Open Journal of Genetics
Vol.2 No.2(2012), Article ID:20391,10 pages DOI:10.4236/ojgen.2012.22017
Genetic diversity and population structure of Prochilodus costatus and Prochilodus argenteus preceding dam construction in the Paraopeba River, São Francisco River Basin, Minas Gerais, Brazil
![]()
1Departamento de Biologia Geral, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, Brazil
2Programa de Pós-Graduação em Zoologia de Vertebrados, Pontifícia Universidade Católica de Minas Gerais, Belo Horizonte, Brazil
3Departamento de Biologia Animal, Centro de Ciências Biológicas e da Saúde, Universidade Federal de Viçosa, Viçosa, Brazil
Email: ekalapo@icb.ufmg.br
Received 24 March 2012; revised 30 April 2012; accepted 22 May 2012
Keywords: Freshwater Fish; Complex Hypervariable Repeats Microsatellite; Genetic Diversity; Population Structure
ABSTRACT
Curimatã-pioa (Prochilodus costatus) and curimatã- pacu (Prochilodus argenteus) are migratory fish species endemic to the São Francisco River Basin in Brazil. Both species play important roles in local fisheries and ecology in the Paraopeba River. A dam was recently constructed on this river and to help in the development and conservation programs, we characterized the genetic variation of both species before dam construction. Complex hypervariable repeats microsatellite was used to asses genetic variation for both species within and between the five collection sites in order to detect population substructuring. Nucleotide substitutions and insertion/deletion polymorphisms (indels) resulted in 35 P. costatus haplotypes (sample size = 89) and 22 P. argenteus haplotypes (sample size = 32). Significant genetic diversity and population differentiation was detected between five sampling sites for both species. Therefore, each of the five sites should be regarded as a group comprising significant genetic differences in species conservation and maintenance plans. Comparing these results to genetic diversity measures after dam construction will be critical for future management in this region.
1. INTRODUCTION
In South American rivers, Prochilodus species are conspicuous, abundant, and widely distributed freshwater fish [1]. They support important fisheries in many parts of the continent. In the lower stretches and flood plain lagoons of the Paraná basin, Prochilodus spp. constitute to 50% - 90% of the total fish biomass. In the Jupiá reservoir in the Paraná River, Prochilodus lineatus (Valenciennes) present significant results in commercial fisheries [2-4]. Prochilodus spp. has high fecundity and spawn all eggs at once in the open waters of the main river channel. Larvae drift passively towards flooded areas where they feed. Juveniles remain on the floodplain for approximately two years before they mature [5]. Prochilodus costatus Valenciennes known as curimatã-pioa and Prochilodus argenteus Agassiz known as curimatã- pacu are endemic to the São Francisco River Basin in Brazil. Within the river, these benthopelagic, detritivorous species play a critical role in cycling organic material [6-8]. As a result, these species must be considered in reproductive management and conservation programs in this basin.
One of the most important tributaries of the São Francisco River is the 510-km long Paraopeba River in Minas Gerais State, southeastern Brazil [9]. The majority (51%) of species endemic to the São Francisco River Basin are present in the Paraopeba River [10]. However, in the Paraopeba River, these species are threatened by human activities, such as mining, discharge of industrial effluxents and domestic sewage, overfishing, destruction of gallery forests, and the draining of wetlands in support of agricultural development [10]. The Paraopeba River has two older dams in its upper section [11] (Igarapé Thermoelectric Dam and Salto do Paraopeba) and a third one built in 2010 in its lower section (Retiro Baixo Hydroelectric Dam; Figure 1).
Molecular markers have enabled large-scale population genetics analyses [12] that address important questions in population biology. Assessing genetic variation
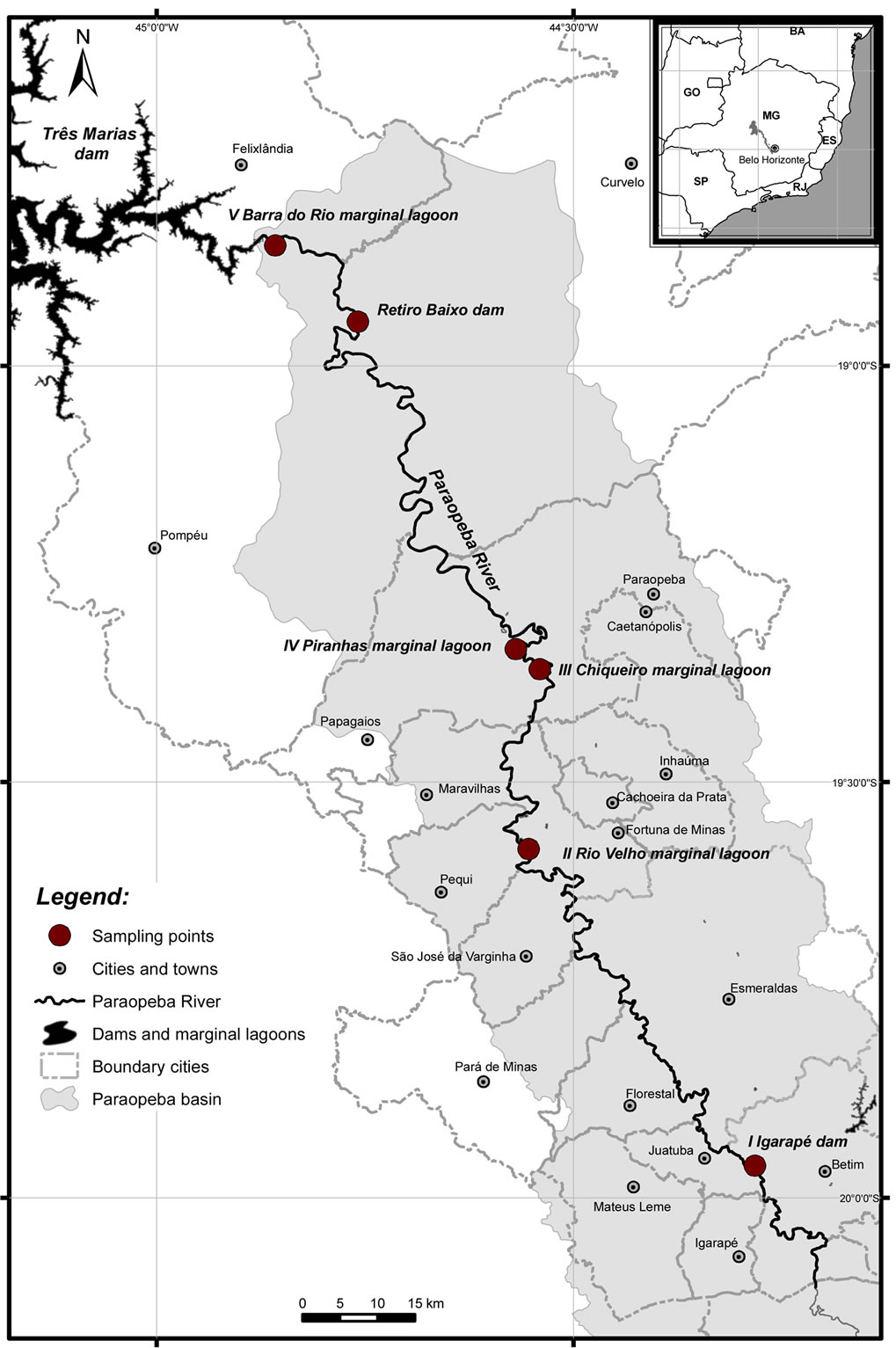
Figure 1. Map of the collection sites in the Paraopeba River, São Francisco River Basin. Site I: Paraopeba River (immediately below the Igarapé Dam); Site II: Rio Velho lagoon; Site III: Chiqueiro lagoon; Site IV: Piranhas lagoon; and Site V: Barra do Rio lagoon.
at a variety of genetic loci, such as random amplification of polymorphic DNA (RAPDs) and mitochondrial and nuclear sequences [13,14] has a wide range of applications in phylogenetic reconstruction, paternity testing, forensic application, and population genetics [15-19]. Microsatellites or short tandem repeats (STRs) are one of the most commonly used molecular markers in population studies present in all prokaryotic and eukaryotic genomes studied to date [20]. STRs are often divided into several categories based on the repeat pattern. Complex hypervariable repeats microsatellite are a category with numerous non-consensus alleles that differ in both size and sequence rates usually being highly polymorphic in natural populations detected and, furthermore, it can be used alone in population studies [21-24].
Dam construction leads to changes in river habitat and flow, which can result in the demise or decline of rheophilic species. Even resilient populations of native species can become isolated, leading to a loss of genetic variability and subsequent fitness reduction and increased risk of extinction [25-27].
In this study, we characterize the standing genetic variation and population structure of P. costatus and P. argenteus prior to the construction of the Retiro Baixo Hydroelectric Dam using complex hypervariable repeats microsatellite. These measurements can then be used as a baseline in future studies investigating changes in the structure, composition, and life cycle of local fish fauna as a result of the dam construction. Our results will be important in the development of effective conservation management programs for these key species.
2. MATERIAL AND METHODS
2.1. Sample Collection
We collected 89 P. costatus and 32 P. argenteus adults’ tissue sampling (1 cm2 of caudal fin) in the Paraopeba River (Minas Gerais, Brazil) during two periods (November 2008 to March 2009 and July 2009 to August 2009). Sampling was conducted using gill nets, casting nets and spearfishing. The fish were collected prior to the construction of the Retiro Baixo Hydroelectric Dam from five distinct sites downstream of the Igarapé Thermoelectric Power Plant (ITH) (Figure 1; Table 1). Site I was at the Paraopeba River immediately downstream the ITH, and the remaining four sites were in marginal lagoons: Rio Velho (site II), Chiqueiro (site III), Piranhas (site IV), and Barra do Rio (site V). Site I was selected as a sampling site to be a place already known as spawning area for migratory fishes such as Prochilodus spp. while the marginal lagoons were selected because they are considered as natural nurseries for migratory species.
2.2. Isolation of Molecular Markers
Genomic DNA was extracted from P. lineatus muscle tissue using the phenolchloroform method [28]. A primary genomic library was constructed from this genomic DNA [16]. A total of 71 positive clones were sequenced with M13 using the BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, CA, USA) on an ABI 3130 automated sequencer (Life Technologies). Repetitive sequences were identified using the Simple Sequence Identification Tool [29]. Clones were sequenced in both directions to build consensus contigs using Phred/Phrap/Consed [30,31]. Primers were designed in PRIMER3 [32].
2.3. Amplification and Analysis of the 2V35 Locus
We amplified the novel marker 2V35 from all 121 specimens by nested polymerase chain reaction (PCR) using the primers: F1 5’-TAA TGA TTC TCT TTG CTT GTG TC-3’, R1 5’-GCA GAC CCC TCA GCA C-3’, F2 5’-TCT TTG CTT GTG TCT TG-3’ and R2 5’-AAG GCC TGA AAT ACA GTG CA-3’. Nested PCR reactions
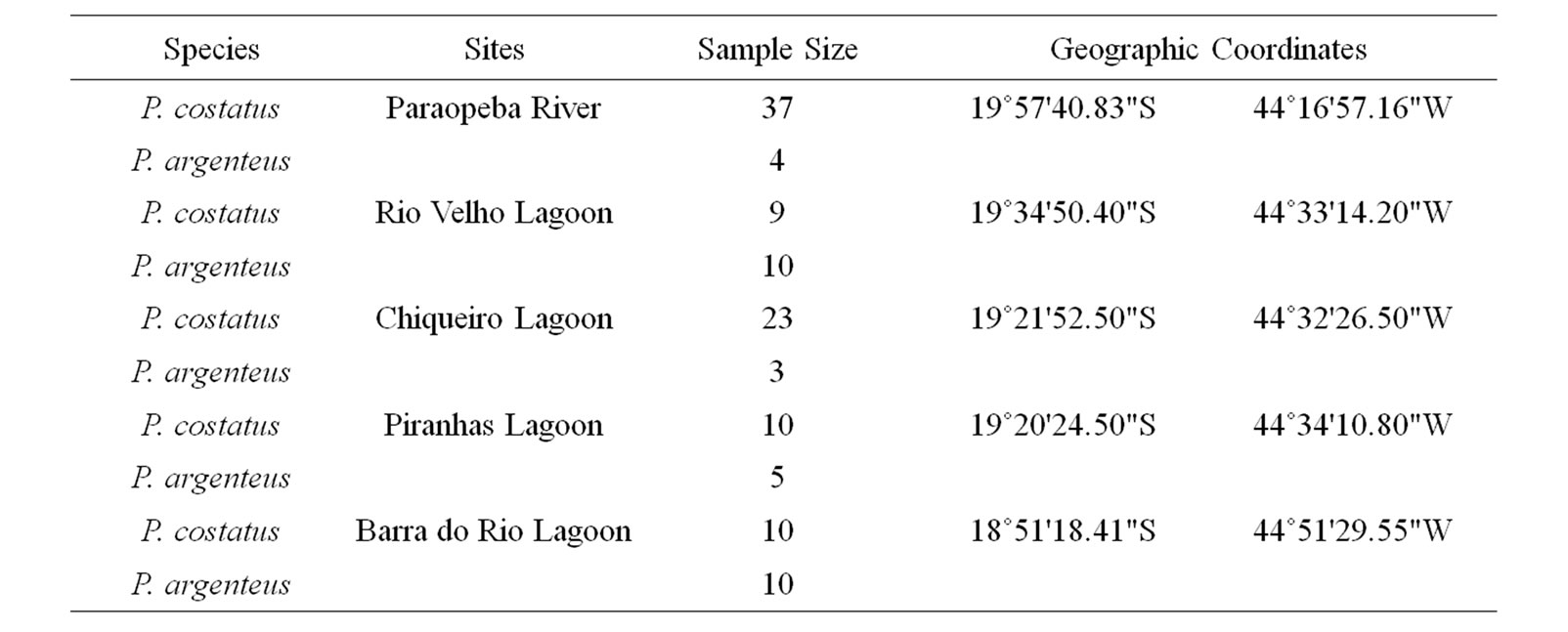
Table 1. Sites, sample sizes, and geographic coordinates of the collected Prochilodus costatus and P. argenteus in the Paraopeba River, São Francisco River Basin. All fishes were collected from 2008 to 2009.
had a total volume of 25 µl, containing 20 - 50 ng of genomic DNA (first round) or 0.5 ng of PCR product (second round), 5 pM of each primer (F1/R1 first round and F2/R2 second round), and a pre-mix containing PCR buffer, dNTPs, and Taq DNA polymerase (Phoneutria Biotecnologia e Serviços, Belo Horizonte, Brazil). The first round of PCR had the following cycling conditions: an initial denaturation at 94˚C for 2 min, followed by 35 cycles of denaturation at 94˚C for 30 s, primer annealing at 54˚C for 30 s, and extension at 72˚C for 40 s, and ending with a final extension step at 72˚C for 5 min. The second PCR round had similar cycling conditions except that the annealing temperature was 51˚C. PCR fragments were sequenced with primers F2 and R2 as described above.
2.4. Statistical Analysis
Arlequin 3.5.1 [33] was used to measure genetic variation at the intra-population level using diversity indices, such as gene diversity, nucleotide diversity, number of observed transitions, transversions, and indels, and the number of haplotypes shared among populations.
The genetic structure of samples was assessed using hierarchical analyses of molecular variance (AMOVA) [34], which was based on the index ΦST and implemented by the program Arlequin v.3.5.1.2, using 16,000 permutations, P < 0.05 [33].
2.5. Isolation by Distance
Isolation by distance was evaluated by testing the correlation between the genetic and geographic distance matrices with a Mantel test [35], using 1000 permutations and logarithmic transformations of both distance measures with the program Alleles in Space [36].
3. RESULTS
3.1. Isolation and Characterization of Molecular Markers
Out of 71 positive clones from the P. lineatus library, 19 (26%) contained repetitive motifs. Approximately 66% of the repeats contained dinucleotide motifs. Of these sequences, 2V35 amplified a fragment with approximately 459-bp in all 121 specimens. Overall, 13 - 17 substitutions and 14 indels resulted in 35 unique P. costatus haplotypes (Table 2) and 4 - 20 substitutions and 2 - 22 indels resulted in 22 unique P. argenteus haplotypes (Table 3).
3.2. Diversity Indices
For each species, most haplotypes were found only at a single site and few haplotypes were shared. For P. costatus, in addition to Haplotype 35 (Hap 35) that was shared among all sites, site III shared Hap 9 with site II, Hap 1 and Hap 2 with site IV, and Hap 3 with sites II and IV. For P. argenteus, sites II, III, IV and V shared Hap 36 and Hap 37, and sites IV and V also shared Hap 46.
Transitions, transversions, and indels in 2V35 and overall diversity indices are given in Table 4 for both species. In P. costatus, genetic diversity was highest (gene diversity = 0.888) in the Paraopeba River population (site I) while in P. argenteus, genetic diversity was highest (gene diversity = 0.942) in the Barra do Rio population (site V).
3.3. Population Differentiation and Structure
Both species showed significant population differentiation at 2V35. The P. costatus populations had a mean Fst of 0.0652. The AMOVA results revealed that 93.48% of genetic variation (P < 0.05) resided within populations. The P. argenteus populations had a mean Fst value of 0.125, and 87.42% of genetic variation resided within populations (P < 0.05) (Table 5).
For P. costatus, pairwise Fst values ranged from 0.009 - 0.12 and were significant in five of ten comparisons (P < 0.05) (Table 6). According to Table 6, site III was significantly different from all other sites, the same is valid for comparison between site I and V (Fst = 0.04, P = 0.17, Table 7). Only site I and site III were not significantly different from each other (Fst = 0.04, P = 0.20).
3.4. Isolation by Distance
Significant isolation by distance was observed for both species. For P. costatus, the correlation coefficient r was 0.115 (P < 0.001) (Figure 2), and for P. argenteus, r was 0.649 (P < 0.001) (Figure 3).
4. DISCUSSION
We characterized the genetic diversity and population structure for P. costatus and P. argenteus, two important endemic fish species of the São Francisco Basin, in one of its most important tributaries, the Paraopeba River. This study is significant in that we captured diversity levels prior to the construction of the Retiro Baixo Hydroelectric Power Plant. We assessed the variation in the novel molecular marker 2V35 with numerous non-consensus haplotypes that differ in both size and sequence from specimens from five sites throughout the basin. The lack of sequence similarity with protein databases (by BLASTX searches; data not shown) and the high level of polymorphism indicate that 2V35 is a non-coding nuclear marker [20,21,37,38]; allowing for the accumulation of mutations in a neutrally evolving region [39].
In the present study, both species had high levels of genetic diversity for the five studied sites and a significant


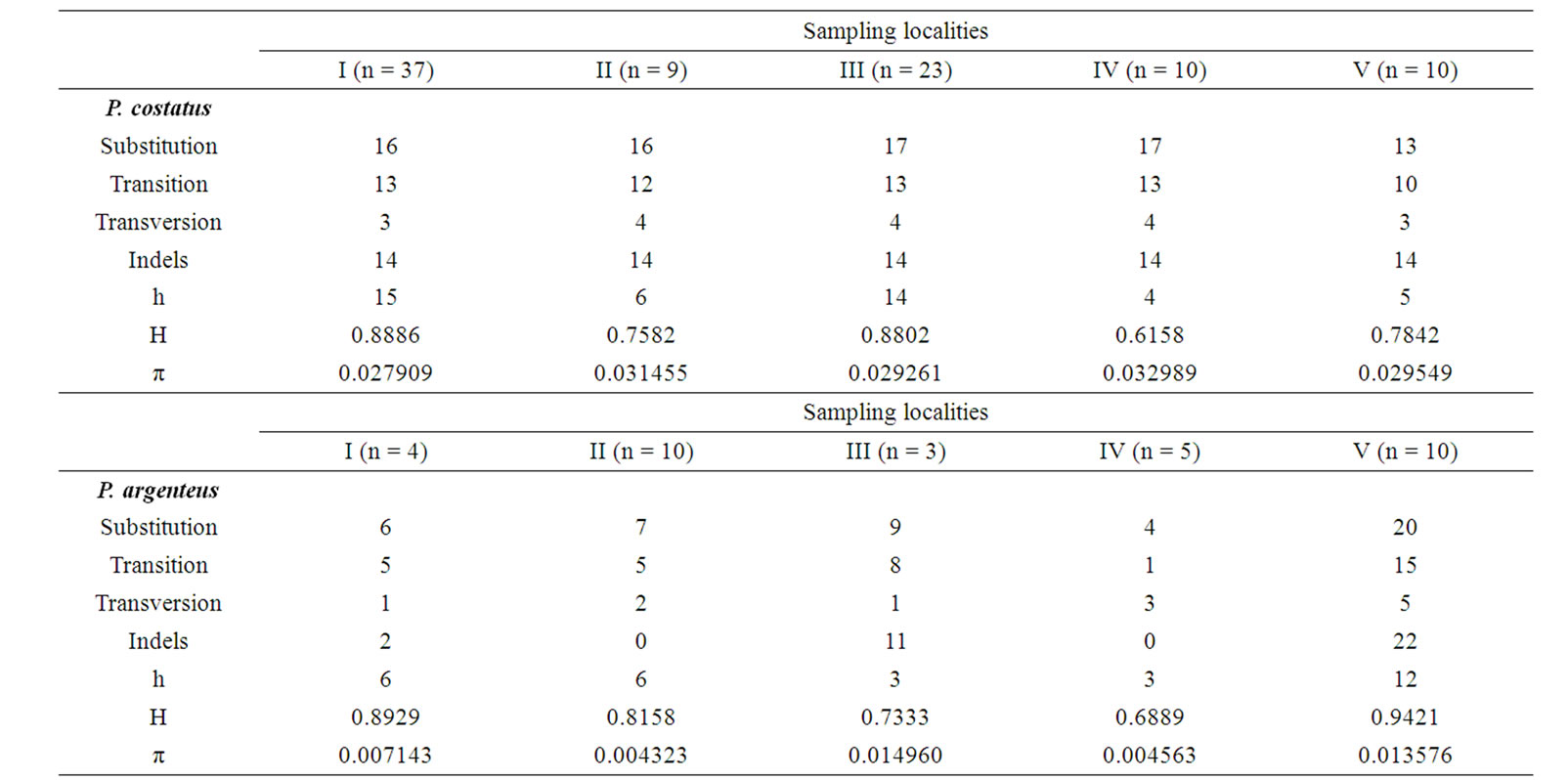
Table 4. Diversity indices at 2V35 in Prochilodus costatus and Prochilodus argenteus in the Paraopeba River, São Francisco River Basin. The diversity indices includes sample size (n); mutations by transitions, transversions and insertion/deletion polymorphisms (indels); number of haplotypes for sampling localities (h); gene diversity (H); nucleotide diversity (π).

Table 5. Analysis of molecular variance (AMOVA) for Prochilodus costatus and Prochilodus argenteus in the Paraopeba River and its marginal lagoons, São Francisco River Basin. *P < 0.05.
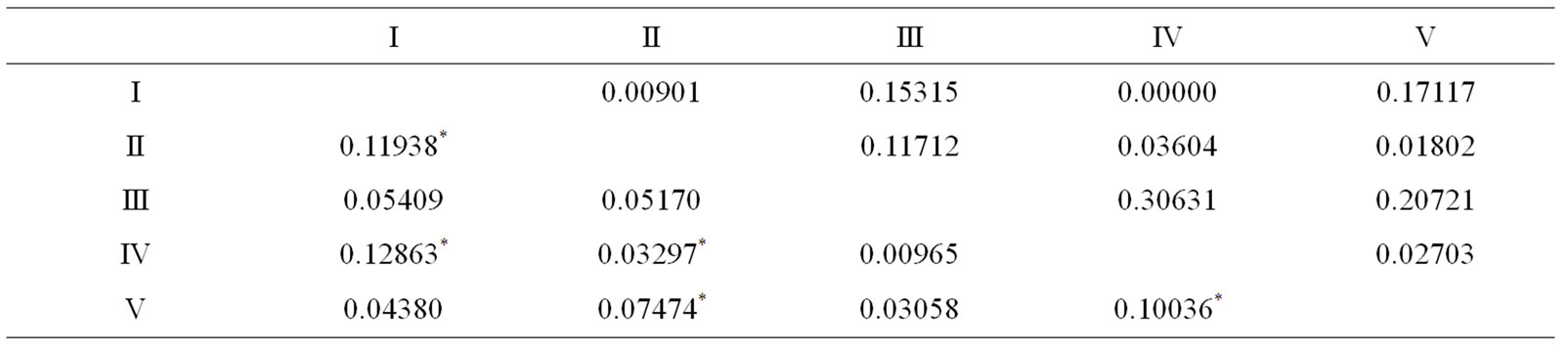
Table 6. Pairwise FST values (below diagonal) and P values (above diagonal) in Prochilodus costatus individuals among the five sampled sites in the Paraopeba River, São Francisco River Basin. *P < 0.05.
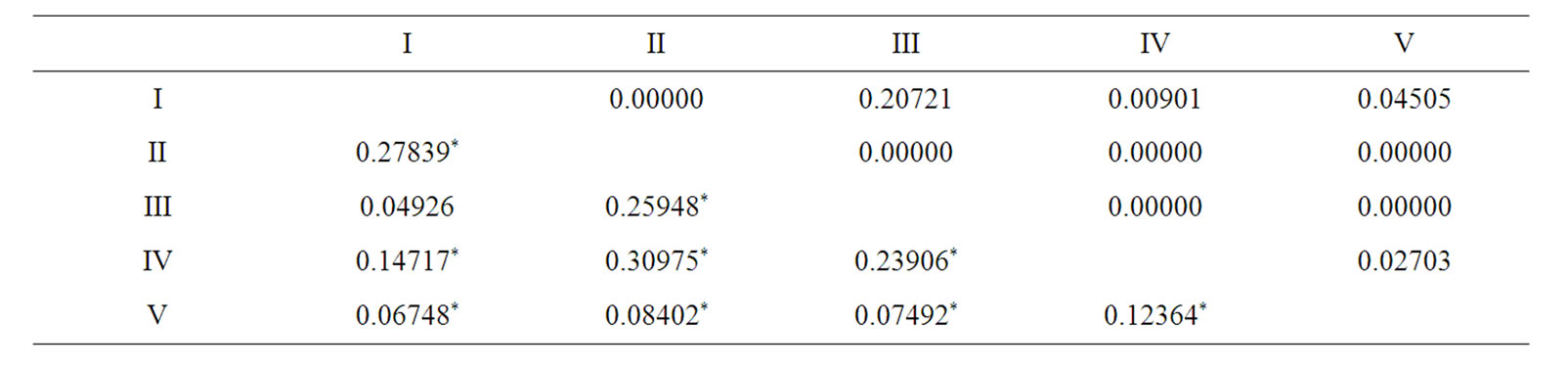
Table 7. Pairwise FST values (below diagonal) and P values (above diagonal) in Prochilodus argenteus individuals among the five sampled sites in the Paraopeba River, São Francisco River Basin. *P < 0.05.
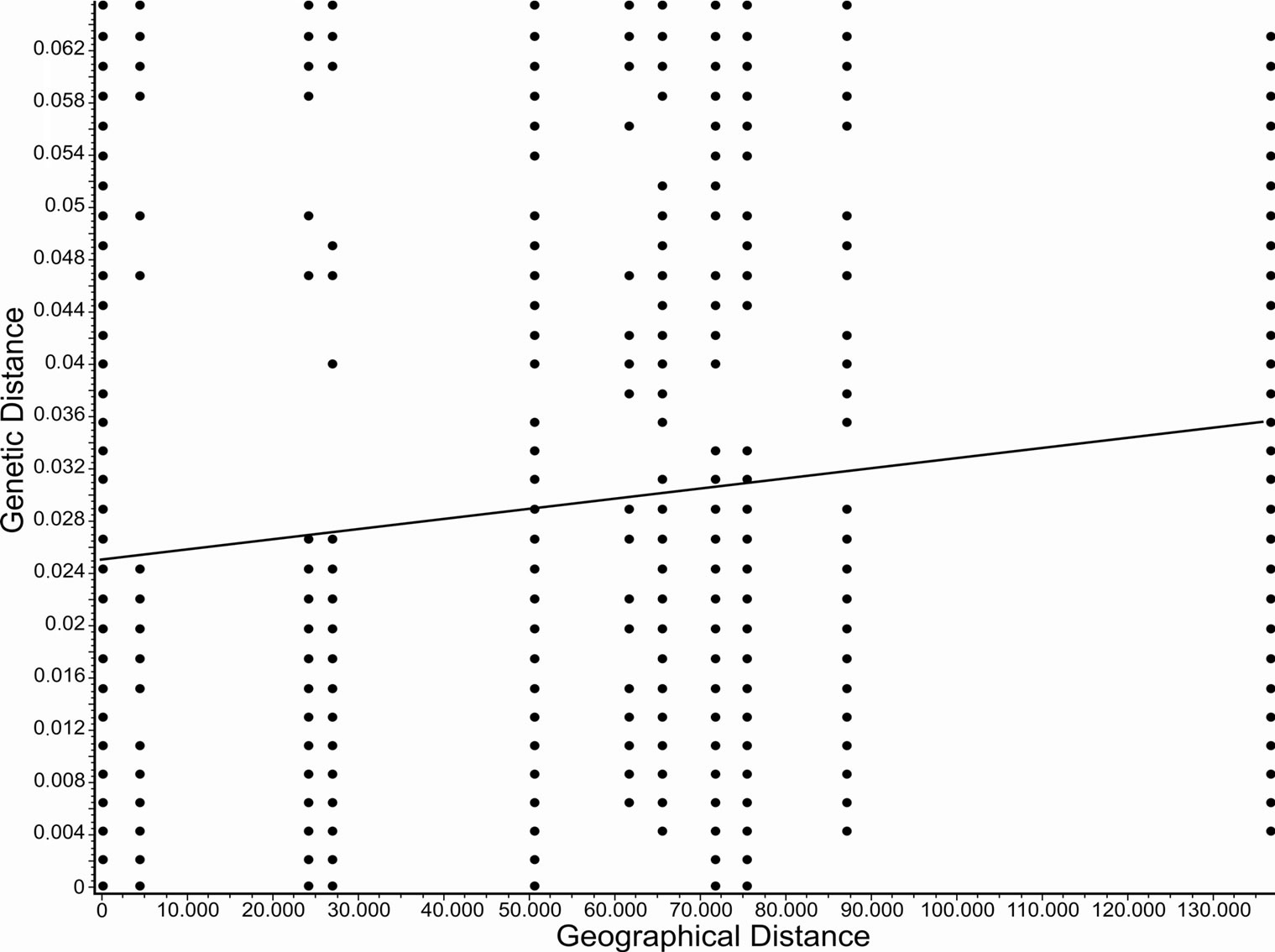
Figure 2. Isolation by distance for Prochilodus costatus (r = 0.115, P < 0.001) calculated using Alleles in Space (Miller, 2005).
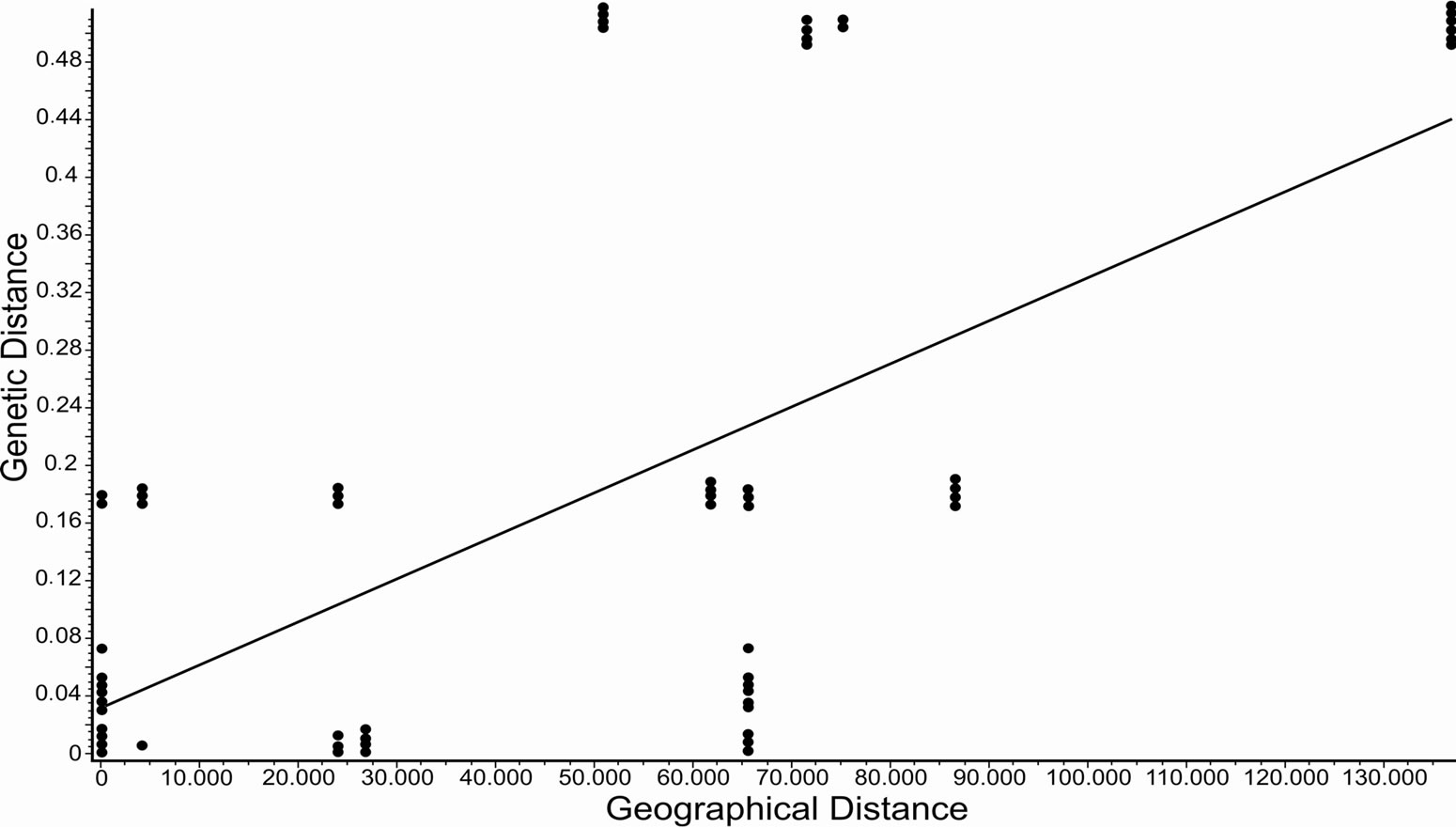
Figure 3. Isolation by distance for Prochilodus argenteus (r = 0.649, P < 0.001) calculated using Alleles in Space (Miller, 2005).
population differentiation at 2V35. Similarly, using microsatellites, P. argenteus populations near the Três Marias Dam were significantly divergent from populations further downstream in the São Francisco River [40].
According to [41], migratory fishes, including Prochilodus spp. generally spawn in the main river channel with the eggs and/or pelagic larvae subsequently concentrated in surrounding wetlands and marginal lagoons. Differences in dispersal ability may affect gene flow, leading to passive drift from some areas of adult migration to sites where pelagic eggs and larvae are found [42]. The observed patterns of genetic diversity, population structure and the significant isolation by distance in these species in the Paraopeba River likely reflect this behaveior. Only half of the Fst pairwise comparisons were significant different in P. costatus. Although, only one comparison was not significant different in P. argenteus. Small sample sizes (n ≤ 4) may overestimate the Fst (e.g. due to rare alleles shared between some populations not sampled). The lowest value in site IV could also be related with small sample sizes.
Thirty-three areas of Minas Gerais State have been recognized as priorities for the conservation of fish biodiversity, and the Paraopeba River was classified in the category of high biological importance [10]. Our genetic data clearly identified five evolutionary significant units (ESUs) for conservation. For both species, the Fst values indicated a significant population differentiation (P. costatus: Fst = 0.065, P < 0.05; P. argenteus: Fst = 0.125, P < 0.05). Similarly, the pattern of few shared 2V35 haplotypes suggests restricted gene flow between populations.
5. CONCLUSION
We conclude that the five study sites are of major importance for conservation and should be managed as a group comprising significant genetic differences. Continued monitoring of these two species post-dam construction will be important to ensure that implemented conservation management plans maintain genetic diversity.
6. ACKNOWLEDGEMENTS
We thank Retiro Baixo Energética and Orteng Equipamentos e Sistemas Ltda. for financial and technical support and biologists V. Torquato, P. Panisa, and LF Ribeiro for the technical support. This research was funded by Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). Dr. E. Kalapothakis is a recipient of a CNPq fellowship.
REFERENCES
- Castro, R.M.C. and Vari, R.P. (2003) Prochilodontidae (Fannel mouth characiforms). In: Reis, R.E., Kullander, S.O. and Ferraris, C.J. Jr., Eds., Checklist of the Freshwater Fishes of South and Central America, Porto Alegre, 65-70.
- Welcomme, R.L. (1979) Fisheries ecology of floodplain rivers. Logman Group Limited, London.
- Bonetto, A.A. (1986) Fish of the Paraná system. In: Davies, B.R. and Walker, K.F., Eds., The Ecology of River Systems, Dordrecht, 573-588.
- CESP (1996) Aspectos limnológicos, ictiológicos e pesqueiros de reservatórios da CESP no período de 1986 a 1994. CESP São Paulo, Série Pesquisa e Desenvolvimento, São Paulo.
- Agostinho, A.A., Vazzoler, A.E.A. de M., Gomes, L.C. and Okada, E.K. (1993) Estratificación y comportamiento de Prochilodus scrofa em distintas fases del ciclo de vida, em la planície de inundação del alto rio Paraná y embalse de Itaipu, Paraná, Brasil. Revue d’Hydrobiolgie Tropicale, 26, 79-90.
- Darnell, R.M. (1961) Trophic spectrum of an estuarine community based upon studies of Lagoon Pontchartrain, Louisiana. Ecology, 42, 552-568. doi:10.2307/1932242
- Darnell, R.M. (1964) Organic detritus in relation to secondary production in aquatic communities. Stuttgart, 15, 462-470.
- Flecker, A.S. (1996) Ecosystem engineering by a dominant detritivore in a diverse tropical stream. Ecology, 77, 1845-1854. doi:10.2307/2265788
- CETEC (Fundação Centro Tecnológico de Minas Gerais) (1983) Diagnóstico ambiental do Estado de Minas Gerais. Série de publicações técnicas/SPT, 010, Minas Gerais.
- Drumond, G.M., Martins, C.S., Machado, A.B.M., Sebaio, F.A. and Antonini, Y. (2005) Biodiversidade em Minas Gerais: um atlas para sua conservação. 2nd Edition, Fundação Biodiversitas, Belo Horizonte.
- Alves, C.B.M. (2007) Evaluation of fish passage through the igarapé dam fish ladder. Neotropical Ichthyology, 5, 233-236. doi:10.1590/S1679-62252007000200019
- Poke, F.S., Vaillancourt, R.E., Potts, B.M. and Reid, J.B. (2005) Genomic research in Eucalyptus. Genetica, 125, 79-101. doi:10.1007/s10709-005-5082-4
- Avise, J.C. (1987) Intraspecific phylogeography: The mitochondrial DNA bridge between population genetics and systematic. Annual Review of Ecology and Systematics, 18, 489-522. doi:10.1146/annurev.es.18.110187.002421
- Parker, P.G., Snow, A.A., Schug, M.D., Booton, G.C. and Fuerst, P.A. (1998) What molecules can tell us about populations: Choosing and using a molecular marker. Ecology, 79, 361-382. doi:10.2307/176939
- Vrijenhoek, R.C. (1998) Conservation genetics of freshwater fish. Journal of Fish Biology, 53, 394-412. doi:10.1111/j.1095-8649.1998.tb01039.x
- Yazbeck, G.M. and Kalapothakis, E. (2007) Isolation and characterization of microsatellite DNA in the piracema fish Prochilodus lineatus (Characiformes). Genetics and Molecular Research, 6, 1026-1034.
- Barbosa, A.C.D.R., Galzerani, F., Corrêa, T.C., Galetti, P.M. Jr. and Hatanaka, T. (2008) Description of novel microsatellite loci in the Neotropical fish Prochilodus argenteus and cross-amplification in P. costatus and P. lineatus. Genetics and Molecular Biology, 31, 357-360. doi:10.1590/S1415-47572008000200032
- Paiva, A.L.B. and Kalapothakis, E. (2008) Isolation and characterization of microsatellite loci in Pimelodus maculatus (Siluriformes: Pimelodidae). Molecular Ecology Resources, 8, 1078-1080.
- Hsu, T.H., Guillén Madrid, A.G., Burridge, C.P., Cheng, H.Y. and Gwo, J.C. (2011) Resolution of the Acanthopagrus black seabream complex based on mitochondrial and amplified fragment-length polymorphism analyses. Journal of Fish Biology, 79, 1182-1192. doi:10.1111/j.1095-8649.2011.03100.x
- Zane, L., Barceloni, L. and Patarnello, T. (2002) Strategies for microsatellite isolation: A review. Molecular Ecology, 11, 1-16. doi:10.1046/j.0962-1083.2001.01418.x
- Urquhart, A., Kimpton, C.P. and Gill, P. (1993) Sequence variability of the tetranucleotide repeat of the human betaactin related pseudogene H-beta-Ac-psi-2 (ACTBP2) locus. Human Genetics, 92, 637-638. doi:10.1007/BF00420957
- Gill, P., Kimpton, C.P., D’Aloja, E., Andersen, J.F., Bar, W., Brinkmann, B., Holgersson, S., Johnsson, V., Kloosterman, A.D., Lareu, M.V., Nellemann, L., Pfitzinger, H., Phillips, C.P., Schmitter, H., Schneider, P.M. and Stenersen, M. (1994) Report of the European DNA profiling group (EDNAP)-towards standardisation of short tandem repeat (STR) loci. Forensic Science International, 65, 51-59. doi:10.1016/0379-0738(94)90299-2
- Reid, T.M., Ingala, D.A., Kraemer, C.M., Dage, W.M., Dieckhoner, C., Fortman, J., Hodge, D.M., Johnson, K.L., Oatman, C., Schlotman, H., Schuh, C. and Baird, M.L. (2003) Distribution of HUMACTBP2 (SE33) alleles in three North American populations. Journal of Forensic Science, 48, 1422-1423.
- Butler, J.M., Hill, C.R., Kline, M.C., Duewer, D.L., Sprecher, C.J., McLaren, R.S., Rabbach, D.R., Krenke, B.E. and Storts, D.R. (2009) The single most polymorphic STR Locus: SE33 performance in US populations. Forensic Science International: Genetics Suppliment Series, 2, 23-24. doi:10.1016/j.fsigss.2009.08.173
- Neraas, L.P. and Spruell, P. (2001) Fragmentation of riverine systems: The genetic effects of dams on bull trout (Salvelinus confluentus) in the Clark Fork River system. Molecular Ecology, 10, 1153-1164. doi:10.1046/j.1365-294X.2001.01269.x
- Frankham, R., Ballou, J.D. and Briscoe, D.A. (2004) A primer of conservation genetics. Cambridge University Press, Cambridge.
- Reid, S.M., Wilson, C.C., Mandrak, N.E. and Carl, L.M. (2008) Population structure and genetic diversity of black redhorse (Moxostoma duquesnei) in a highly fragmented watershed. Conservation Genetics, 9, 531-546. doi:10.1007/s10592-007-9367-2
- Sambrook, J. and Russell, D. (2001) Molecular cloning: A laboratory manual. 3rd Edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Temnykh, S., De Clerck, G., Lukashova, A., Lipovich, L., Cartinhour, S. and McCouch, S. (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): Frequency, length variation, transposon associations, and genetic marker potential. Genome Research, 11, 1441-1452. doi:10.1101/gr.184001
- Ewing, B. and Green, P. (1998) Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Research, 8, 186-194.
- Gordon, D., Abajian, C. and Green, P. (1998) Consed: A graphical tool for sequence finishing. Genome Research, 8, 195-202.
- Rozen, S. and Skaletsky, H.J. (2000) Primer3 on the www for general users and for biologist programmers. In: Krawetz, S. and Misener, S., Eds., Bioinformatics Methods and Protocols: Methods in Molecular Biology, Humana Press, Totowa, 365-386.
- Excoffier, L. and Lischer, H.E.L. (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567. doi:10.1111/j.1755-0998.2010.02847.x
- Excoffier, L., Smouse, P.E. and Quattro, J.M. (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics, 131, 479-491.
- Mantel, N. (1967) The detection of disease clustering and a generalized regression approach. Cancer Research, 27, 209-220.
- Miller, M.P. (2005) Alleles in space (AIS): Computer software for the joint analysis of interindividual spatial and genetic information. Journal of Heredity, 96, 722- 724. doi:10.1093/jhered/esi119
- Schlötterer, C. (2000) Evolutionary dynamics of microsatellite DNA. Chromosoma, 109, 365-371. doi:10.1007/s004120000089
- Ellegren, H. (2004) Microsatellites: Simple sequences with complex evolution. Nature Reviews Genetics, 5, 435-445. doi:10.1038/nrg1348
- Chenuil, A. (2006) Choosing the right molecular markers for studying biodiversity: From molecular evolution to practical aspects. Genetica, 127, 101-120. doi:10.1007/s10709-005-2485-1
- Hatanaka, T., Henrique-Silva, F. and Galetti, P.M. Jr. (2006) Population substructutring in a migratory freshwater fish Prochilodus argenteus (Characiformes, Prochilodontidae) from the São Francisco River. Genetica, 126, 1-7. doi:10.1007/s10709-005-1445-0
- Nakatani, K., Baumgarter, G. and Cavicchioli, M. (1997) Ecologia de ovos e larvas de peixes. In: Vazzoler, A.E., A.M., Agostinho, A.A. and Hahn, N.S., Eds., A Planície de Inundação do Alto rio Paraná: Aspectos Físicos, Biológicos e Socioeconômicos, Maringá, 281-306.
- Maggio, T., Lo Brutto, S., Garoia, F., Tinti, F. and Arculeo, M. (2009) Microsatellite analysis of red Mullus barbatus (Perciformes, Mullidae) reveals the isolation of the Adriatic Basin in the Mediterranean Sea. ICES Journal of Marine Science, 66, 1883-1891. doi:10.1093/icesjms/fsp160

