255-261. doi:10.1146/annurev.physiol.63.1.289
- Y. Q. Zhang, J. Roote, S. Brogna, A. W. Davis, D. A. Barbash, D. Nash and M. Ashburner, “Stress Sensitive B Encodes an Adenine Nucleotide Translocase in Drosophila melanogaster,” Genetics, Vol. 153, No. 2, 1999, pp. 891-903.
- M. D. Brand, J. L. Pakay, A. Ocloo, J. Kokoszka, D. C. Wallace, P. S. Brookes and E. J. Cornwall, “The Basal Proton Conductance of Mitochondria Depends on Adenine Nucleotide Translocase Content,” Biochemical Journal, Vol. 392, Pt. 2, 2005, pp. 353-362. doi:10.1042/BJ20050890
- R. Rikhy, M. Ramaswami and K. S. Krishnan, “A Temperature-Sensitive Allele of Drosophila sesB Reveals Acute Functions for the Mitochondrial Adenine Nucleotide Translocase in Synaptic Transmission and Dynamin Regulation,” Genetics, Vol. 165, No. 3, 2003, pp. 1243- 1253.
- N. Trotta, C. K. Rodesch, T. Fergestad and K. Broadie, “Cellular Bases of Activity-Dependent Paralysis in Drosophila Stress-Sensitive Mutants,” Journal of Neurobiology, Vol. 60, No. 3, 2004, pp. 328-347. doi:10.1002/neu.20017
- Y. Q. Zhang, J. Roote, S. Brogna, A. W. Davis, D. A. Barbash, D. Nash and M. Ashburner, “Stress Sensitive B Encodes an Adenine Nucleotide Translocase in Drosophila melanogaster,” Genetics, Vol. 153, No. 2, 1999, pp. 891-903.
- A. Louvi and S. G. Tsitilou, “A cDNA Clone Encoding the ADP/ATP Translocase of Drosophila melanogaster Shows a High Degree of Similarity with the Mammalian ADP/ATP Translocases,” Journal of Molecular Evolution, Vol. 35, No. 1, 1992, pp. 44-50. doi:10.1007/BF00160259
- A. Grado, C. Manchado, R. Iglesias, M. Giralt, F. Villarroya, T. Mampel and O. Vinas, “Muscle/Heart Isoform of Mitochondrial Adenine Nucleotide Translocase (ANT1) Is Transiently Expressed during Perinatal Development in Rat Liver,” FEBS Letters, Vol. 421, No. 3, 1998, pp. 213- 216. doi:10.1016/S0014-5793(97)01563-9
- P. Barthmaier and E. Fyrberg, “Monitoring Development and Pathology of Drosophila Indirect Flight Muscles Using Green Fluorescent Protein,” Developmental Biology, Vol. 169, No. 2, 1995, pp. 770-774. doi:10.1006/dbio.1995.1186
- M. Vendelin, M. Lemba and V. A. Saks, “Analysis of Functional Coupling: Mitochondrial Creatine Kinase and Adenine Nucleotide Translocase,” Biophysical Journal, Vol. 87, No. 1, 2004, pp. 696-713. doi:10.1529/biophysj.103.036210
- R. Ventura-Clapier, A. Kaasik and V. Veksler, “Structural and Functional Adaptations of Striated Muscles to CK Deficiency,” Molecular and Cellular Biochemistry, Vol. 256-257, No. 1-2, 2004, pp. 29-41. doi:10.1023/B:MCBI.0000009857.69730.97
- D. G. Brdiczka, D. B. Zorov and S. S. Sheu, “Mitochondrial Contact Sites: Their Role in Energy Metabolism and Apoptosis,” Biochimica et Biophysica Acta, Vol. 1762, No. 2, 2006, pp. 148-163.
- A. P. Halestrap and C. Brennerb, “The Adenine Nucleotide Translocase: A Central Component of the Mitochondrial Permeability Transition Pore and Key Player in Cell Death,” Current Medicinal Chemistry, Vol. 10, No. 16, 2003, pp. 1507-1525. doi:10.2174/0929867033457278
- S. M. Claypool, Y. Oktay, P. Boontheung, J. A. Loo and C. M. Koehler, “Cardiolipin Defines the Interactome of the Major ADP/ATP Carrier Protein of the Mitochondrial Inner Membrane,” The Journal of Cell Biology, Vol. 182, No. 5, 2008, pp. 937-950. doi:10.2174/0929867033457278
- R. Liu, A. L. Strom, J. Zhai, J. Gal, S. Bao, W. Gong and H. Zhu, “Enzymatically Inactive Adenylate Kinase 4 Interacts with Mitochondrial ADP/ATP Translocase,” The International Journal of Biochemistry & Cell Biology, Vol. 41, No. 6, 2009, pp. 1371-1380. doi:10.1016/j.biocel.2008.12.002
- F. Verrier, A. Deniaud, M. Lebras, D. Metivier, G. Kroemer, B. Mignotte, G. Jan and C. Brenner, “Dynamic Evolution of the Adenine Nucleotide Translocase Interactome during Chemotherapy-Induced Apoptosis,” Oncogene, Vol. 23, No. 49, 2004, 8049-8064. doi:10.1038/sj.onc.1208001
- G. Csordas, C. Renken, P. Varnai, L. Walter, D. Weaver, K. F. Buttle, T. Balla, C. A. Mannella and G. Hajnoczky, “Structural and Functional Features and Significance of the Physical Linkage between ER and Mitochondria,” The Journal of Cell Biology, Vol. 174, No. 7, 2006, pp. 915- 921. doi:10.1083/jcb.200604016
- R. Rizzuto, P. Pinton, W. Carrington, F. S. Fay, K. E. Fogarty, L. M. Lifshitz, R. A. Tuft and T. Pozzan, “Close Contacts with the Endoplasmic Reticulum as Determinants of Mitochondrial Ca2+ Responses,” Science, Vol. 280, No. 5370, 1998, pp. 1763-1766. doi:10.1126/science.280.5370.1763
- J. Dai, K. H. Kuo, J. M. Leo, C. van Breemen and C. H. Lee, “Rearrangement of the Close Contact between the Mitochondria and the Sarcoplasmic Reticulum in Airway Smooth Muscle,” Cell Calcium, Vol. 37, No. 4, 2005, pp. 333-340. doi:10.1016/j.ceca.2004.12.002
- K. C. Rowland, N. K. Irby and G. A. Spirou, “Specialized Synapse-Associated Structures within the Calyx of Held,” The Journal of Neuroscience, Vol. 20, No. 24, 2000, pp. 9135-9144.
- X. Jiang and X. Wang, “Cytochrome C-Mediated Apoptosis,” Annual Review of Biochemistry, Vol. 73, 2004, pp. 87-106. doi:10.1146/annurev.biochem.73.011303.073706
- L. O. Martinez, S. Jacquet, J. P. Esteve, C. Rolland, E. Cabezon, E. Champagne, T. Pineau, V. Georgeaud, J. E. Walker, F. Terce, et al., “Ectopic Beta-Chain of ATP Synthase Is An Apolipoprotein A-I Receptor in Hepatic HDL Endocytosis,” Nature, Vol. 421, No. 6918, 2003, pp. 75-79. doi:10.1038/nature01250
- R. Buettner, G. Papoutsoglou, E. Scemes, D. C. Spray and R. Dermietzel, “Evidence for Secretory Pathway Localization of a Voltage-Dependent Anion Channel Isoform,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 97, No. 7, 2000, pp. 3201-3206. doi:10.1073/pnas.97.7.3201
- J. Sylvestre, A. Margeot, C. Jacq, G. Dujardin and M. Corral-Debrinski, “The Role of the 3’ Untranslated Region in mRNA Sorting to the Vicinity of Mitochondria Is Conserved from Yeast to Human Cells,” Molecular Biology of the Cell, Vol. 14, No. 9, 2003, pp. 3848-3856. doi:10.1091/mbc.E03-02-0074
- R. Shalgi, M. Lapidot, R. Shamir and Y. Pilpel, “A Catalog of Stability-Associated Sequence Elements in 3’ UTRs of Yeast mRNAs,” Genome Biology, Vol. 6, 2005, p. R86. doi:10.1186/gb-2005-6-10-r86
- Y. Adereth, V. Dammai, N. Kose, R. Li and T. Hsu, “RNA-Dependent Integrin Alpha3 Protein Localization Regulated by the Muscleblind-Like Protein MLP1,” Nature Cell Biology, Vol. 7, No. 12, 2005, pp. 1240-1247.
- L. Machuca-Tzili, H. Thorpe, T. E. Robinson, C. Sewry and J. D. Brook, “Flies Deficient in Muscleblind Protein Model Features of Myotonic Dystrophy with Altered Splice Forms of Z-Band-Associated Transcripts,” Human Genetics, Vol. 120, No. 4, 2006, pp. 487-499. doi:10.1007/s00439-006-0228-8
- A. J. Pineda and W. R. Ellington, “Immunogold Transmission Electron Microscopic Localization of Arginine Kinase in Arthropod Mitochondria,” Journal of Experimental Zoology, Vol. 281, No. 2, 1998, pp. 73-79. doi:10.1002/(SICI)1097-010X(19980601)281:2<73::AID-JEZ1>3.0.CO;2-7
- T. Wallimann, M. Wyss, D. Brdiczka, K. Nicolay and H. M. Eppenberger, “Intracellular Compartmentation, Structure and Function of Creatine Kinase Isoenzymes in Tissues with High and Fluctuating Energy Demands: The ‘Phosphocreatine Circuit’ for Cellular Energy Homeostasis,” The Biochemical Journal, Vol. 281, Pt. 1, 1992, pp. 21-40.
- W. R. Ellington and A. C. Hines, “Mitochondrial Activities of Phosphagen Kinases Are Not widely Distributed in the Invertebrates,” Biological Bulletin, Vol. 180, No. 3, 1991, pp. 505-507. doi:10.2307/1542352
- L. R. Munneke and G. E. Collier, “Cytoplasmic and Mitochondrial Arginine Kinases in Drosophila: Evidence for a Single Gene,” Biochemical Genetics, Vol. 26, No. 1-2, 1988, pp. 131-141. doi:10.1007/BF00555494
- A. B. Lang, C. Wyss and H. M. Eppenberger, “Localization of Arginine Kinase in Muscles Fibres of Drosophila melanogaster,” Journal of Muscle Research & Cell Motility, Vol. 1, No. 2, 1980, pp. 147-161. doi:10.1007/BF00711796
- C. Ferguson, A. Lakey, A. Hutchings, G. W. Butcher, K. R. Leonard and B. Bullard, “Cytoskeletal Proteins of Insect Muscle: Location of Zeelins in Lethocerus Flight and Leg Muscle,” Journal of Cell Science, Vol. 107, Pt. 5, 1994, pp. 1115-1129.
- T. Hornemann, M. Stolz and T. Wallimann, “IsoenzymeSpecific Interaction of Muscle-Type Creatine Kinase with the Sarcomeric M-Line Is Mediated by Nh2-Terminal Lysine Charge-Clamps,” The Journal of Cell Biology, Vol. 149, 2000, pp. 1225-1234. doi:10.1083/jcb.149.6.1225
- T. Wallimann, M. Wyss, D. Brdiczka, K. Nicolay and H. M. Eppenberger, “Intracellular Compartmentation, Structure and Function of Creatine Kinase Isoenzymes in Tissues with High and Fluctuating Energy Demands: The ‘Phosphocreatine Circuit’ for Cellular Energy Homeostasis,” Biochemical Journal, Vol. 281, Pt. 1, 1992, pp. 21- 40.
- P. W. Hochachka, “Muscles as Molecular and Metabolic Machines,” CRC Press, Boca Raton, 1994.
- B. Sacktor, “Utilization of Fuels by Muscle,” In: D. J. Candy and B. A. Kilby, Eds., Insect Biochemistry and Function, Chapman and Hall, London, 1975, pp. 1-81. doi:10.1007/978-94-009-5853-1_1
- B. Saktor, “Utilization of Fuels by Muscle,” Chapman and Hall, London, 1975.
- M. Novotova, M. Pavlovicova, V. I. Veksler, R. VenturaClapier and I. Zahradnik, “Ultrastructural Remodeling of Fast Skeletal Muscle Fibers Induced by Invalidation of Creatine Kinase,” American Journal of Physiology Cell Physiology, Vol. 291, No. 6, 2006, pp. C1279-C1285. doi:10.1152/ajpcell.00114.2006
- A. G. Jimenez, B. R. Locke and S. T. Kinsey, “The Influence of Oxygen and High-Energy Phosphate Diffusion on Metabolic Scaling in Three Species of Tail-Flipping Crustaceans,” The Journal of Experimental Biology, Vol. 211, 2008, pp. 3214-3225. doi:10.1242/jeb.020677
- K. Yoshizaki, H. Watari and G. K. Radda, “Role of Phosphocreatine in Energy Transport in Skeletal Muscle of Bullfrog Studied by 31P-NMR,” Biochimica et Biophysica Acta, Vol. 1051, No. 2, 1990, pp. 144-150. doi:10.1016/0167-4889(90)90186-H
- R. A. de Graaf, A. van Kranenburg and K. Nicolay, “In Vivo 31P-NMR Diffusion Spectroscopy of ATP and Phosphocreatine in Rat Skeletal Muscle,” Biophysical Journal, Vol. 78, No. 4, 2000, pp. 1657-1664. doi:10.1016/S0006-3495(00)76717-8
- A. Kaasik, V. Veksler, E. Boehm, M. Novotova, A. Minajeva and R. Ventura-Clapier, “Energetic Crosstalk between Organelles: Architectural Integration of Energy Production and Utilization,” Circulation Research, Vol. 89, 2001, pp. 153-159. doi:10.1161/hh1401.093440
- H. R. Ramay and M. Vendelin, “Diffusion Restrictions Surrounding Mitochondria: A Mathematical Model of Heart Muscle Fibers,” Biophysical Journal, Vol. 97, No. 2, 2009, pp. 443-452. doi:10.1016/j.bpj.2009.04.062
- V. Saks, A. Kuznetsov, T. Andrienko, Y. Usson, F. Appaix, K. Guerrero, T. Kaambre, P. Sikk, M. Lemba and M. Vendelin, “Heterogeneity of ADP Diffusion and Regulation of Respiration in Cardiac Cells,” Biophysical Journal, Vol. 84, No. 5, 2003, pp. 3436-3456. doi:10.1016/S0006-3495(03)70065-4
- K. Ashman, T. Houthaeve, J. Clayton, M. Wilm, A. Podtelejnikov, O. N. Jensen and M. Mann, “The Application of Robotics and Mass Spectrometry to the Characterisation of the Drosophila melanogaster Indirect Flight Muscle Proteome,” Letters in Peptide Science, Vol. 4, No. 2, 1997, pp. 57-65. doi:10.1007/BF02443516
- D. K. Nam, S. Lee, G. Zhou, X. Cao, C. Wang, T. Clark, J. Chen, J. D. Rowley and S. M. Wang, “Oligo (dT) Primer Generates a High Frequency of Truncated c DNAs through Internal Ploy(A) Priming during Reverse Transcription,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 99, No. 9, 2002, pp. 6152-6156. doi:10.1073/pnas.092140899
- J. O. Vigoreaux, J. D Saide and M. L. Pardue, “Structurally Different Drosophila Striated Muscles Utilize Distinct Variants of Z-Band-Associated Proteins,” Journal of Muscle Research & Cell Motility, Vol. 12, No. 4, 1991, pp. 340-354. doi:10.1007/BF01738589
- U. K. Laemmli, “Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4,” Nature, Vol. 277, 1970, pp. 680-685. doi:10.1038/227680a0
- G. Ayer and J. O. Vigoreaux, “Flightin Is a Myosin Rod Binding Protein,” Cell Biochemistry and Biophysics, Vol. 38, No. 1, 2003, pp. 41-54. doi:10.1385/CBB:38:1:41
NOTES
*Corresponding author.

