Natural Resources
Vol. 3 No. 4 (2012) , Article ID: 26278 , 7 pages DOI:10.4236/nr.2012.34028
Preliminary Comparative Physiological and Biochemical Study of Five Different Goat Breeds Inhabiting Saudi Arabia
![]()
1Department of Biology, Faculty of Science, Taif University, Taif, Saudi Arabia; 2Department of Zoology, Faculty of Science, Cairo University, Giza, Egypt.
Email: *yasser92us@yahoo.com
Received September 20th, 2012; revised October 27th, 2012; accepted November 13th, 2012
Keywords: Goat Breeds; Electrophoresis; Isoenzymes; Physiology; Saudi Arabia
ABSTRACT
Three arbitrary chosen enzymes were examined by native-polyacrylamide gel electrophoresis to investigate physiological and genetic variations among five different goat breeds inhabiting Saudi Arabia. The goat breeds were Pakistani, Tihami, Syrian, Masri and Aardi while the investigated enzymes were alkaline phosphatase (ALP), malate dehydrogenase (Mdh) and malic enzyme (ME). Six polymorphic loci with six monomeric alleles have been recorded in all studied breeds. The second locus of ME was characteristic of Syrian breed where it showed dimeric alleles. The similarity matrix that has been calculated according to the number of sharing bands indicated that these breeds have been divided into two groups: Pakistani and Tihami as one group while the other three breeds clustered in another group. The activity of the metabolic enzymes (Mdh and ME) showed concordance with the constructed relationship where the percentage amounts of these enzymes showed significant variations between the two groups more than that occurred within each group. Mdh was expressed in the second group more than in the first while ME showed, nearly, equal expression in the different breeds. Both genetic and physiological results agreed in clustering the studied breeds into Pakistani and Tihami in one group and the other three breeds in another group. This division was based on a few gene loci and a few sampling size and it needs to be supported by collecting more genetic data and more sampling size in a further molecular study.
1. Introduction
Goat (Capra aegagrus), as one of the ancient livestock, was first domesticated in western Asia in the period of 9000 - 7000 BC [1]. It was considered that the domestic goat breeds in the world were from a single origin and their separations in different ecological areas followed [2]. It was also suggested by Zhang and Li [3] that the goat in the world could be divided into three populations according to the Hb gene frequency [4]. Generally, goat breeds are named by their places of origin and classified by their uses of products. As the effect of different ecological conditions and long-term artificial selection, goat breeds with different genetic characteristics and product orientation are formed [5]. Eighty eight percentages of the world goat populations are located in Asia and Africa, mostly in the tropics and sub-tropics [6]. Goats are one of the worlds most adaptable and widespread livestock species, and are one of the main economic recourses in many developing countries [7].
The available literature related to the field of metabolism in goats is scarce [8]. Some investigations have studied healthy goats and presented biochemical indicators in relation to breeds [9-12]. Goat living in a desert environment, like Arabian goat, is characterized by low body mass and low metabolic requirements [13].
Alkaline phosphates are a group of isoforms which hydrolyze many types of phosphate esters, whose natural substrate or substrates are unknown [14]. In both humans and animals, the major sources of ALPs are the liver, bone, kidney and placenta. Total serum ALP activity has diagnostic value in the hepatic and bone diseases in dogs and cats [15]. It is of little value in hepatic diseases of horses and ruminants. The range of serum ALP value in goats may be 10-fold with no evidence of hepatic damage. Values within the individual are fairly constant for sequential evaluation [15].
Malate dehydrogenase and malic enzyme are among the tricarboxylic acid cycle enzymes that are responsible for lipid and energy metabolism [16,17]. These two enzymes are commonly expressed in most eukaryotic organisms as two fractions, one is cytosolic and the other is mitochondrial [18]. Both enzymes have been studied in many ruminants [19].
Genetic diversity is defined as the sum of genetic differences in multiple loci among individuals in a population, and is most readily reflected in the phenotypic variation seen in many populations. Genetic diversity is a valuable asset as the adaptability of a population, that is the population’s ability to adapt to changes, depends on it [20]. It is well known that species can face great environmental changes over time, such as in climate, pollution and in diseases, and genetic diversity is required for populations to adapt to these changes [21].
In recent years, molecular genetics have experienced considerable advances and offer a convenient way for characterization of population structure. Nowadays, DNA molecular markers are extensively applied to the goat breed classification instead of blood protein and isoenzyme [4]. MtDNA (mitochondrial DNA) is an important maternal genetic marker in studying the genetic relationships of livestock species and breeds [22]. The mtDNA has proven to be especially valuable in the study of genetic variation because it shows the maternal inheritance and changes much more rapidly than single copy nuclear DNA in mammals [23]. Compared with analyses of allozymic variations, analyses of mtDNA polymorphism at the nucleotide level have shown to be very useful for detection of genetic variation.
The Saudi Arabian goat breeds are widely diversified and unfortunately they are rarely studied physiologically and genetically. So far, few investigations have been conducted for some Arabian goat breeds [24,25]. The present study therefore aimed to investigate the physiological and genetic background for the most popular goat breeds in Saudi Arabia as a trial for addressing their physiological adaptations and the degree of kinship.
2. Materials and Methods
2.1. Sample Preparation and Isoenzyme Assay
Twenty four blood samples were freshly obtained from five different goat breeds (approximately five males from each breed) from private owners in Taif city. These goat breeds were as follows: Pakistani, Tihami, Syrian, Masri and Aardi. The blood samples were placed in tubes with an anticoagulant and immediately centrifuged at 3000 rpm for approximately 3 - 5 minutes. The plasmatic supernatants were stored at –80˚C for further isoenzyme assay.
For electrophoresis, 30 μL of the plasma was mixed with 10 μL of treatment buffer and 35 μL of this mixture was loaded into the well. Isoenzymes were electrophorased in 10% native-polyacrylamide gel as described by Stegemann et al. [26]. After electrophoresis, the gels were stained according to their enzyme system with the appropriate substrate and chemical solutions then incubated at room temperature in dark for complete staining. In most cases the incubation for about 1 to 2 hours is enough.
2.2 Alkaline Phosphatase (ALP)
After electrophoresis, the gel was soaked in 100 mL of 50 mM Na-acetate buffer pH 5.0 containing 100 mg Fast blue BB salt, 100 mg a-naphthyl phosphate, 100 mg MgCl2 100 mg MnCl2 and 1 g NaCl in 100 ml of 0.1 M Tris HCl pH 8.5 [27].
2.3. Malate Dehydrogenase (Mdh)
After electrophoresis, the gel was soaked in 100 mL of 0.05 M Tris HCl pH 8.5 containing 25 mg NBT, 25 mg EDTA, 25 mg NAD, 10 mg malic acid and 3 mg PMS [28]. 0.05 M Tris HCl pH 8.5 was prepared by dissolving 0.605 g Tris in 50 mL distilled water. The pH was adjusted to 8.5 by HCl. Then the solution was completed to 100 mL with distilled water.
2.4. Malic Enzyme (ME)
After electrophoresis, the gel was soaked in 100 mL of 0.05 M Tris HCl pH 8.5 containing 25 mg NBT, 25 mg EDTA, 25 mg NADP, 10 mg malic acid, 100 MgCl2 and PMS [28].
2.5. Gel Fixation
After the appearance of the enzyme bands, the reaction was stopped by washing the gel two or three times with tap water. This was followed by adding the fixative solution, which consists of ethanol and 20% glacial acetic acid (9:11 v/v). The gel was kept in the fixative solution for 24 hours and then was photographed.
2.6. Statistics
All gels were scanned using Gel Doc-2001 Bio-Rad system. The gels were analyzed by Pro Gel analyzer ver. 3 for obtaining the optical density and the percentage amounts for each band. For isoenzymes, the bands of enzyme activity were designated using the known system of nomenclature [29]. An abbreviation which corresponds to the name of the enzyme designated each locus. When multiple loci were involved, the fastest anodal protein band was designated as locus one, the next as locus two and so on. Student t-test in the SPSS package v. 13 was used to calculate the significant differences of the percentage amounts for isoforms among the different studied breeds.
A similarity matrix of allele presence (1) and absence (0) was constructed and the dendrogram defining the relationship among the different breeds was obtained by the UPGMA method from NTSYS-pc package [30].
3. Results and Discussion
Table 1 showed the mean and standard error for the percentage amount of the studied isoenzymes in the different goat breeds. The two ALP isoforms did not show significant variations among most studied breeds. For this isoenzyme, the values with zero SE mean that the corresponding isoform has been expressed in one sample of the corresponding breed. Figure 1 shows in which samples these values have been recorded. The total ALP showed significant increase in Pakistani (P < 0.05) compared to Syrian while ALP-2 was significantly higher (P < 0.05) in Aardi compared to Masri. The percentage amount for the total Mdh showed significant increase in Masri (P < 0.05) and in Aardi (P < 0.01) compared to Tihami. The activity of ME showed marked differentiation among the studied breeds. ME-1 showed a significant increase (P < 0.05) in Pakistani compared to Aardi. ME-2 was significantly higher (P < 0.05) in Syrian compared to Tihami and Aardi and this isoenzyme was also significantly higher in Masri (P < 0.01) compared to Tihami. With respect to the percentage amount of total ME, Syrian breed showed higher significance compared to Masri (P < 0.05) and Aardi (P < 0.01). Masri breed showed a significant higher percentage amount (P < 0.05) of total ME compared to Aardi.
The electrophoretic behaviors for the studied isoenzymes are shown in Figures 1-3 (ALP, Mdh, ME, respectively). These isoenzymes recorded 6 polymorphic loci and 6 monomeric alleles. In all zymograms, the origin of the electrophoretic patterns is at the top and the anode is at the bottom.
The banding pattern and the recoded alleles for ALP isoforms are shown in Figure 1 and Table 2. Two isoforms were recorded with the second being more common. ALP-1 was expressed commonly in Tihami breed while ALP-2 was fixed in Syrian and Masri breeds. ALP isoforms did not show a clear expression in Pakistani and Aardi breeds. Total serum ALP activity has diagnostic value in the hepatic and bone diseases in dogs and cats [15] but it may be not an indication of diseases in goat. As serum ALP values may show an increase in goats

Table 1. Mean ± SE of the percentage amount for the studied isoenzymes in Plasma of different goat breeds. The significance level was estimated by Student t-test. *significance level between Masri and Aardi; +significance level between Pakistani and Syrian; #significance level between Tihami and Masri; $significance level between Tihami and Aardi; &significance level between Tihami and Syrian; ᴥsignificance level between Syrian and Aardi; £significance level between Pakistani and Aardi and ■significance level between Syrian and Masri.
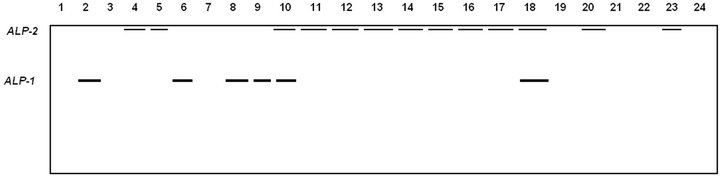
Figure 1. Electrophoretic banding pattern of ALP isoforms for Pakistani (1 - 4), Tihami (5 - 9), Syrian (10 - 14), Masri (15 - 19) and Aardi (20 - 24) goat breeds.
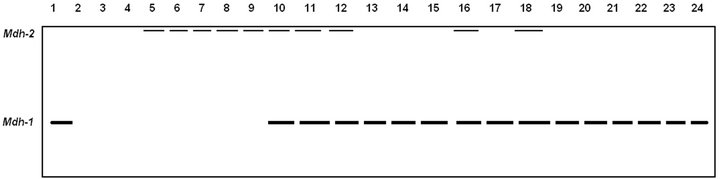
Figure 2. Electrophoretic banding pattern of MDH isoenzymes for Pakistani (1 - 4), Tihami (5 - 9), Syrian (10 - 14), Masri (15 - 19) and Aardi (20 - 24) goat breeds.

Figure 3. Electrophoretic banding pattern of ME isoenzymes for Pakistani (1 - 4), Tihami (5 - 9), Syrian (10 - 14), Masri (15 - 19) and Aardi (20 - 24) goat breeds.
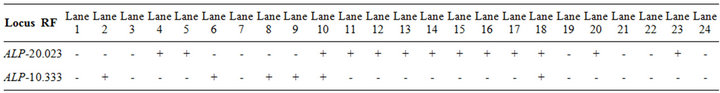
Table 2. The recorded alleles and their relative mobilities of ALP isoforms in Pakistani (1 - 4), Tihami (5 - 9), Syrian (10 - 14), Masri (15 - 19) and Aardi (20 - 24) goat breeds.
with no evidence of hepatic damage [15], the significant increase and the expression of this enzyme or any of its isoforms, in some of the studied breeds, could be considered as a normal case.
Among the five breeds studied herein, the cytosolic Mdh-1 isozymes did not express in Pakistani (except only in one individual) and Tihami breeds but it were approximately fixed in Syrian, Masri and Aardi breeds (Figure 2, Table 3). The mitochondrial Mdh-2 was fixed in all breeds but it did not show any expression in Pakistani breed. As Mdh enzyme catalyses the reversible shuttle down of malate-oxalloactate pathway [31], the metabolic rate of the Syrian, Masri and Aardi breeds may be more energetic than the other two breeds.
The two fractions of ME showed expression in most of the individuals of the all studied breeds (Figure 3 and Table 4). In spite of the monomeric exhibition of all studied phenotypes, the second fraction (ME-2) showed double bands in Syrian breed only and therefore this fraction is characteristic of this breed. Since ME has a role in lipid biosynthesis [32,33], it seems that all breeds may possess similar adaptability of lipid biosynthesis, particularly, in the desert environment of Saudi Arabia.
The dendrogram obtained by the UPGMA clustering method, according to the presence and the absence of bands, addressed the genetic relationship among the different breeds (Figure 4). The breeds examined were divided into two groups: the first group contained Pakistani and Tihami breeds, whereas the second group included Syrian, Masri and Aardi breeds. Therefore, Figure 4 agreed with the physiological data (Table 1) where the enzymatic activity differentiated clearly the studied breeds into two groups: one with a low metabolic rate (Pakistani and Tihami) and the other with a high metabolic rate (Syrian, Masri and Aardi). Saber et al. [25] used RAPD-PCR to construct the genetic relationship of four Saudi goat breeds among which Aardi and Masri clustered in two different groups without any bootstrap support. We therefore considered that the genetic relationship among Saudi Arabian goat breeds needs to be addressed

Table 3. The recorded alleles and their relative mobilities of MDH isoenzymes in Pakistani (1 - 4), Tihami (5 - 9), Syrian (10 - 14), Masri (15 - 19) and Aardi (20 - 24) goat breeds.
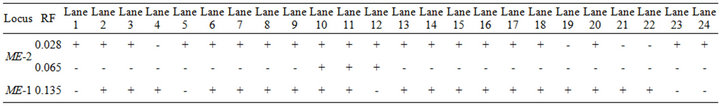
Table 4. The recorded alleles and their relative mobilities of ME in Pakistani (1 - 4), Tihami (5 - 9), Syrian (10 - 14), Masri (15 - 19) and Aardi (20 - 24) goat breeds.
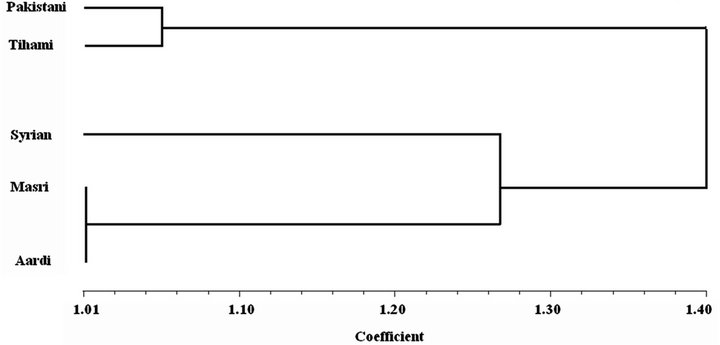
Figure 4. Dendrogram based on UPGMA analysis of genetic similarity obtained from isozyme data showing the relationships among the studied goat breeds.
on the molecular level.
4. Conclusion
In this work it has been shown that the goat breeds examined were differentiated genetically into Pakistani and Tihami as one group and the other three breeds in another group. The physiological data, similarly, differentiated the studied breeds into a low metabolic rate Pakistani and Tihami group and a high metabolic rate Syrian, Masri and Aardi group. This division was based on few gene loci and small sampling size and it needs to be supported by collecting more genetic data and more sampling size in further study. We therefore considered this conclusion as a preliminary evidence for goat breeds systematics and physiology.
5. Acknowledgement
We thank Dr. Shawkat at Ain Shams University of Egypt for his technical support in conducting the practical part of this work.
REFERENCES
- F. E. Zenuner, “A History of Domesticated Animals,” Hutchinson, London, 1963.
- K. Nozawa, “Coat-Color Polymorphism in the Black Bengal Goats,” Report of the Society for Researches on Native Livestock, Vol. 12, 1988, pp.187-198.
- C. J. Zhang and Q. J. Li, “The Study on Polymorphism of Qinhai Goat,” Journal of Qinhai Animal Veterinary Science, Vol. 4, 1990, pp. 5-6.
- M.H. Li, K. Li and S.H. Zhao, “Diversity of Chinese Indigenous Goat Breeds: A Conservation Perspective, A Review,” Diversity and Conservation of Chinese Goat, Vol. 17, No. 5, 2004, pp. 726-732.
- W. B. Yue, “Modern Goat and Sheep Farming,” Chinese Agricultural Press, Beijing, 2000.
- M. Knight and G. W. Garcia, “Characteristics of the Goat (Capra hircus) and Its Potential Role as a Significant Milk Producer in the Tropics: A Review,” Small Ruminant Research, Vol. 26, No. 3, 1977, pp. 203-215. doi:10.1016/S0921-4488(96)00977-7
- G. Luikart, L. Gielly, L. Excoffier, J. D. Vigne, J. Bouvet and P. Taberlet, “Multiple Maternal Origins and Weak Phylogeographic Structure in Domestic Goats,” Proceeding of the Natural Academy of Sciences USA, Vol. 98, No. 10, 2001, pp. 5927-5932. doi:10.1073/pnas.091591198
- D. Žubčić, “Some Biochemical Parameters in the Blood of Grazing German Improved Fawn Goats from Istria, Croatia,” Journal of Veterinarski Arhiv, Vol. 71, No. 5, 2001, pp. 237-244.
- A. Castro, D.S. Dhindsa, A.S. Hoversland, H. Malkus, C. Rosenthiel and J. Metcalfe, “Serum Biochemistry Values in Normal Pygmy Goats,” American Journal of Veterinary Research, Vol. 38, No. 12, 1977, pp. 2085-2087.
- S. Jana, B. Bhattaacharyya, R. Duttagupta and D. N. Moitra, “A Note of Some Biochemical Constituents of Blood in Pregnant Goats Reared on Extensive Management System,” Indian Veterinary Journal, Vol. 68, No. 6, 1991, pp. 592-594.
- M. E. Azab and H. A. Abdel-Maksoud, “Changes in Some Haematological and Biochemical Parameters during Pregnancy and Post-Partum Periods in Female Baladi Goats,” Small Ruminant Research, Vol. 34, No. 1, 1999, pp. 77- 85. doi:10.1016/S0921-4488(99)00049-8
- F. M. Tambuwal, B. M. Agale and A. Bangana, “Haematological and Biochemical Values of Apparently Healthy Red Sokoto Goats,” Proceeding of the 27th Annual Conference of the Nigeria Society of Animal Production (NSAP), Akure, 17-21 March 2002, pp. 50-53.
- N. Silanikove, “The Physiological Basis of Adaptation in Goats to Harsh Environments,” Small Ruminant Research, Vol. 35, No. 3, 2000, pp. 181-193. doi:10.1016/S0921-4488(99)00096-6
- K. O. Soetan, O. O. Aiyelaagbe and C. O. Olaiya, “A Review of the Biochemical, Biotechnological and Other Applications of Enzymes,” African Journal of Biotechnology, Vol. 9, No. 4, 2010, pp. 382-393.
- J. J. Kaneko, “Clinical Biochemistry of Domestic Animals,” Academic Press Inc., London, 1989.
- M. J. MacDonald, “Feasibility of a Mitochondrial Pyruvate Malate Shuttle in Pancreatic Islets. Further Implication of Cytosolic NADPH in Insulin Secretion,” Journal of Biological Chemistry, Vol. 270, No. 34, 2005, pp. 1-8.
- P. Daniš and R. Farkas, “Hormone-Dependant and Hormone-Independant Control of Metabolic and Developmental Functionals of Malate Dehydrogenase—Review,” Endocrine Reviews, Vol. 43, 2009, pp. 39-52.
- A. U. Wurochekke, A. E. Anthony and W. Obidah, “Biochemical Effects on the Liver and Kidney of Rats Administered Aqueous Stem Bark Extract of Xemenia americana,” African Journal of Biotechnology, Vol. 7, No. 16, 2008, pp. 2777-2780.
- R. A. Nafikov and D. C. Beitz, “Carbohydrate and Lipid Metabolism in Farm Animals. Symposium: History of Nutrition: Impact of Research with Cattle, Pigs, and Sheep on Nutritional Concepts,” Journal of Nutrition, Vol. 137, No. 3, 2007, pp. 702-705.
- J. Woolliams, P. Berg, A. Mäki-Tanila, T. Meuwissen and E. Fimland, “Sustainable Management of Animal Genetic Resources,” Nordic Genbank Husdyr, 2005.
- R. Frankham, J. D. Ballou and D. A. Briscoe, “Introduction to Conservation Genetics,” Cambridge University Press, Cambridge, 2002. doi:10.1017/CBO9780511808999
- J. C. Awise, “Ten Unorthodox Perspectives on Evolution Prompted by Comparative Population Genetic Findings on mtDNA,” Annual Review of Genetics, Vol. 25, No. 1, 1991, pp. 45-69. doi:10.1146/annurev.ge.25.120191.000401
- W. M. Brown, E. M. Prager, A. Wang and A. C. Wilson, “Mitochondria DNA Sequences of Primates: Tempo and Mode of Evolution,” Journal of Molecular Evolution, Vol. 18, No. 4, 1982, pp. 225-239. doi:10.1007/BF01734101
- M. Alamer, “Physiological Responses of Saudi Arabia Indigenous Goats to Water Deprivation,” Small Ruminant Research, Vol. 63, No. 1, 2006, pp. 100-109. doi:10.1016/j.smallrumres.2005.02.004
- J. S. M. Sabir, M. H. Z. Mutawakil, A. A. El-Hanafy and M. M. Ahmed, “Genetics Similarity among Four Breeds of Goat in Saudi Arabia Detected by Random Amplified Polymorphic DNA Marker,” African Journal of Biotechnology, Vol. 11, No. 17, 2012, pp. 3958-3963.
- H. Stegemann, A. El-Moneim R. Afify and K. R. F. Hussein, “Identification of Dates (Phoenix dactylifera) by Protein Patterns,ˮ Phytochemistry, Vol. 26, No. 1, 1986, pp. 149-153.
- J. F. Wendel and N. F. Weeden, “Visualization and Interpretation of Plant Isozymes,ˮ In: D. E. Soltis and P. S. Soltis, Eds., Isozymes in Plant Biology, Dioscorides Press, Portland, 1989, pp. 5-45. doi:10.1007/978-94-009-1840-5_2
- F. W. Jonathan and N. F. Wendell, “Visualization and Interpretation of Plant Isozymes,ˮ In: D. E. Soltis and P. S. Soltis, Ed., Isozymes in Plant Biology, Champan and Hall, London, 1990, pp. 5-45.
- W. Allendorff and F. M. Utter, “Population Genetics,ˮ In: W. S. Hoaran and D. J. Randalal, Eds., Fish Physiology, Academic Press, New York, 1978, pp. 407-454.
- F. J. Rohlf, “On the Use of Shape Spaces to Compare Morphometric Methods, Hystrix,” Italian Journal of Mammology (n.s.), Vol. 11, No. 1, 2000, pp. 9-25.
- S. Zhao, W. Xu, W. Jiang, W. Yu, Y. Lin, T. Zhang, J. Yao, L. Zhou, Y. Zeng, H. Li, Y. Li, J. Shi, W. An, S. M. Hancock, F. He, L. Qin, J. Chin, P. Yang, X. Chen, Q. Lei, Y. Xiong and K. L. Guan, “Regulation of Cellular Metabolism by Protein Lysine Acetylation,” Science, Vol. 327, No. 5968, 2010, pp.1000-1004. doi:10.1126/science.1179689
- J. P. Wynn and C. Ratledge, “Malic Enzyme Is a Major Source of NADPH for Lipid Accumulation by Aspergillus nidulans,” Microbiology, Vol. 143, No. 1, 1997, pp. 253-257. doi:10.1099/00221287-143-1-253
- Y. Zhang, I. P. Adams and C. Ratledge, “Malic Enzyme: The Controlling Activity for Lipid Production? Overexpression of Malic Enzyme in Mucor Circinelloides Leads to a 2.5-Fold Increase in Lipid Accumulation,” Microbiology, Vol. 153, No. 7, 2007, pp. 2013-2025. doi:10.1099/mic.0.2006/002683-0
NOTES
*Corresponding author.

