Paper Menu >>
Journal Menu >>
 J. Biomedical Science and Engineering, 2011, 4, 552-561 doi:10.4236/jbise.2011.48071 Published Online August 2011 (http://www.SciRP.org/journal/jbise/ JBiSE ). Published Online August 2011 in SciRes. http://www.scirp.org/journal/JBiSE Segmentation of MS lesions using entropy-based EM algorithm and Markov random fields Ahmad Bijar1, Mahdi Mohamad Khanloo1, Antonio Peñalver Benavent2, Rasoul Khayati1 1Department of Biomedical Engineering, Shahed University, Tehran, Iran; 2Department de Estadística, Matemáticas e Informática, Universidad Miguel Hernández, Elche, Spain. Email: ahmad_bijar@live.com; mahdi.khanlo@gmail.com; a.penalver@umh.es; khayati@shahed.ac.ir Received 25 June 2011; revised 28 July 2011; accepted 8 August 2011. ABSTRACT This paper presents an approach for fully automatic segmentation of MS lesions in fluid attenuated inver- sion recovery (FLAIR) Magnetic Resonance (MR) images. The proposed method estimates a gaussian mixture model with three kernels as cerebrospinal fluid (CSF), normal tissue and Multiple Sclerosis le- sions. To estimate this model, an automatic Entropy based EM a lgo r it h m is u s ed to f ind th e b est es t ima ted Model. Then, Markov random field (MRF) model and EM algorithm are utilized to obtain and upgrade the class conditional pro bability density function and the apriori probability of each class. After estimation of Model parameters and apriori probability, brain tissues are classified using bayesian classification. To evaluate the result of the proposed method, similarity criteria of different slices related to 20 MS patients are calculated and compared with other methods which include manual segmentation. Also, volume of segmented lesions are computed and compared with gold standard using correlation coefficient. The pro- posed method has better performance in comparison with previous works which are reported here. Keywords: Gaussian Mixture Model; EM; Entropy; Markov Random Field; Multiple Sclerosis 1. INTRODUCTION Magnetic Resonance Imaging (MRI) permits the non- invasive detailed visualization of internal anatomical structures. The segmentation of MR images into ana- tomical tissues, fluids, and structures is an area of inter- est in MRI, and is increasingly being used to assess the progression of the disease and to evaluate the effect of drug therapy, supplementing traditional neurological disability scales such as the extended disability status scale (EDSS) [1]. Multiple Sclerosis (MS) is a chronic, inflammatory, demyelinating disease of the Central Nervous System (CNS) that primarily affects the white matter (WM) of the central nervous system. MS lesions exhibit hypersignals in T2 weighted and hyposignals in T1 weighted, with respect to normal white matter inten- sities. Previous research has shown that the FLAIR (Fluid Attenuated Inversion Recovery) sequence con- tains the most distinctive lesion-healthy tissue differen- tiation for segmentation of white matter lesions. The radiological criteria for MS include the number of lesion on the MRI, their locations and their sizes, and these quantitative information is also crucial for studying the progression of MS lesions and the effect of drug treat- ments. To make a quantitative analysis of the brain scans of the patient, human experts are required to identify the multiple sclerosis lesions that are present in those scans as manual segmentation which is extremely time con- suming. So, automatic and semi-automatic methods for segmentation of MS lesions are recommended and have been investigated extensively. There are many proposed approaches, automatic and semi-automatic, for segmen- tation of brain into different tissues, including MS le- sions. These approaches include a variety of methods such as statistical, fuzzy, neural networks, and fuzzy neural networks. In this paper, we develop a fully auto- matic method for segmentation of MS lesions that re- quires no training, atlas, or thresholding steps. Our method can be divided into two main sections: at first, intensities of brain pixels are modeled using a gaussian mixture model which consists of three kernels as CSF, normal tissue and MS lesions classes. This step starts with only one kernel and uses an entropy based EM al- gorithm to estimate three kernels as three mentioned classes in an automatic manner which does not need initial values for parameter estimation. After estimation of the model, we would be able to classify brain pixels by knowing apriori probabilities of the classes. The next step is obtaining these apriori probabilities with no training and atlas. So, a MRF model and EM algorithm  A. Bijar et al. / J. Biomedical Science and Engineering 4 (2011) 552-561 553 are applied to update and attain apriori probabilities and means and variances of each class. 2. METHODS 2.1. Patients and MR Imaging The proposed method is evaluated on a dataset which is obtained and used in [2]. This dataset contains 16 female and 4 male with average age of 29 ± 8 years old, this dataset is selected according to the revised Mc Donald criteria 2005 [3]. All images were acquired according to full field MRI criteria of MS [3] in T2-weighted (T2-w), T1-weighted (T1-w), Gadolinium enhanced T1-weighted, and FLAIR in axial, sagittal and coronal surfaces. We selected the FLAIR images, especially axial ones, with lesions in deep, priventricular, subcortical, and cortical white matters (supratentorial lesions). More lesion load and higher accuracy of FLAIR in revealing of these MS lesions were the reason for this selection [4]. Each image volume (patient data) consisted of averagely 40 slices with a 256 × 256 scan matrix. The pixel size was 1mm2, and the slice thickness was 3mm without any gap. 2.2. Manual Segmentation of MS Lesions and Brain Segmentation The segmentation of MS lesions was performed manu- ally by neurologist and radiologist in Flair images with visual inspection of corresponding T1-w and T2-w im- ages. These manually segmented images were used as Gold standard [5] to evaluate the performance of pro- posed method. To evaluate the proposed method, differ- ent types of images which have different lesion volumes were applied. Also, the brain segmentation was per- formed using a fully automatic object-oriented approach [6]. This method was based on the regional-spatial char- acteristics of brain in MR images. At first, original im- age is converted to a binary image. Then, morphological opening on the binary image is performed and tiny re- gions are eliminated. Three rectangular masks showing the cerebral regions are produced and the regions in the binary image which have overlap with these rectangles are preserved and, the rest are eliminated. Final mask is generated by dilation of selected regions and filling tiny holes. Finally, an image, which includes only cerebral tissues, is obtained by applying the resulted mask on the original image. 3. PROBLEM FORMULATION 3.1. Gaussian Mixture Model A finite mixture model is the weighted sum of M > 1 components in for xp n n 1 1 xπxx Mn m m ppm M (1) where corresponds to the weight of each component which satisfies π0,11,2,, mm 1mπ1 M m . For the Gaussian mixtures model, each component den- sity xpm is a Gaussian probability density given by xm 1 2 2 1 x 2πdet 1 exp x 2 mn m T mm p (2) where T denotes the transpose operation, m is the mean vector and m is the covariance matrix which is assumed positive definite. Here we encapsulate these parameters into a parameter vector, writing the parame- ters of each component asmmm to get ,, 12 π,π,,π, ,. 12 ,, m m Eq.1 can be rewritten as 1 xπx mm m pp M (3) If we knew the component from which x came, then it would be simple to determine the parameters . Simi- larly, if we knew the parameters , we could determine the component that would be most likely to have pro- duced x. The difficulty is that we know neither. 3.2. Bayesian Classification Bayesian Classification is a probabilistic technique of pattern recognition and is based on the principle of Bayes decision theory [7], given in Eq.4 below j j j px P Px px (4) where, x is a given feature vector, j denotes a class, or state of nature, j P is the prior probability of class j , px is prior probability of the feature vec- tor x, j px is aposteriori probability, which a fea- ture vector should be classified as belonging to class j , j P x is the conditional probability that a feature vector occurs in a given class j . For the approach here, the feature x shall consist of one component, intensity of brain pixels. The quantity is known as the evi- dence, and serves only as a scale factor, such that the quantity in Eq.4 is indeed a true probability, with values between zero and one. So, the maximum a posteriori (MAP) estimate of Eq.4 is used as below px j jj Pxpx P (5) According to Bayesian theory [8], the feature vector x is classified to j of which the aposteriori probability given x is the largest between the classes. max jj iii px P px P j x (6) C opyright © 2011 SciRes. JBiSE  A. Bijar et al. / J. Biomedical Science and Engineering 4 (2011) 552-561 554 Bayes decision rule is optimal in the sense of minimi- zation of the probability of error. It is quite obvious that such an Ideal Bayesian solution can be used only if dis- tributions j px , and the apriori probabilitles j P are known. In the context of classification of brain tissue, the probability models are not known, and therefore, must be approximated. The performance of the Bayesian classifier is directly related to how well these distribu- tions can be modeled. 3.3. EM Algorithm The Expectation-Maximization (EM) algorithm [9] is a general-purpose iterative algorithm for maximum likeli- hood (ML) estimation.The EM algorithm consists of two steps, E-step (expectation) and M-step (maximization) which are alternately used until the 1 log n i ipx converges to a local optimum. Let ,1,, i X xi n be a set of n realizations of a random d-dimensional vector χ with a pdf . Thus the parameterized pdf can be written as a combination of pdfs of the M com- ponents m characterizing the finite mixture model i px ,M 1,wm 1π M im m px px im (7) The likelihood can be expressed as 1 11 π nn M im m ii Lpx px im (8) E-step: the data χ are assumed to be incomplete and the complete data set is determined by es- timating the set of variables ,Y Z 12 ,, M Z ZZ Z, where each M Z is an N-dimensional vector . The log likelihood of the complete data Y is T , N m z 12 ,, mm zz 11 loglog πx NM ii mm m im pYz p (9) where 1 πx x, πx tit mm iit mMtit ll l p zPm p (10) is the posterior probability and is the parameter estimate obtained after t iterations. t M-step: in this step, the parameters are deter- mined according to the estimate of the variables . For Gaussian mixture models this corresponds to reestimat- ing the , the 1t i m z 1 πt m 1t m and 1t m for each m according to: 1 1 1 πN t m iz N i m (11) 11 1 x Ni mi ti mNi m i z z (12) 111 11 1 xx T tit t mi mi m ti N mi m i z z (13) 4. ENTROPY In the information theory, information entropy is a measurement of the system uncertain degree. The term of entropy in the theory of information was first intro- duced by Shannon, in 1948, in [10]. 4.1. Shannon Entropy Let 1,, n X xx be a discrete random variable with values in S and probability mass function px, the information (or uncertainty) of each possible event is log i i I xp x (14) The mean of I X is introduced as Shannon en- tropy and is denoted by H X log xs H XEI Xpxpx (15) H X varies from zero to log S zero meaning that there is no uncertainty, whereas log S is reached when all elements of X have equal probabilities, in this case, the uncertainty is at its maximum [11]. 4.2. Rényi Entropy Rényi further expanded the concept of Sannon entropy, and defined the 0, 1qq q order Rényi [12] en- tropy of probability density function as px 1log, 1 1 q qxs HXpx q q (16) Rényi entropy is a non-increasing function of q and It can be verified, by applying the 1’Hôpital rule that Shannons entropy is the Rényi entropy of order 1. 5. ENTROPY ESTIMATION In this paper, Rényi entropy [12] is selected as entropy measure and estimation of this type of entropy which will be express, is based on Leonenko and et al. [13]. Let n X be a random vector with probability measure μ having the density p. The proposed method by Leonenko estimates q H from a sample of N inde- pendent and identically distributed (i.i.d.) random vari- ables 12 N based on the nearest neigh- bor distances in the sample. This work is resumption of the method which estimates ,,XX,X N 2 1 H and proposed by Ko- zachenko and Leonenko [14]. Leonenko’s method estimates q, Eq.17, for I1q through the computation of conditional moments of nearest-neighbor distances. 1, q q qq xs IIppxpx q 1 (17) C opyright © 2011 SciRes. JBiSE  A. Bijar et al. / J. Biomedical Science and Engineering 4 (2011) 552-561 555 5.1. Estimators Suppose that 12 , are i.i.d. with a probability measure μ having a density p with respect to the Lebesgue measure. Let ,,, 2 N XXX N , x y denote the Euclid- ean distance between two points , x y of . For a given sample m 1, N X X and a given i X in the sample, from the distances 1N ij ,, 1,,, X Xj N i the order statistics 1, 12, 11, 1 NN ,ji ii NN are formed, so that is the kth nearest-neighbor distance from i ,1 i kN X to some other j X in the sample, .jiq I in Eq.18 is estimated for , by 1q 1 ,, ,, 1 ˆq Nkq Nik IN ζ (18) where ,,, 1 1 m i Nikk mkN NCV ζ 2 π21 m m Vm ( is the volume of the unit ball in ) m V 0, 1 m 11q 1 k Ckkq So, q H in Eq.17 is estimated as below ,, ,, ˆ log 1 NKq Nkq H Iq (19) 5.2. Maximum Entropy Suppose that we have a group of variables which have obtained from different types of distributions and have equal variance. According to the second Gibbs theorem [15], among all of them, Gaussian variables have maxi- mum entropy. Therefor, by using this theorem, we would be able to understand that if the modeling of the variable with a gaussian kernel is acceptable or not. This procedure is done through the comparing of the theo- retical maximum entropy and the real one of the under- lying data. Let X be a d dimensional variable, the theo- retical maximum entropy is computed as below max 1log 2π 2 d HX (20) where, is covariance of observations. There would be no difference between the theoretical maximum en- tropy and the real one. If the data were truly Gaussian. We have used this property to determine the worst esti- mated kernel in the mixture. 6. SELECTION AND SPLIT OF THE WORST ESTIMATED KERNEL For every kernel in the mixture, the theoretical maxi- mum entropy and the real one of the underlying data is computed and evaluated in the normalized weighted form [16] to find the worst estimated kernel. The worst estimated kernel would be selected through max real max arg maxπ kk H kH k kHk (21) where, k indicates the worst estimated kernel, real H k is the real entropy of the data under the kth kernel, and max H k is the maximum entropy of the kth kernel. Suppose that a special kernel has been se- lected to split, during the split step, overall dispersion and two new covariance matrices have to be constant and positive define, respectively. If the kth component had to split into the k1th and k2th components, the first two moments, the mean vectors and the covariance ma- trices should satisfy the following split equations which are proposed by Richardson and Green [17] 12 112 2 πππ ππ π kkk kkkkkk 11112 222 π ππ T kk kk TT kkkkk kkk (22) Solving the split equations is an ill-posed problem because the number of the equations is less than the number of the unknowns. To solve this ill-posed prob- lem, we have used the method which is proposed by Dellaportas and Papageorgiou in [18]. They have used the spectral decomposition of the current covariance matrix, and the original problem is replaced by estimat- ing the new eigenvalues and eigenvectors of the new covariance matrices without the previously cited con- straint. Let k the spectral decomposition of the covariance matrix VV T kkk k , with , the eigenvector matrix, Vk 1 diag ,d kkk a diagonal matrix con- taining the eigenvalues of k with increasing order, and d the problem dimension. Let also D be a d × d rota- tion matrix with orthonormal unit vectors as columns. D is built by generating its lower triangular matrix inde- pendently from 12dd different uniform U(0,1) densities. The proposed split operation is given by 11 21 2 1 1 1 2 2 1 2 2 1 2 1 322 322 ππ π1π π Vπ π Vπ diag diagdiag diagdiagdiag 1 kkk kkk dk iii kk kk i k dk iii kk kk i k kk kk u u uuu uuu C opyright © 2011 SciRes. JBiSE  A. Bijar et al. / J. Biomedical Science and Engineering 4 (2011) 552-561 556 1 2 VV VV k T kk D D k (23) where is a d × 1 vector of ones, and T 12 12 222 ,,,, d uuu uu, and are T 12 333 3 ,,, d uuu u 2d + 1 random variables needed to build priors, means, and eigenvalues for the new component in the mixture. They are calculated as 1 12 1 233 2, 21,2 1,1 1,0,1 j uud uUu duU j d (24) with , and and denoting beta and uniform distributions, respectively. The expres- sions for 1 2, ,j .,.U .,. and 2 establish that the new means are obtained by moving across the axes formed by the ei- genvectors with sizes 11 2k u and 22 2k u . We must keep positive but let vary in [–1, 1] the interval because the moves in both directions across the large eigenvector are achieved via expressions for 1 1 2 u2 2 u and 2 , but the moves across the shorter axis require both positive and negative . 2 2 u 7. ESTIMATION OF PROBABILITY DENSITY FUNCTION This section describes the proposed method for fitting Gaussian mixture models in depth. The algorithm starts with only one kernel, whose initial parameters are given by the whole samples. As is shown in [19], if we denote 12, a set of samples of a pdf, the ML hypothesis for the mean and covariance matrix are ,, d N yy y 11 T 1 11 1 1 1 N i i N i i y N y N (25) Furthermore, the initial value for the first prior 1 is 1. EM algorithm computes a fitted model which consists only one kernel by these initial values. Whereas the model has only one kernel, this kernel should be split and replace by two other ones. This work is done by Eq.23. The new model with two kernels has to fit on samples. EM algorithm with initial values which are obtained by previous split, is applied for this reason. Now, the fitted model has two kernels as two classes and it is less than our interested number of kernels which is three, csf, normal tissue and MS lesions. So, the worst fitted kernel among these two kernels has to be selected and split. As mentioned before, the worst fitted kernel has big difference between the theoretical maximum entropy and the real one against other kernels. So, we could select the worst kernel by means of maximum entropy and real one using Eq.21. π The worst fitted kernel is replaced by two new kernels through split (Eq.23) and the other kernel is remained with no variation. To find the final fitted model which has three kernels, another EM algorithm is applied, the initial parameters for EM algorithm are obtained by split (split of the worst fitted kernel) and from previous EM algorithm (for remained kernel). Figure 1 shows the Entropy-based EM algorithm to fit the best fitted mix- ture to barin samples. 8. MARKOV RANDOM FIELDS An advantage of MRF models [20] is the use of neighborhood information to improve the apriori prob- abilities P [21]. The intuition behind the MRF model is that most pixels belong to the same class as their neighbors, and it is a powerful tool to describe the class assigning or labeling dependence between adjacent pixels. Suppose a digital image bases on a M N lattice, so image describes as ,1 ,1SsxyxM yN (26) Fit a mixture with three kernels to brain samples Brain Image Fit a kernel to all brain samples through EM algorithm. Split the obtained kernel into two new kernels and use the obtained result as initial values to fit a new model with two kernels to all samples through the EM algorithm. Find the worst estimated kernel in the mixture which has two kernels by comparing the maximum entropy and real entropy, and split it into two new kernels and use its result as initial values for the next level , the other kernel has no variation . Fit a model w ith three kernels to samples through the EM algorithm, initial values are prepared from previous step. Figure 1. The Entropy-based EM algorithm to find the best fitted model to barin samples. C opyright © 2011 SciRes. JBiSE 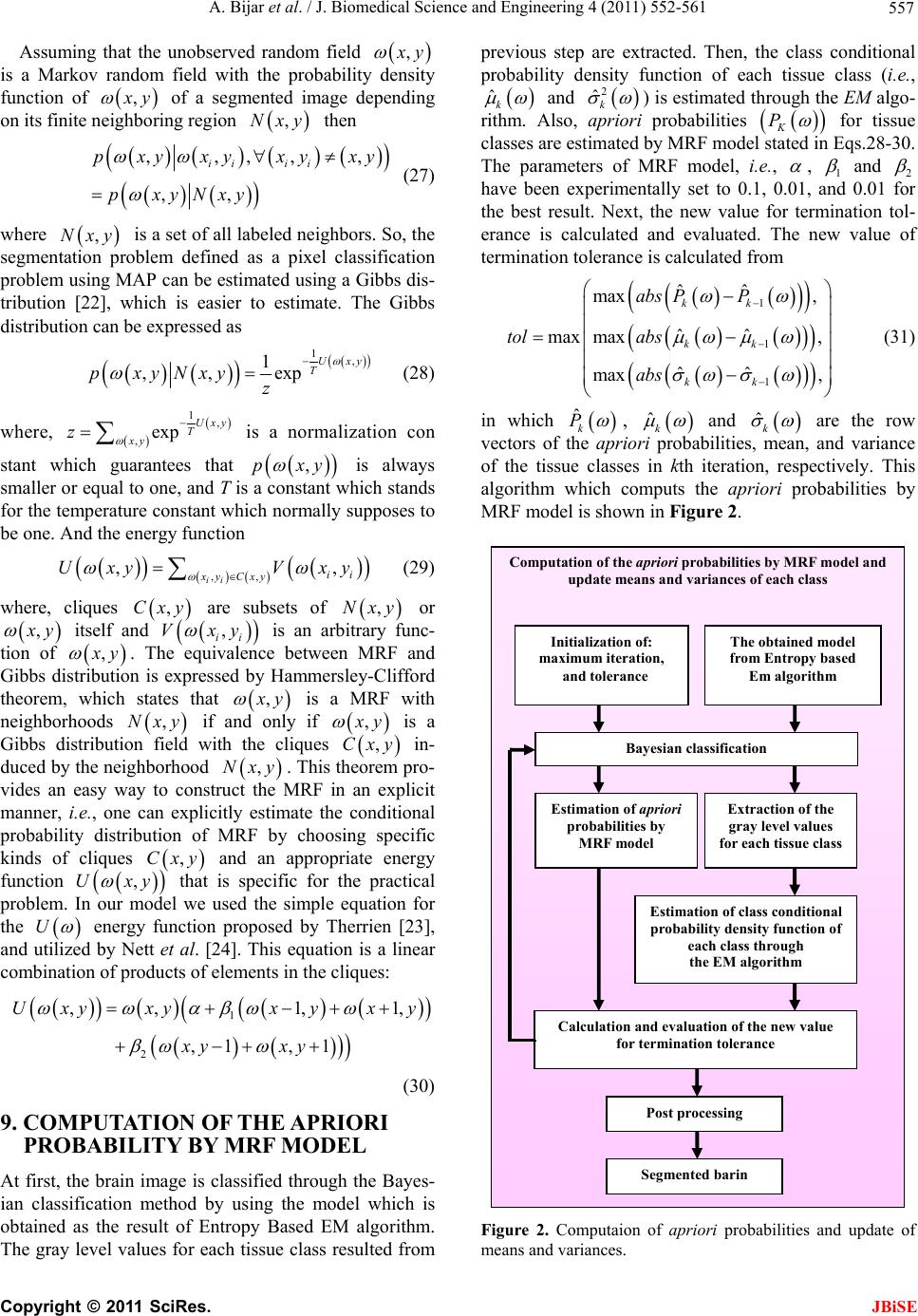 A. Bijar et al. / J. Biomedical Science and Engineering 4 (2011) 552-561 557 Assuming that the unobserved random field , x y is a Markov random field with the probability density function of , x y of a segmented image depending on its finite neighboring region then ,Nxy ,,,,, ,, ii ii pxy xyxyxy pxyNxy (27) where is a set of all labeled neighbors. So, the segmentation problem defined as a pixel classification problem using MAP can be estimated using a Gibbs dis- tribution [22], which is easier to estimate. The Gibbs distribution can be expressed as ,Nxy 1, 1 ,,exp Uxy T pxyNxy z (28) where, 1, ,exp Uxy T xy z is a normalization con stant which guarantees that is always smaller or equal to one, and T is a constant which stands for the temperature constant which normally supposes to be one. And the energy function ,pxy ,, ,, ii ii xy Cxy Uxy Vxy (29) where, cliques are subsets of or ,Cxy ,Nxy , x y itself and , ii Vxy is an arbitrary func- tion of , x y . The equivalence between MRF and Gibbs distribution is expressed by Hammersley-Clifford theorem, which states that , x y is a MRF with neighborhoods if and only if ,Nxy , x y ,Cxy is a Gibbs distribution field with the cliques in- duced by the neighborhood y,Nx . This theorem pro- vides an easy way to construct the MRF in an explicit manner, i.e., one can explicitly estimate the conditional probability distribution of MRF by choosing specific kinds of cliques and an appropriate energy function that is specific for the practical problem. In our model we used the simple equation for the ,Cxy ,yUx U energy function proposed by Therrien [23], and utilized by Nett et al. [24]. This equation is a linear combination of products of elements in the cliques: 1 2 , ,1,1, ,1 ,1 Uxyxyxy xy xy xy (30) 9. COMPUTATION OF THE APRIORI PROBABILITY BY MRF MODEL At first, the brain image is classified through the Bayes- ian classification method by using the model which is obtained as the result of Entropy Based EM algorithm. The gray level values for each tissue class resulted from previous step are extracted. Then, the class conditional probability density function of each tissue class (i.e., ˆk and 2 ˆk ) is estimated through the EM algo- rithm. Also, apriori probabilities K P for tissue classes are estimated by MRF model stated in Eqs.28-30. The parameters of MRF model, i.e., , 1 and 2 have been experimentally set to 0.1, 0.01, and 0.01 for the best result. Next, the new value for termination tol- erance is calculated and evaluated. The new value of termination tolerance is calculated from 1 1 1 ˆˆ max , ˆˆ max max, ˆˆ max , kk kk kk abs PP tol abs abs (31) in which ˆk P , ˆk and ˆk are the row vectors of the apriori probabilities, mean, and variance of the tissue classes in kth iteration, respectively. This algorithm which computs the apriori probabilities by MRF model is shown in Figure 2. Computation of the apriori probabilities by MRF model and update means and variances of each class The obtained model from Entropy based Em algorithm Initialization of: maximum iteration, and tolerance Bayesian classification Extraction of the gray level values for each tissue class Estimation of apriori probabili ties by MRF model Estimation of class conditional probabili ty density function of each class through the EM algorithm Calculation and evaluation of the new value for termination tolerance Post processing Segmented barin Figure 2. Computaion of apriori probabilities and update of means and variances. C opyright © 2011 SciRes. JBiSE  A. Bijar et al. / J. Biomedical Science and Engineering 4 (2011) 552-561 Copyright © 2011 SciRes. 558 10. POSTPROCESSING In manual segmentation, the possible lesions with sizes as small as one or two pixels are not usually considered as MS lesions by experts. However, as the gray level of neighboring pixels in an image usually are highly corre- lated, in our approach, the one-pixel object is considered as a possible noise if its gray level is higher than the average gray levels of its neighbor pixels (in 3 × 3 neighborhood) plus a margin of 25 (which is selected by trial). This object is recognized as noise and removed in the next step. Regarding the two pixel objects, in con- trast with the manual procedure, these objects may be identified as seed points of MS lesions as recommended by the neurologist. JBiSE 11. SIMULATION RESULTS The results of proposed method, for two slices which contain different lesion loads are shown in Figure 3 and Figure 4. Segmented brain of typical original FLAIR images are shown in Figure 3(a) and Figure 4(a). Mul- tiple sclerosis pixels and the resulting segmented slices are shown in Figure 3(b) and Figure 4(b). Figure 3(c) and Figure 4(c) show the histogram of brain pixels and estimated distribution through the proposed method. Also, three estimated kernels which constitute gaussian mixture model are shown in Figure 3(d) and Figure 4(d). 12. EVALUATION To evaluate the proposed method, SI [25], OF and EF [26] criteria are considered and computed over all 20 patients. 2TP SI 2TPFPFN (32) Figure 3. Result of applying the proposed algorithm to the image of a patient: (a) brain image; (b) result of proposed method; (c) histogram of brain, overlaid.  A. Bijar et al. / J. Biomedical Science and Engineering 4 (2011) 552-561 559 Figure 4. Result of applying the proposed algorithm to the image of a patient: (a) brain image; (b) result of proposed method; (c) histogram of brain, overlaid. TP OF TP FN (33) FP EF TP FN (34) where, TP stands for true positive voxels, FP for false positive voxels, and FN for false negative voxels. SI and OF for a good segmentation should be close to 1 and EF should be close to 0. As mentioned before, patients are categorized into 3 groups. The proposed method has been applied to all patients and SI, OF and EF criteria are computed and mean values of each group are illus- trated in Table 1. It is noticeable that SI, OF and EF are improved with an increase in lesion load. Also, the volumetric comparison of lesions between the proposed method and gold standard using correlation coefficient (CC) for each patient group is shown in Table 1. By attention to correlation coefficient values, accuracy of the proposed method is increased for patients with large lesion load. 13. CONCLUSIONS In this paper, we have described and validated a fully automatic method for classification of brain tissues in MR FLAIR images of MS patients. A gaussian mixture model which has three kernels as brain tissues is ob- tained through entropy based EM algorithm. This task starts with only one kernel and finally finds three kernels with no initially supplied training samples. Then, apriori probabilities of classes are estimated using MRF. At last, brain tissues are classified through Bayesian Classifica- tion. This framework has been validated on a data set of C opyright © 2011 SciRes. JBiSE  A. Bijar et al. / J. Biomedical Science and Engineering 4 (2011) 552-561 560 Table 1. Similarity criteria and volumetric comparison of lesions for each patient group. Correlation analysis Similarity criteria Correlation CoefficientM ± SD (CC) MGS ± SDGS (CC) EF OF SI N Patient category 0.93 1.87 ± 1.11 1.89 ± 1.13 0.27230.7277 0.7262 7 Small lesion load 0.95 11.58 ± 3.08 11.59 ± 3.10 0.22010.7395 0.7531 10 Moderate lesion load 0.98 22.23 ± 3.52 22.21 ± 3.54 0.15410.7857 0.8101 3 Large lesion load N: number of patients in each group, M: mean, SD: standard deviation Table 2. Similarity index (SI) values for the proposed method and the other methods. Method Similarity index (SI) Johnston et al. [3] 0.65 Boudraa et al. [13] 0.62 Leemput et al. [14] 0.51 Zijdenbos et al. [31] 0.68 Proposed Method 0.75 MR FLAIR images of 20 MS patients via similarity cri- teria (i.e., SI, OF, and EF). There are other segmentation methods which have been evaluated via similar ways (i.e., SI), and have used manual segmentation for evaluation of them, such as Johnston et al. [27], Boudraa et al. [28], Leemput et al. [29], and Zijdenbos et al. [30]. Comparison of proposed method with these methods is shown in Table 2. Johnston et al. [31] presented a semi- automatic segmentation approach based on stochastic relaxation method (SRM) and iterated conditional mode. Boudraa et al. [28] performed an automatic detection of MS lesions by using fuzzy C-means (FCM) algorithm. Leemput et al. [29] proposed the use of an intensity- based and stochastic model-aided segmentation ap- proach for fully automatic segmentation of MS lesions. Zijdenbos et al. [30] developed an automatic pipeline based on a supervised artificial neural network (ANN) classifier. As it is seen in Table 2, the proposed method has improved the results reported in [26-29]. Also, in [31], it is mentioned that a value for SI more than 0.7 represents a very good segmentation in this field. Future work will include improving accuracy and correlation value. REFERENCES [1] Kurtzke, J.F. (1983) Rating neurologic impairment in multiple sclerosis: An expanded disability status scale (EDSS). Neurology, 33, 1444-1452. [2] Rasoul, K., Mansur, V., Farzad, T., Massood Nabavi., S. (2008) Fully automatic segmentation of multiple sclero- sis lesions in brain MR FLAIR images using adaptive mixtures method and Markov random field model. Com- puters in Biology and Medicine, 38, 379-390. doi:10.1016/j.compbiomed.2007.12.005 [3] Polman, C.H., Reingold, S.C., Edan, G., Fillippi, M., Hartung, HP. and Kappos, L. (2005) Diagnostic criteria for MS 2005 revisions to the MC Donald criteria. Annals of Neurology, 58, 840-846. doi:10.1002/ana.20703 [4] Edelman, R.R., Hesselink, J.R. and Zlatkin, M.B. (2006) Clinical magnetic resonance imaging. 3rd Edition, Saun- ders, Philadelphia, 1571-1615. [5] Anbeek, P., Vincken, K.L., van Osch, M.J.P., Bisschops, R.H.C. and Van der Grond, J. (2004) Probabilistic seg- mentation of white matter lesions in MR imaging, NeuroImage, 21, 1037-1044. doi:10.1016/j.neuroimage.2003.10.012 [6] Khayati, R. (2006) Quantification of multiple sclerosis lesions based on fractal analysis. PhD Thesis, Technical Report No. 1: Fully automatic object oriented brain seg- mentation in MRI. Amirkabir University of Technology, Tehran. [7] Duda, R.O., Hart, P.E. and Stork, D.G. (2001) Pattern classification. 2nd Edition, Wiley, New York. [8] Bernardo, J.M. and Smith, A.F.M. (1994) Bayesian the- ory. Wiley, New York. doi:10.1002/9780470316870 [9] Dempster, A.P., Laird, N.M. and Rubin, D.B. (1997) Maximun likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Soceity B, 39, 1-38. doi:10.1.1.133.4884 [10] Shannon, C. (1948) A mathematical theory of communi- cation. Bell System Technical Journal, 27, 379-423. [11] Cover, T.M. and Thomas, J.A. (1991) Elements of infor- mation theory. Wiley & Sons, New York. doi:10.1002/0471200611 [12] Renyi, A. (1961) On measures of entropy and informa- tion. University California Press, Berkeley, 547-561. [13] Leonenko, N., Pronzato, L. and Savani, V. (2008) A class of Rényi information estimators for multidimensional densities. Annals of Statistics, 36, 2153-2182. doi:10.1214/07-AOS539 [14] Kozachenko, L. and Leonenko, N. (1987) On statistical estimation of entropy of a random vector. Problems In- formation Transmission, 23, 95101. [15] Jones, M. and Sibson, R. (1987) What is projection pur- suit. Journal of the Royal Statistical Society: Series A, 150, 136. http://www.jstor.org/stable/2981662 [16] Benavent, A.P. Ruiz, F.E. and Sáez, J.M. (2009) Learning Gaussian mixture models with entropy-based criteria. IEEE Transactions on Neural Networks, 20, 1756-1771. doi:10.1109/TNN.2009.2030190 [17] Richardson, S. and Green, P. (1991) On Bayesian analy- sis of mixtures with an unknown number of components (with discussion). Journal of the Royal Statistical Society: C opyright © 2011 SciRes. JBiSE  A. Bijar et al. / J. Biomedical Science and Engineering 4 (2011) 552-561 561 Series B, 59, 731-792. doi:10.1111/1467-9868.00095 [18] Dellaportas, P. and Papageorgiou, I. (2006) Multivariate mixtures of normals with unknown number of compo- nents. Statistics and Computing, 16, 57-68. doi:10.1007/s11222-006-5338-6 [19] Mitchell, T.M. (1997) Machine Learning. McGraw-Hill, Boston. doi:10.1036/0070428077 [20] Li, S.Z. (2001) Markov random field modeling in image analysis. Springer, Tokyo. [21] Held, K., Kops, E.R., Krause, B.J., Wells, W.M., Kikinis, R. and Mller-Grtner, H.W. (1997) Markov random field segmentation of brain MR images. IEEE Transactions on Medical Imaging, 16, 878-886. doi:10.1109/42.650883 [22] Geman, D., Geman, S., Graffigne, C. and Dong, P. (1990) Boundary detection by constrained optimization, IEEE Transactions on Pattern Analysis and Machine Intelli- gence, PAMI-12, 609-628. doi:10.1109/34.56204 [23] Therrien, C.W. (1989) Decision, estimation, and classifi- cation. Wiley, New York. [24] Nett, J.M. (2001) The study of MS using MRI, image processing, and visualization. Master’s Thesis, Univer- sity of Louisville, Louisville. [25] Zijdenbos, A.P., Dawant, B.M., Margolin, R.A. and Palmer, A.C. (1994) Morphometric analysis of white matter lesions in MR images: Method and validation. IEEE Transactions on Medical Imaging, 13, 716-724. doi:10.1109/42.363096 [26] Stokking, R., Vincken, K.L. and Viergever, M.A. (2000) Automatic morphologybased brain segmentation (MBRASE) from MRI-T1 data. Neu roImage, 12, 726-738. doi:10.1006/nimg.2000.0661 [27] Johnston, B., Atkins, M.S., Mackiewich, B. and Ander- son, M. (1996) Segmentation of multiple sclerosis lesions in intensity corrected multispectral MRI. IEEE Transac- tions on Medical Imaging, 15, 154-169. doi:10.1109/42.491417 [28] Boudraa, A.O., Dehakb, S.M.R., Zhu, Y.M., Pachai, C., Bao, Y.G. and Grimaud, J. (2000) Automated segmenta- tion of multiple sclerosis lesions in multispectral MR imaging using fuzzy clustering. Computers in Biology and Medicine, 30, 23-40. doi:10.1016/S0010-4825(99)00019-0 [29] Leemput, K.V., Maes, F., Vandermeulen, D., Colchester, A. and Suetens, P. (2001) Automated segmentation of multiple sclerosis lesions by model outlier detection. IEEE Transactions on Medical Imaging, 20, 677-688. doi:10.1109/42.938237 [30] Zijdenbos, A.P., Forghani, R. and Evans. A.C. (2002) Automatic pipeline analysis of 3-D MRI data for clinical trials: Application to multiple sclerosis. IEEE Transac- tions on Medical Imaging, 21, 1280-1291. doi:10.1109/TMI.2002.806283 [31] Bartko, J.J. (1991) Measurement and reliability: statisti- cal thinking considerations. Schizophrenia Bulletin, 17, 483-489. doi:10.1093/schbul/17.3.483 C opyright © 2011 SciRes. JBiSE |

