Open Journal of Animal Sciences
Vol.3 No.2(2013), Article ID:29917,5 pages DOI:10.4236/ojas.2013.32019
Tyrosine phosphorylation of monocyte-derived macrophage proteins in buffalo (Bubalus bubalis): A potential phenotype of natural resistance
![]()
Consiglio per la Ricerca e la Sperimentazione in Agricoltura (Agricultural Research Council), Monterotondo, Italy; *Corresponding Author: maria.miarelli@entecra.it
Copyright © 2013 Maria Miarelli et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 18 January 2013; revised 1 March 2013; accepted 5 April 2013
Keywords: Buffalo; Macrophage; Natural Resistance; Tyrosine Phosphorylation
ABSTRACT
The aim of this work was to explore the possibility of using the presence of tyrosine-phosphorylated macrophage proteins as a phenotype of natural resistance. Tyrosine-phosphorylation of macrophage proteins was investigated in 18 buffaloes, that carried either the resistant, or the non-resistant, Natural Resistance-Associated Macrophage Protein one (NRAMP1) genotype, that various authors have associated with susceptibility to intracellular bacterial diseases. Monocyte-derived macrophages were Interferon-gamma (IFN-γ) stimulated and tyrosine-phosphorylation was assessed by Western blotting. Evidence of phosphorylation after IFN-γ stimulation was shown by 75% of the buffaloes carriers of the resistant genotype, and by 20% of the carriers of the non-resistant genotype (Chisquare value between the groups = 5.44; P = 0.02). The study of the Proteoma of monocyte-derived macrophages might open the way to the genetic control of disease resistance.
1. INTRODUCTION
Breeding objectives relating to health, functional traits and welfare need to receive priority in the research programs and selection schemes, but very few reports are available on natural resistant genotype in buffalo [1], where some important diseases cause severe economic losses and pose serious zoonotic threats.
Macrophages are key cells in innate immune response against pathogens; macrophage activation, defined as “acquisition of competence to execute a complex function”, plays an important role in influencing natural resistance to infection. During the innate immune response, macrophage receptors recognize either conserved pathogen-associated molecular patterns or several cytokines, e.g. interferon-γ (IFN-γ). Cellular activation involves a variety of post-translational modifications of cytoplasmatic macrophage proteins, that regulate a wide range of cellular functions. The phosphorylation state, in particular, makes the proteins highly dynamic in controlling the biochemical pathways [2], i.e. the responsible pathways of host innate response in immune-competent cells [3]. Immune system activity is under control of host genetic background [4] and the resistant allelic form of NRAMP1 gene in mouse was positively correlated with a heightened state of phosphorylation of tyrosine residues of protein kinase on macrophages exposed to various biochemical stimuli [5,6].
In the present study we wanted to explore the possibility of using the presence of tyrosine-phosphorylated macrophage proteins as a phenotype of natural resistance in water buffalo, both in native or IFN-γ stimulated monocyte-derived macrophages (MDM).
We chose two groups of buffaloes, carrying the NRAMP1 genotypes associated with either resistance or susceptibility to Brucella abortus [7,8]; we used an in vitro system to obtain MDM and we analyzed the presence of protein tyrosine phosphorylation either in un-stimulated or in IFN-γ stimulated cells.
This in vitro system, that reproduces several intracellular significant events happening during in vivo bacterial contacts, was suggested by various authors [5,9] and we used this model to estimate the cellular immune activation of MDM as an indicator of natural resistance, to be related to the NRAMP1 genotype.
2. MATERIALS AND METHODS
2.1. Animal Sampling
The trial was carried out on 18 buffaloes of two different genotypes at the NRAMP1 gene. The NRAMP1 locus in buffalo, consists of a polymorphic microsatellite located in the 3’UTR [8] and the two animal groups were composed respectively of N = 8 heterozygous and N = 10 homozygous animals. The buffaloes were reared in the same experimental farm and the animals were maintained under uniform feeding and housing conditions; they were negative to the following infections: brucellosis (Brucella abortus), Johne’s disease (M. aviumsobsp. paratubercolosis), leukosis (Bovine leukemia virus) and tubercolosis (M. bovis).
2.2. Monocytes Extraction-MDM Culture-MDM Activation
Peripheral blood mononuclear cells (PBMC) were isolated by collecting aseptically 40 mL of blood from the jugular vein of each animal using vacuum tubes containing tripotassium ethylene diamine tetraacetic acid (Vacu-tek-K3 EDTA 15% solution-ratio of 1.8 mg/mL of blood). After centrifugation (20 min - 600 g - 4˚C), buffy coats were collected, added with 30 mL of cold HBSS and 10 mL of LymphoprepTM (1.077 g/mL; AXIS-SHIELD) were underlain. Tubes were centrifuged (40 min - 500 g - room temperature). Mononuclear cell band was removed, washed twice with 30 mL of cold Hank’s Buffered Salt Solution (HBSS) and cells were pelleted by centrifugation (10 min - 340 g - 4˚C). PBMC were resuspended in RPMI1640 medium (Sigma-Aldrich) supplemented with 10% of heat inactivated fetal bovine serum (FBS; EuroClone), 2 mM L-glutamine (Sigma-Aldrich); 0.05 mg/mL gentamicin (Sigma-Aldrich); 1% fungizone (EuroClone), 0.05 mM mercaptoethanol (SigmaAldrich).
PBMC microscopic count was performed by Thoma chamber. The cell viability was checked by Trypan blue exclusion test. The cells were distributed at a density of 10 × 106 cells/4mL medium/well, in 6-well tissue culture plate and maintained at 37˚C in a humidified atmosphere under 5% CO2. Monocytes were obtained by adherence. Following 24 hours incubation, each well was washed four times with 39˚C PBS for lymphocytes and platelets elimination and the medium was replaced.
Monocytes culture homogeneity was assessed by detecting monocyte marker CD14 antigen. Adherent cells were fixed 15 minutes with paraformaldehyde (SigmaAldrich, 1% PBS) and incubated 2 h at room temperature with a monoclonal mouse anti bovine antibody CD14 (PBS-bovine serum albumine (BSA) 4% diluted 1:50 AbD Serotec). For the following steps, cells were washed three times for 5 min with PBS pH 7.3, incubated 1 h at room temperature with a secondary polyclonal antibody goat anti mouse IGg fluorescein isothiocyanate-conjugated (PBS-BSA 4% diluted 1:200 AbD Serotec) and washed three times for 5 min with pH 7.3 PBS. Nuclear stain was performed with 4’6-diamidino-2-phenylindole dihydrochloride DAPI (0.1 g/mL AbD Serotec). After mounting (Mounting Medium R&D Systems), cells were visualized by fluorescence microscopy (Zeiss System) (Figure 1).
Monocyte cells were allowed to differentiate into MDM in the 6-well tissue culture plates containing 5 mL/well of RPMI 1640 supplemented culture medium, at 37˚C in a humidified atmosphere under 5% CO2. Seven days later cells were checked for morphology (larger size and a surface extension [10]) under a light microscope [11] (Figure 2). The cell viability was checked by Try pan blue exclusion test.
Macrophage activation was performed by IFN-γ in half of wells. Cells were washed once and treated for 30 min with Recombinant Bovine IFN-γ (R&D Systems; 250 ng/mL in RPMI 1640 medium 1 mL/well). Concentration of this cytokine and cellular stimulation time was established on the basis of experimental protocols involving murine macrophages [5]. Stimulated and unstimulated MDM were separately harvested by scraping
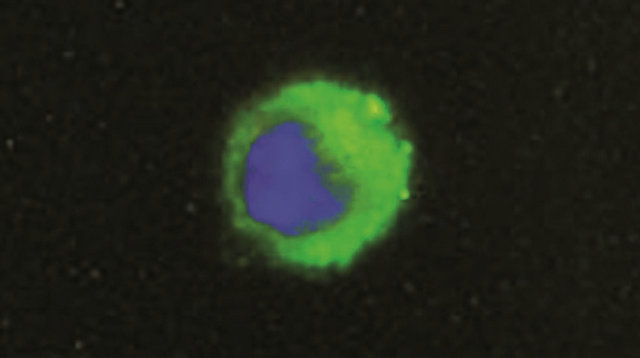
Figure 1. Representative immunofluorescence (fluorescein isothiocyanate) showing one CD14 cell (monocyte). cellular nuclei were stained with DAPI. The samples were viewed and photographed with a microscope equipped with epifluorescence illumination at a magnification of ×1000.
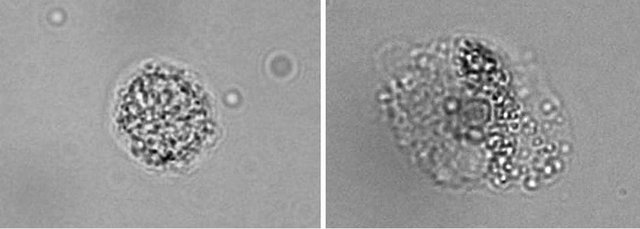
Figure 2. Morphological characterization of blood monocytes and monocyte-derived macrophages. Images were obtained by light transmitted microscopy, at a magnification of ×1000 change in the morphology of monocytes was evident by the larger size and the surface extension, that indicate the differentiation of monocytes into macrophages.
with cold PBS and pelleted by centrifugation for 10 min - 3000 g - 4˚C.
2.3. Protein Separation
Cells pellet was lysed with NP-40 buffer (1% NP-40, 0.5% sodium deoxycholate, 2 mM EDTA, 150 mM NaCl, 50 mM Tris pH 7.5, 50 mM NaF, 0.1 mM sodium orthovanadate, 1 ug/mL leupeptin and aprotinin, 1 mM PMSF) and centrifuged according to [5].
The total protein concentration in the samples was quantified using the 2-D Quant Kit (GE Healthcare, Niskayuna, NY). All samples were diluted with Laemmli sample buffer (1:1), boiled at 95˚C for 4 min and equivalent amount (50 μg) of proteins were separated by 1D SDS-PAGE on a 10% acrylamide minigel at 200 V for 50 min.
2.4. Western Blotting
Separated molecules were transferred onto a PVDF (polyvinylidene difluoride) membrane, using a discontinuous buffer system, at 15 V for 50 min. Membranes were blocked overnight at 4˚C in TTBS with 3% BSA. Primary antibody was phospho-tyrosine mouse monoclonal antibody (P-Tyr-100) (diluted 1:1000—Cell Signaling Technologies). Incubation was carried on for 1 h at room temperature. After extensive washing with Tris-Tween Buffered Saline, tyrosine phosphorylated proteins were detected by stabilized goat anti-mouse horseradish peroxide conjugated antibody (diluted 1:1500— Thermo scientific) and an ECL (Bio-Rad). Monoclonal mouse anti-actin antibody was used as a loading control.
3. RESULTS
The PBMC obtained from 40 mL of blood ranged from 4 to 9 × 107 cells and the cells viability, assessed by Trypan blue exclusion test, was higher than 85% for all samples. On day one, after colture washing, CD14 immunofluorescence test assessed that the MDM culture purity was about 95%. The final yield of MDM cells ranged from 1.8 to 4.3 × 104 cells/blood mL (Automated Cell Counter-Bio-Rad).
Due to the lack of cytokines and primary monoclonal antibody specific for buffalo species, we used recombinant IFN-γ bovine to MDM stimulating, because of the high level of similarity between bovine and buffalo IFN-γ [12]. Moreover we also used a mouse anti bovine CD14 monoclonal antibody as specific monocyte marker and a multi-species cross reacting P-Tyr-100 monoclonal antibody, not specifically tested from the producer on buffalo species. The bovine IFN-γ had effect on MDM activation and the two antibodies used recognized both CD14 and P-Tyr-100 buffalo antigens.
The unstimulated MDM were used to check the presence of tyrosine phosphorylated endogenous proteins.
Western blot analysis using the P-Tyr-100 antibody revealed a 60 KDa protein band.
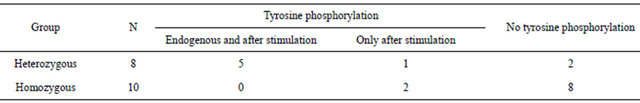
Table 1 reports the results of the Western blot analysis for the two groups of buffaloes.
Of the eight heterozygous animals, five revealed the tyrosine phosphorylation both in unstimulated and in IFN-γ stimulated cells; one animal showed the phosphorylation only after stimulation; the remaining two animals did not show tyrosine phosphorylation.
Eight of the ten homozygous animals did not show the endogenous tyrosine phosphorylation, and remained negative after IFN-γ stimulation. Two homozygous animals showed the phosphorylation, but only after stimulation.
The two groups therefore showed to differ significantly (Chi-square value = 5.44; P = 0.02). We noted, besides, that the phosphorylation signal was higher after IFN-γ stimulation, compared to the endogenous presence (Figure 3) (actine normalized photodensity values).
4. DISCUSSION
Natural resistance to diseases has a central role in early disease defence. Current literature reports a wide range of studies on human and mouse innate immunity, but fewer on livestock, in spite of the increased interest for animal health, for the control of the zoonotic risks and for the production systems that take into account animal welfare.
The genetic control of innate immune system is associated in several species with NRAMP1 gene, also known as solute carrier family 11 member a1 (Slc11a1), as responsible for natural resistance to unrelated intracellular pathogen infections. Even if it is unclear how this gene works in antimicrobial function, it has many pleiotropic effects on macrophage activation. Different allelic variants of NRAMP1 were associated to a wide range of intracellular effects on human and mouse macrophage activation, including an increased protein kinase tyrosine phosphorylation as expression of the intracellular transduction of signal [5,6] and of cellular activation.
Similarly to previous studies performed on healthy buffaloes, where the immune response was assessed after
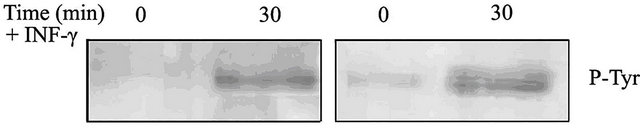
Figure 3. Tyrosine phosphorylation evidence of MDM from two different animals (Western blot analysis). On the left an example showing phosphorylation only after IFN-γ stimulation; on the right an example of both endogenous and after stimulation phosphorylation.
Table 1. Evidence of tyrosine phosphorylated proteins in the two buffalo groups (number of animals).
in vitro stimulation [13], in this trial the immune response to a potential infection was created by stimulating the macrophages with IFN-γ, a key cytokine that plays a pivotal role in the modulation of immune response [14]. Interferons (IFN-α/IFN-β secreted by virus-infected cells and IFN-γ secreted by T Lymphocytes) were originally exclusively considered as agents interfering with viral replication, but IFN-γ is among the most important activator of microbicidal functions by stimulating the bactericidal activity of phagocyte cells [15]. This molecule, moreover, demonstrated to be a great powerful macrophage response enhancer, just by triggering intracellular tyrosine phosphorylation [16] and inducing the expression of a set of early genes by tyrosine phosphorylation of latent transcription factors [17]. On the other hand, IFN-γ exhibited its antimicrobial function through NRAMP1 up regulation [15].
Our experimental model consisted in showing evidence of tyrosine phosphorylated proteins in monocytederived macrophages from healthy buffaloes with a different immune genotype, stimulated or not by IFN-γ.
This approach might only quite partially reproduce the whole biomolecular events occurring during the pathogen aggression because of the wide range of different agents activating macrophages, and also for the complexity of the genetic patterns regulating immune system and involving several protein pathways. Although the complexity of the macrophage function in innate immunity cannot be certainly limited to the IFN-γ stimulation and the study of natural resistance cannot be certainly limited to the expression of a single gene [18], nevertheless our results revealed an association of frequency between animals with different NRAMP1 genotypes and tyrosine phosphorylated proteins in macrophages.
In fact, 100% of animals carrying homozygous didn’t revealed endogenous phosphorylation and 80% of them remained negative also after stimulation, whereas the 20% responded to IFN-γ activation; regarding the heterozygous genotype, on the contrary, the majority of the animals (62.5%) showed endogenous phosphorylation, that enhanced after IFN-γ activation, whereas the remaining 37.5% did not reveal endogenous phosphorylation and, after stimulation, the 25% out them remained negative, whereas the remaining 12.5% reacted to the IFN-γ stimulus.
5. CONCLUSIONS
The present work, that explores the possibility to approach the natural resistance in buffalo at a biomolecular level, must be considered an example of how the study of a protein pattern could improve genetic knowledge.
Our study must be considered a preliminary study and could represent an experimental model useful in further experiments for the achievement of different protein phenotypes, either activating macrophages with different stimulators (i.e. the extracts of specific bacteria) or exploring the specific protein pathways. The next generation sequencing techniques will then give the opportunity to detect the candidate genes related to healthy traits, opening the way to the genetic control of disease resistance.
6. ACKNOWLEDGEMENTS
This study is part of the “NATRES” research programme, funded by the Consiglio per la ricerca e la sperimentazione in agricoltura (CRA).
REFERENCES
- Malik, Y.P.S., Chakravarti, S., Sharma, K., Vaid, N., Rajak, K.K., Balamurugan, V., Viswas, S.K., Mondal, B., Kataria, R.S. and Singh, R.K. (2011) Genomic analyses of toll-like receptor 4 and 7 exons of Bos indicus from tem-perate sub-himalayan region of India. Asian Australasian Journal of Animal Sciences, 24, 1019-1025. doi:10.5713/ajas.2011.10342
- Kovářová, H., Hajduch, M., Livingstone, M., Dzubak, P. and Lefkovits, I. (2003) Analysis of state-specific phosphorylation of proteins by two-dimensional gel electrophoresis approach. Journal of Chromatography B, 787, 53-61. doi:10.1016/S1570-0232(02)00729-8
- Nag, K. and Chaudhary, A. (2009) Mediators of tyrosine phosphorylation in innate immunity: From host defense to inflammation onto oncogenesis. Current Signal Transduction Therapy, 4, 76-81. doi:10.2174/157436209788167501
- Wells, C.A., Ravasi, T., Faulkner, G.J., Carninci, P., Okazaki, Y., Hayashizaki, Y., Sweet, M., Wainwright, B.J. and Hume, D.A. (2003) Genetic control of the innate immune response. BMC Immunology, 4, 5. doi:10.1186/1471-2172-4-5
- Kovářová, H., Nečasová, R., Porkertová, S., Radzioch, D. and Macela, A. (2001) Natural resistance to intracellular pathogens: Modulation of macrophage signal transduction related to the expression of the Bcg locus. Proteomics, 1, 587-596. doi:10.1002/1615-9861(200104)1:4<587::AID-PROT587>3.0.CO;2-T
- Gomez, M.A., Li, S., Trembly, L. and Olivier, M. (2007) NRAMP-1 expression modulates protein-tyrosine phosphatase activity in macrophages. Journal of Biological Chemistry, 282, 36190-38198. doi:10.1074/jbc.M703140200
- Borriello, G., Capparelli, R., Bianco, M., Fenizia, D., Alfano, F., Capuano, F., Ercolini, D., Parisi, A., Roperto, S. and Iannelli, D. (2006) Genetic resistance to Brucella abortus in the water buffalo (Bubalus bubalis). Infection and Immunity, 74, 2115-2120. doi:10.1128/IAI.74.4.2115-2120.2006
- Capparelli, R., Alfano, F., Amoroso, M.G., Borriello, G., Fenizia, D., Bianco, A., Roperto, S., Roperto, F. and Iannelli, D. (2007) Protective effect of the NRAMP-1 BB genotype against Brucella abortus in the water buffalo (Bubalus bubalis). Infection and Immunity, 75, 988-996. doi:10.1128/IAI.00948-06
- Taraktsoglou, M., Szalabska, U., Magee, D.A., Browne, J.A., Sweeney, T., Gormley, E. and MacHugh, D.E. (2011) Transcriptional profiling of immune genes in bovine monocyte-derived macrophages exposed to bacterial antigens. Veterinary Immunology and Immunopathology, 140, 130-139. doi:10.1016/j.vetimm.2010.12.002
- Pinet, F., Dupont, A., Bencherif, N., Guihot, A.L., Quatannens, B. and Amouyel, P. (2003) Morphology, homogeneity and functionality of human monocyte-derived macrophages. Cellular and Molecular Biology, 49, 899- 905.
- Murphy, J.T., Sommer, S., Kabara, E.A., Verman, N., Kuelbs, M.A., Saama, P., Halgren, R. and Coussens, P.M. (2006) Gene expression profiling of monocyte-derived macrophages following infection with Mycobacterium avium subspecies avium and Mycobacterium avium subspecies paratuberculosis. Physiological Genomics, 28, 67-75. doi:10.1152/physiolgenomics.00098.2006
- Premraj, A., Sreekumar, E. and Rasool, T.J. (2006) Cloning and biological characterization of buffalo (Bubalus bubalis) interferon-γ. Molecular Immunology, 43, 717- 724. doi:10.1016/j.molimm.2005.03.012
- Shah, S.M., Ravi Kumar, G.V.P.P.S., Brah, G.S., Santra, L. and Pawar, H. (2012) Differential expression of Th1- and Th2-type cytokines in peripheral blood mononuclear cells of murrah buffalo (Bubalus bubalis) on TLR2 induction by B. Subtilis peptidoglycan. Asian-Austalasian Journal of Animal Sciences, 25, 1021-1028. doi:10.5713/ajas.2012.12033
- Farrar, M.A. and Schreibe, R.D. (1993) The molecular cell biology of interferon-gamma and its receptor. Annual Review of Immunology, 11, 571-611. doi:10.1146/annurev.iy.11.040193.003035
- Schroder, K., Hertzog, P.J., Ravasi, T. and Hume, D.A. (2004) Interferon-γ: An overview of signals, mechanisms and functions. Journal of Leukocyte Biology, 75, 163- 188. doi:10.1189/jlb.0603252
- English, B.K., Orlicek, S.L., Mei, Z. and. Meals, E.A. (1997) Bacterial LPS and IFN-γ trigger the tyrosine phosphorylation of vav in macrophages: Evidence for involvement of the Hck tyrosine kinase. Journal of Leukocyte Biology, 62, 859-864.
- Igarashi, K., Garotta, G., Ozmen, L., Ziemiecki, A., Harpur, A.F., Larner, A.C. and Finbloom, D.S. (1994) Interferon-γ induces tyrosine phosphorylation of interferon-γ receptor and regulated association of protein tyrosine kinase, Jak1 and Jak2, with its receptor. Journal of Biological Chemistry, 269, 14333-14336.
- Ganguly, I., Sharma, A., Singh, R., Deb, S.M., Singh, D.K. and Mitra, A. (2008) Association of microsatellite (GT) polymorphism at 3’UTR of NRAMP-1 with the macrophage function following challenge with Brucella LPS in buffalo (Bubalus bubalis). Veterinary Microbiology, 129, 23-26.

