 Vol.3, No.8, 702-722 (2011) Natural Science http://dx.doi.org/10.4236/ns.2011.38094 Copyright © 2011 SciRes. OPEN ACCESS Growth dynamics of individual clones of normal human keratinocytes: observations and theoretical considerations John Jacob Wille Department of Cell Biology, Mayo Medical School, Rochester, USA; jjwille@aol.com Received 22 March 2011; revised 28 April 2011; accepted 3 May 2011. ABSTRACT The life histories of 429 individual epidermal keratinocyte clones picked at random were studied. Individual basal keratinocytes were derived from asynchronous rapidly proliferating subconfluent cultures propagated in either a low calcium (0.1 mM) or a high calcium (2 mM) serum-free medium. Single-celled clones were isolated by seeding trypsin-EDTA dissociated cells into a Petri dish containing cloning chips. Chips with only one cell per chip were trans- ferred into dishes containing either low calcium or high calcium growth factor replete serum- free medium. Clone formation was monitored microscopically and the number of cells in each colony tallied at least twice daily for further analysis. A total of 369 clones were established from seven different neonatal foreskin cell strains (A-F), and 60 clones were derived from one adult human skin cell strain (G). During a five-day culture interval, among 32 clones of strain A, 83% divided at least once, 50% divided once in 24 hours, 86% divided at least three times within three days, and more than 50% di- vided at least four to five times in five days. Of 231 clones amongst the other five cell strains (B-F), an average of 63% (±12 S,E) divided more than three times in an eight day period, the re- mainder divided either once, twice or not at all. Of the 106 clones of strain G, reared in high cal- cium serum-free medium, 67% divided more than three times in a six-day period, and 55% divided five or more times in 6 days. Clones derived from adult skin strain H had a lower clone forming potential with 70% dividing at least once in seven days, and only 30% dividing three or more times. By contrast, the average generation time (AvGT) for second and third passage keratinocytes derived from neonatal foreskin cultures was 24 hrs. Detailed dendro- grams were constructed for many of the prolif- erating clones. The majority of clones ex- pressed a synblastic division pattern with every cell dividing at least once per day. A fraction of clones either exceeded this circadian division rate or displayed a biphasic division pattern with all cells initially dividing once a day and then abruptly slowing to once every other day or to an intermediate rate. A minority of clones was committed to a few terminal divisions. The division patterns of the non-synblastic clones fit an alternating bifurcated branching mode of clonal expansion expressed by the Fibonacci sequence for numbers of accumulated cells per clone per day. These results were analyzed in terms of deterministic, probabilistic and a limit cycle oscillator models of cell division timing. Keywords: Biochemical Oscillators; Clonal Growth Dynamics; Human Keratinocytes; Interdivision Times; Temporal Control of Mitosis Models 1. INTRODUCTION Previously the life histories of individual keratino- cytes were examined by the technique of time-lapse cin- emacrography [1]. These authors reported on pedigree analyses of only eight guinea pig keratinocyte (GPK) cells using time-lapse filming. They recorded the gen- eration times and position in the field of observations up to the sixth generation in subconfluent cultures formed from cells that were serially passaged by trypsin disso- ciation using a split ratio (1:500) through twenty-eight serial subcultures in a serum-containing medium. The results indicated inter-clonal variation in growth rates. More than half the clone displayed a broad unimodal distributioin of GT clustered between 1200 to 1400 min- utes; the remaining clones displayed bimodal distribu-  J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 703703 tions with the larger fraction clustered around 1800 to 2200 minutes. In fact, the most proliferating of the eight cell clones only produced 22 cells after five days of cul- ture, while the least proliferating clones produced a total of only 7 cells. A statistically significant positive corre- lation was demonstrated between parental generation time and filial generation time. No evidence was pre- sented that these were normal diploid cells. No mention was made about the propensity of these cultures to un- dergo commitment and terminal differentiation under the serum culture growth conditions. Kitano et al. [2] stud- ied cell proliferation patterns and interdivision times of primary cultures of normal human keratinocytes in Ea- gle’s minimal essential medium (2 mM calcium) con- taining 20% fetal bovine serum. Cultures were filmed between 8 - 20 days after seeding and small polygonal groups of cells were followed up to six days. Six differ- ent cell strains were established from donors of both genders, ranging in age from 5 to 55 years. The average inter-division time for the six strains was about 26 hours and ranged from 15.1 (±4.1) for cells from a 10-year old female to 27.6 (±13.5) for a 5-year old male. Six den- drograms were made from the six cultures of keratino- cytes filmed. Detailed genealogies of the clones within each culture were presented. Most of the cells in the field divided two to three times. Sister-sister pairs of the second and third generations divided after approximately the same inter-division times. Some cells never divided, some sister-sister pairs differed substantially in their inter-division time, and in some clones synchronous divisions were observed. The results were interpreted as showing that patterns of cell proliferation do not support a statistical scheme for maintenance of a steady state, but rather a typical pattern of logarithmic growth. In other studies [3,4], the clone-forming ability of normal human keratinocyte was examined by isolating individual cells either directly from the epidermis or from cell culture. Keratinocytes isolated directly from the epidermis and subsequently cultured from clones only if they were <11 μm or less in diameter. The colony-forming ability of clone-founding cells derived from cell culture was also shown to be a function of cell size. The colony-forming efficiency was >80% for founding cells with a cell size below 12 μm in diameter. Interestingly, they reported that there was no significant difference in the distribu- tion of cell size in progeny founding cells derived from large or small founding cells, and large clone-forming cells were shown to give rise to colonies with cell size distributions that contained small progeny cells. These results were interpreted to mean that cells in various stage of cell cycle can regenerate the average cell size distribution if they have not left the cell cycle, while cells that attain a cell size larger than the cell volume predicted for G2/M cells in the cell cycle are irreversibly committed to terminal differentiation and exit the cell cycle. These authors also reported that the colony form- ing capacity of individual clones is heterogenous and can be assigned to three different classes, holoclones, para- clones and meroclones. Holoclones are defined as hav- ing the highest reproductive potential, paraclones have the shortest lifespans of less than 15 cell generations, and meroclones that are a mixture of the former two. The incidence of these different types is affected by age of donors such that older donors yield a lower proportion of holoclones than younger skin donors. In this study, observations were made of the clone forming potential of individual keratinocyte clones iso- lated during the rapid logarithmic growth mode of sub- confluent cutures propagated in serum-free medium. The growth kinetics of over four hundred different keratino- cyte clones derived from either neonatal foreskin or adult skin cell strains were followed daily for up to eight days of culture. In this manner, the individual life histo- ries of each clone initiated from among numerous ran- domly selected cells could be compared for their clone forming potential and for possible differences in their patterns of cell proliferation. Preliminary results were reported earlier [5] 2. MATERIALS & METHODS 2.1. Cell Culture Human epidermal keratinocytes were derived either from neonatal foreskin and adult skin as previously de- scribed [6], using a modified MCDB 153 serum-free medium, containing 110 mM NaCl and 20 mM HEPES, pH 7.2. The basal medium was supplemented with ethanolamine (0.1 mM), phophoethanolamine (0.1 mM), hydrocortisone (0.1 μM), human recombinant EGF (5 ng/ml, and human recombinant IGF-1, both obtained from Sigma Chemical Co., St. Louis, MO. 2.2. Isolation of Single-Celled Clones Parent secondary cultures of keratinocytes were estab- lished in complete serum-free growth medium and the cultures trypsinized in mid-logarithmic mode of growth as established by daily cell counts of parallel cultures. The steps of enzymatic cell dissociation and harvesting procedures were those previously described [6]. Ap- proximately 1000 cells were seeded into sterile100mm diameter Petri dishes containing 5 ml of pre-warmed complete modified serum-free medium. Petri dishes were pre-seeded with Belco (New Brunswick, NJ) clon- ing chips. The cells were allowed to attach for 30 min- utes and individual cloning chips with only one cell per 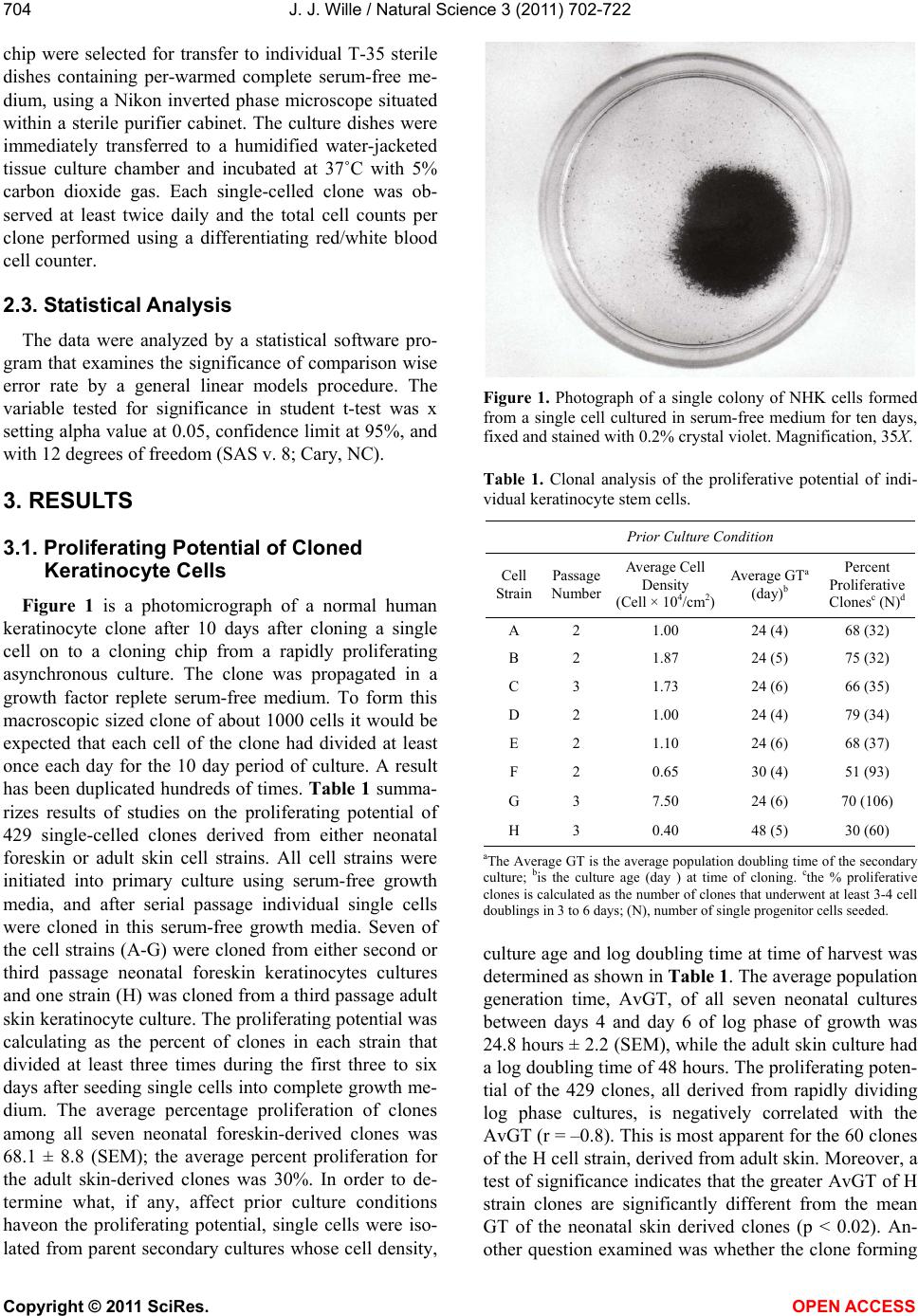 J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 704 chip were selected for transfer to individual T-35 sterile dishes containing per-warmed complete serum-free me- dium, using a Nikon inverted phase microscope situated within a sterile purifier cabinet. The culture dishes were immediately transferred to a humidified water-jacketed tissue culture chamber and incubated at 37˚C with 5% carbon dioxide gas. Each single-celled clone was ob- served at least twice daily and the total cell counts per clone performed using a differentiating red/white blood cell counter. 2.3. Statistical Analysis The data were analyzed by a statistical software pro- gram that examines the significance of comparison wise error rate by a general linear models procedure. The variable tested for significance in student t-test was x setting alpha value at 0.05, confidence limit at 95%, and with 12 degrees of freedom (SAS v. 8; Cary, NC). 3. RESULTS 3.1. Proliferating Potential of Cloned Keratinocyte Cells Figure 1 is a photomicrograph of a normal human keratinocyte clone after 10 days after cloning a single cell on to a cloning chip from a rapidly proliferating asynchronous culture. The clone was propagated in a growth factor replete serum-free medium. To form this macroscopic sized clone of about 1000 cells it would be expected that each cell of the clone had divided at least once each day for the 10 day period of culture. A result has been duplicated hundreds of times. Table 1 summa- rizes results of studies on the proliferating potential of 429 single-celled clones derived from either neonatal foreskin or adult skin cell strains. All cell strains were initiated into primary culture using serum-free growth media, and after serial passage individual single cells were cloned in this serum-free growth media. Seven of the cell strains (A-G) were cloned from either second or third passage neonatal foreskin keratinocytes cultures and one strain (H) was cloned from a third passage adult skin keratinocyte culture. The proliferating potential was calculating as the percent of clones in each strain that divided at least three times during the first three to six days after seeding single cells into complete growth me- dium. The average percentage proliferation of clones among all seven neonatal foreskin-derived clones was 68.1 ± 8.8 (SEM); the average percent proliferation for the adult skin-derived clones was 30%. In order to de- termine what, if any, affect prior culture conditions haveon the proliferating potential, single cells were iso- lated from parent secondary cultures whose cell density, Figure 1. Photograph of a single colony of NHK cells formed from a single cell cultured in serum-free medium for ten days, fixed and stained with 0.2% crystal violet. Magnification, 35X. Table 1. Clonal analysis of the proliferative potential of indi- vidual keratinocyte stem cells. Prior Culture Condition Cell Strain Passage Number Average Cell Density (Cell × 104/cm2) Average GTa (day)b Percent Proliferative Clonesc (N)d A 2 1.00 24 (4) 68 (32) B 2 1.87 24 (5) 75 (32) C 3 1.73 24 (6) 66 (35) D 2 1.00 24 (4) 79 (34) E 2 1.10 24 (6) 68 (37) F 2 0.65 30 (4) 51 (93) G 3 7.50 24 (6) 70 (106) H 3 0.40 48 (5) 30 (60) aThe Average GT is the average population doubling time of the secondary culture; bis the culture age (day ) at time of cloning. cthe % proliferative clones is calculated as the number of clones that underwent at least 3-4 cell doublings in 3 to 6 days; (N), number of single progenitor cells seeded. culture age and log doubling time at time of harvest was determined as shown in Table 1. The average population generation time, AvGT, of all seven neonatal cultures between days 4 and day 6 of log phase of growth was 24.8 hours ± 2.2 (SEM), while the adult skin culture had a log doubling time of 48 hours. The proliferating poten- tial of the 429 clones, all derived from rapidly dividing log phase cultures, is negatively correlated with the AvGT (r = –0.8). This is most apparent for the 60 clones of the H cell strain, derived from adult skin. Moreover, a test of significance indicates that the greater AvGT of H strain clones are significantly different from the mean GT of the neonatal skin derived clones (p < 0.02). An- other question examined was whether the clone forming 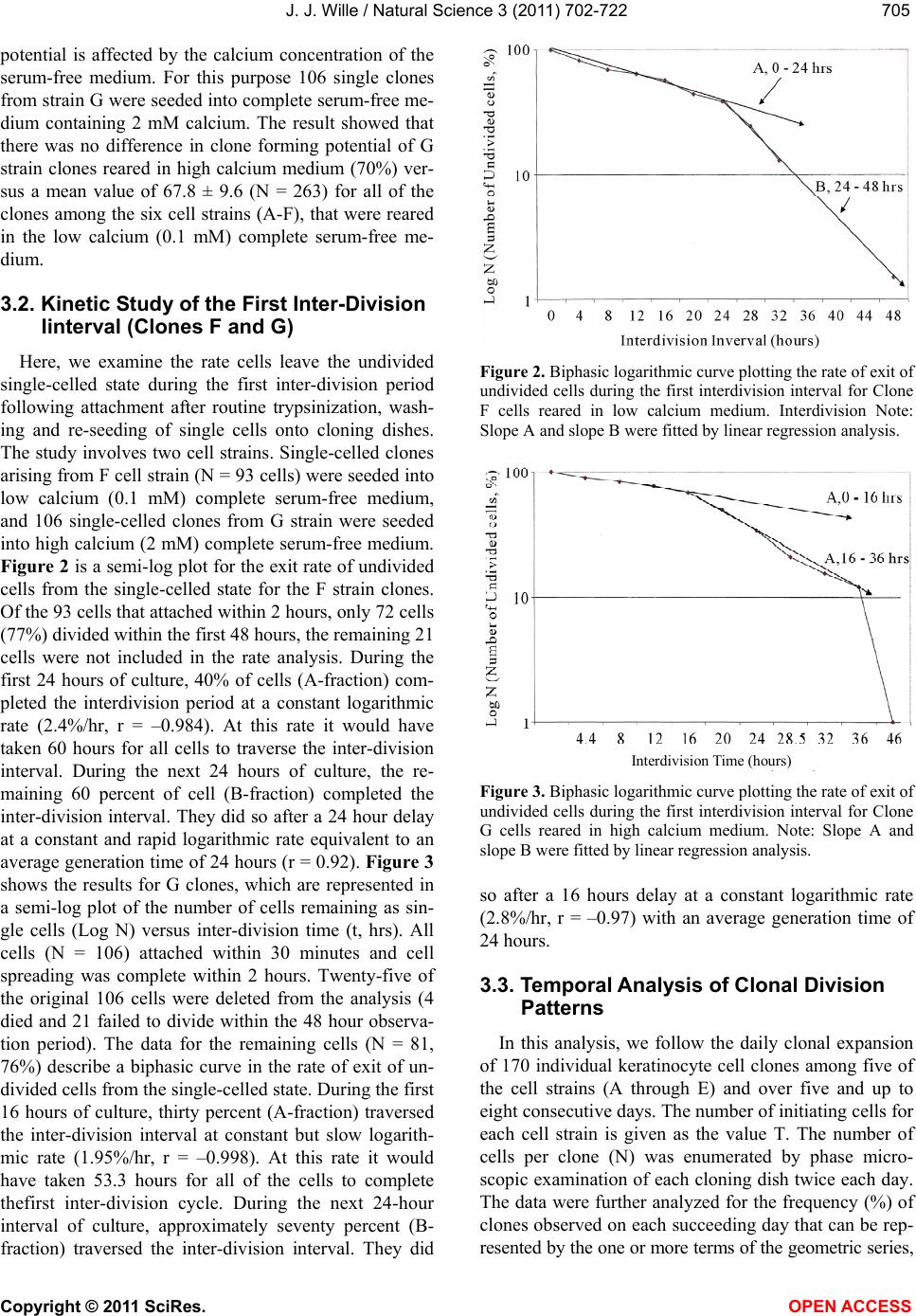 J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 705705 potential is affected by the calcium concentration of the serum-free medium. For this purpose 106 single clones from strain G were seeded into complete serum-free me- dium containing 2 mM calcium. The result showed that there was no difference in clone forming potential of G strain clones reared in high calcium medium (70%) ver- sus a mean value of 67.8 ± 9.6 (N = 263) for all of the clones among the six cell strains (A-F), that were reared in the low calcium (0.1 mM) complete serum-free me- dium. 3.2. Kinetic Study of the First Inter-Division Iinterval (Clones F and G) Here, we examine the rate cells leave the undivided single-celled state during the first inter-division period following attachment after routine trypsinization, wash- ing and re-seeding of single cells onto cloning dishes. The study involves two cell strains. Single-celled clones arising from F cell strain (N = 93 cells) were seeded into low calcium (0.1 mM) complete serum-free medium, and 106 single-celled clones from G strain were seeded into high calcium (2 mM) complete serum-free medium. Figure 2 is a semi-log plot for the exit rate of undivided cells from the single-celled state for the F strain clones. Of the 93 cells that attached within 2 hours, only 72 cells (77%) divided within the first 48 hours, the remaining 21 cells were not included in the rate analysis. During the first 24 hours of culture, 40% of cells (A-fraction) com- pleted the interdivision period at a constant logarithmic rate (2.4%/hr, r = –0.984). At this rate it would have taken 60 hours for all cells to traverse the inter-division interval. During the next 24 hours of culture, the re- maining 60 percent of cell (B-fraction) completed the inter-division interval. They did so after a 24 hour delay at a constant and rapid logarithmic rate equivalent to an average generation time of 24 hours (r = 0.92). Figure 3 shows the results for G clones, which are represented in a semi-log plot of the number of cells remaining as sin- gle cells (Log N) versus inter-division time (t, hrs). All cells (N = 106) attached within 30 minutes and cell spreading was complete within 2 hours. Twenty-five of the original 106 cells were deleted from the analysis (4 died and 21 failed to divide within the 48 hour observa- tion period). The data for the remaining cells (N = 81, 76%) describe a biphasic curve in the rate of exit of un- divided cells from the single-celled state. During the first 16 hours of culture, thirty percent (A-fraction) traversed the inter-division interval at constant but slow logarith- mic rate (1.95%/hr, r = –0.998). At this rate it would have taken 53.3 hours for all of the cells to complete thefirst inter-division cycle. During the next 24-hour interval of culture, approximately seventy percent (B- fraction) traversed the inter-division interval. They did Figure 2. Biphasic logarithmic curve plotting the rate of exit of undivided cells during the first interdivision interval for Clone F cells reared in low calcium medium. Interdivision Note: Slope A and slope B were fitted by linear regression analysis. Interdivision Time (hours) Figure 3. Biphasic logarithmic curve plotting the rate of exit of undivided cells during the first interdivision interval for Clone G cells reared in high calcium medium. Note: Slope A and slope B were fitted by linear regression analysis. so after a 16 hours delay at a constant logarithmic rate (2.8%/hr, r = –0.97) with an average generation time of 24 hours. 3.3. Temporal Analysis of Clonal Division Patterns In this analysis, we follow the daily clonal expansion of 170 individual keratinocyte cell clones among five of the cell strains (A through E) and over five and up to eight consecutive days. The number of initiating cells for each cell strain is given as the value T. The number of cells per clone (N) was enumerated by phase micro- scopic examination of each cloning dish twice each day. The data were further analyzed for the frequency (%) of clones observed on each succeeding day that can be rep- resented by the one or more terms of the geometric series, 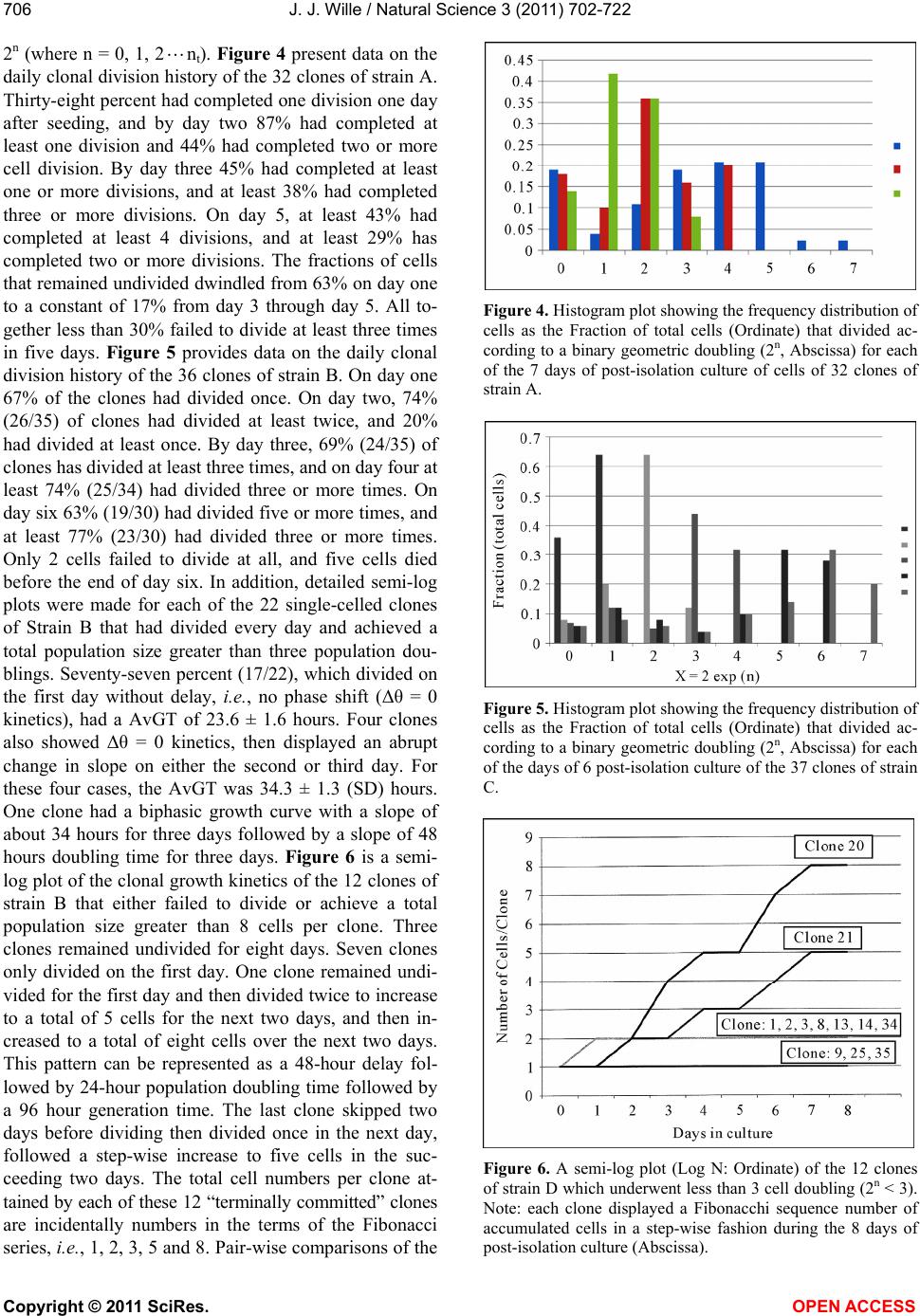 J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 706 2n (where n = 0, 1, 2nt). Figure 4 present data on the daily clonal division history of the 32 clones of strain A. Thirty-eight percent had completed one division one day after seeding, and by day two 87% had completed at least one division and 44% had completed two or more cell division. By day three 45% had completed at least one or more divisions, and at least 38% had completed three or more divisions. On day 5, at least 43% had completed at least 4 divisions, and at least 29% has completed two or more divisions. The fractions of cells that remained undivided dwindled from 63% on day one to a constant of 17% from day 3 through day 5. All to- gether less than 30% failed to divide at least three times in five days. Figure 5 provides data on the daily clonal division history of the 36 clones of strain B. On day one 67% of the clones had divided once. On day two, 74% (26/35) of clones had divided at least twice, and 20% had divided at least once. By day three, 69% (24/35) of clones has divided at least three times, and on day four at least 74% (25/34) had divided three or more times. On day six 63% (19/30) had divided five or more times, and at least 77% (23/30) had divided three or more times. Only 2 cells failed to divide at all, and five cells died before the end of day six. In addition, detailed semi-log plots were made for each of the 22 single-celled clones of Strain B that had divided every day and achieved a total population size greater than three population dou- blings. Seventy-seven percent (17/22), which divided on the first day without delay, i.e., no phase shift (Δθ = 0 kinetics), had a AvGT of 23.6 ± 1.6 hours. Four clones also showed Δθ = 0 kinetics, then displayed an abrupt change in slope on either the second or third day. For these four cases, the AvGT was 34.3 ± 1.3 (SD) hours. One clone had a biphasic growth curve with a slope of about 34 hours for three days followed by a slope of 48 hours doubling time for three days. Figure 6 is a semi- log plot of the clonal growth kinetics of the 12 clones of strain B that either failed to divide or achieve a total population size greater than 8 cells per clone. Three clones remained undivided for eight days. Seven clones only divided on the first day. One clone remained undi- vided for the first day and then divided twice to increase to a total of 5 cells for the next two days, and then in- creased to a total of eight cells over the next two days. This pattern can be represented as a 48-hour delay fol- lowed by 24-hour population doubling time followed by a 96 hour generation time. The last clone skipped two days before dividing then divided once in the next day, followed a step-wise increase to five cells in the suc- ceeding two days. The total cell numbers per clone at- tained by each of these 12 “terminally committed” clones are incidentally numbers in the terms of the Fibonacci series, i. e. , 1, 2, 3, 5 and 8. Pair-wise comparisons of the Figure 4. Histogram plot showing the frequency distribution of cells as the Fraction of total cells (Ordinate) that divided ac- cording to a binary geometric doubling (2n, Abscissa) for each of the 7 days of post-isolation culture of cells of 32 clones of strain A. Figure 5. Histogram plot showing the frequency distribution of cells as the Fraction of total cells (Ordinate) that divided ac- cording to a binary geometric doubling (2n, Abscissa) for each of the days of 6 post-isolation culture of the 37 clones of strain C. Figure 6. A semi-log plot (Log N: Ordinate) of the 12 clones of strain D which underwent less than 3 cell doubling (2n < 3). Note: each clone displayed a Fibonacchi sequence number of accumulated cells in a step-wise fashion during the 8 days of post-isolation culture (Abscissa). 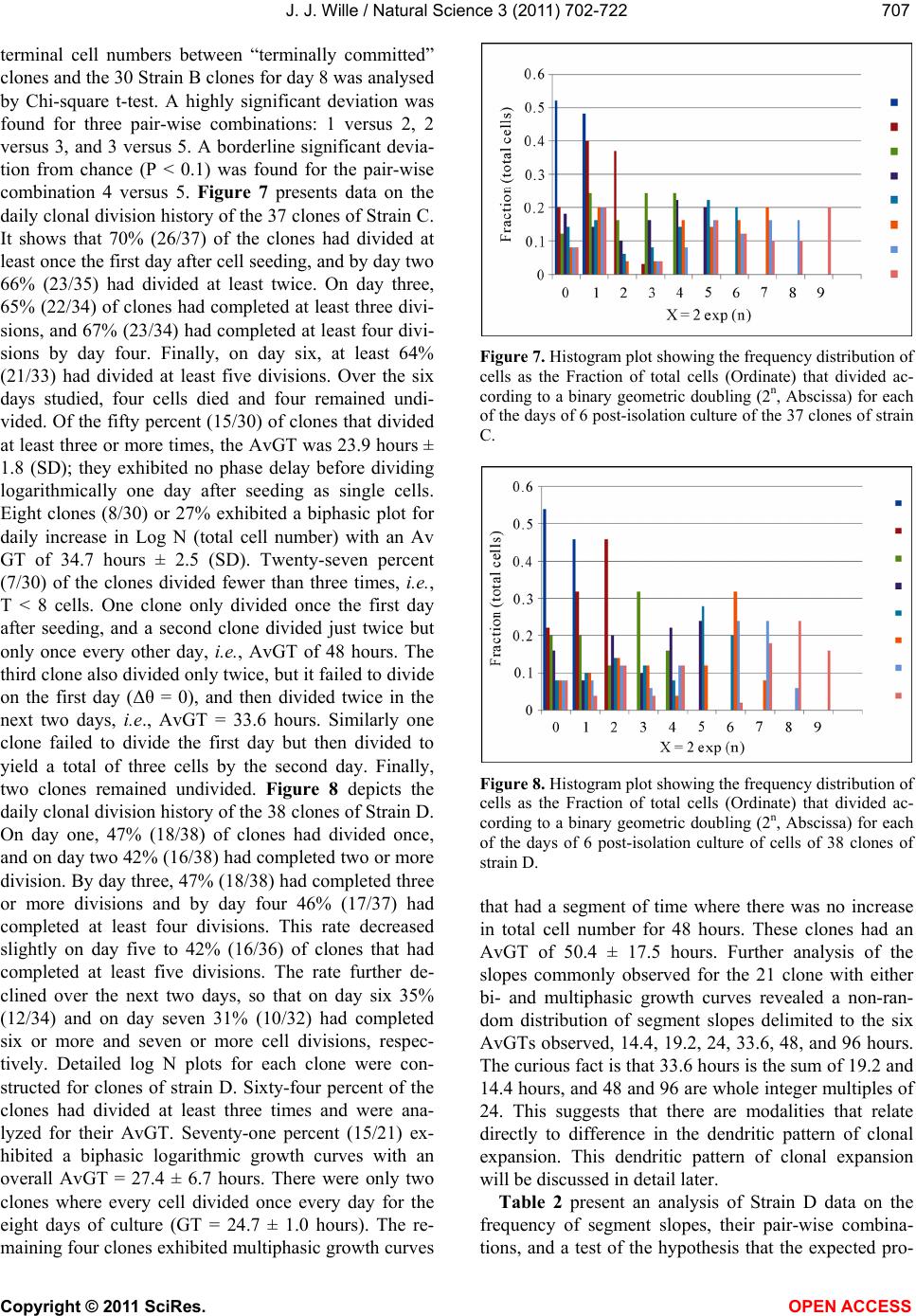 J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 707707 terminal cell numbers between “terminally committed” clones and the 30 Strain B clones for day 8 was analysed by Chi-square t-test. A highly significant deviation was found for three pair-wise combinations: 1 versus 2, 2 versus 3, and 3 versus 5. A borderline significant devia- tion from chance (P < 0.1) was found for the pair-wise combination 4 versus 5. Figure 7 presents data on the daily clonal division history of the 37 clones of Strain C. It shows that 70% (26/37) of the clones had divided at least once the first day after cell seeding, and by day two 66% (23/35) had divided at least twice. On day three, 65% (22/34) of clones had completed at least three divi- sions, and 67% (23/34) had completed at least four divi- sions by day four. Finally, on day six, at least 64% (21/33) had divided at least five divisions. Over the six days studied, four cells died and four remained undi- vided. Of the fifty percent (15/30) of clones that divided at least three or more times, the AvGT was 23.9 hours ± 1.8 (SD); they exhibited no phase delay before dividing logarithmically one day after seeding as single cells. Eight clones (8/30) or 27% exhibited a biphasic plot for daily increase in Log N (total cell number) with an Av GT of 34.7 hours ± 2.5 (SD). Twenty-seven percent (7/30) of the clones divided fewer than three times, i.e., T < 8 cells. One clone only divided once the first day after seeding, and a second clone divided just twice but only once every other day, i.e., AvGT of 48 hours. The third clone also divided only twice, but it failed to divide on the first day (Δθ = 0), and then divided twice in the next two days, i.e., AvGT = 33.6 hours. Similarly one clone failed to divide the first day but then divided to yield a total of three cells by the second day. Finally, two clones remained undivided. Figure 8 depicts the daily clonal division history of the 38 clones of Strain D. On day one, 47% (18/38) of clones had divided once, and on day two 42% (16/38) had completed two or more division. By day three, 47% (18/38) had completed three or more divisions and by day four 46% (17/37) had completed at least four divisions. This rate decreased slightly on day five to 42% (16/36) of clones that had completed at least five divisions. The rate further de- clined over the next two days, so that on day six 35% (12/34) and on day seven 31% (10/32) had completed six or more and seven or more cell divisions, respec- tively. Detailed log N plots for each clone were con- structed for clones of strain D. Sixty-four percent of the clones had divided at least three times and were ana- lyzed for their AvGT. Seventy-one percent (15/21) ex- hibited a biphasic logarithmic growth curves with an overall AvGT = 27.4 ± 6.7 hours. There were only two clones where every cell divided once every day for the eight days of culture (GT = 24.7 ± 1.0 hours). The re- maining four clones exhibited multiphasic growth curves Figure 7. Histogram plot showing the frequency distribution of cells as the Fraction of total cells (Ordinate) that divided ac- cording to a binary geometric doubling (2n, Abscissa) for each of the days of 6 post-isolation culture of the 37 clones of strain C. Figure 8. Histogram plot showing the frequency distribution of cells as the Fraction of total cells (Ordinate) that divided ac- cording to a binary geometric doubling (2n, Abscissa) for each of the days of 6 post-isolation culture of cells of 38 clones of strain D. that had a segment of time where there was no increase in total cell number for 48 hours. These clones had an AvGT of 50.4 ± 17.5 hours. Further analysis of the slopes commonly observed for the 21 clone with either bi- and multiphasic growth curves revealed a non-ran- dom distribution of segment slopes delimited to the six AvGTs observed, 14.4, 19.2, 24, 33.6, 48, and 96 hours. The curious fact is that 33.6 hours is the sum of 19.2 and 14.4 hours, and 48 and 96 are whole integer multiples of 24. This suggests that there are modalities that relate directly to difference in the dendritic pattern of clonal expansion. This dendritic pattern of clonal expansion will be discussed in detail later. Table 2 present an analysis of Strain D data on the frequency of segment slopes, their pair-wise combina- tions, and a test of the hypothesis that the expected pro- 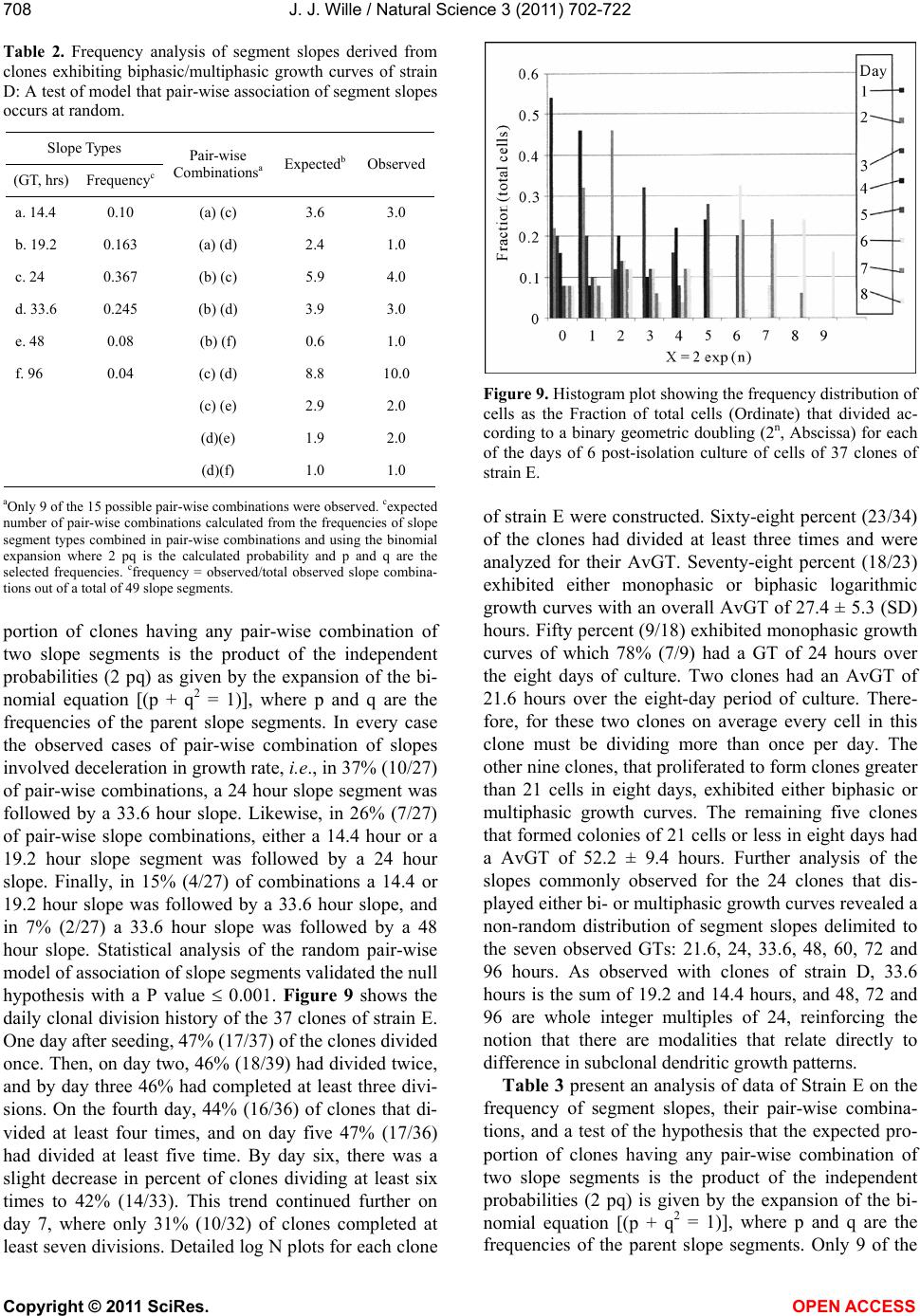 J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 708 Table 2. Frequency analysis of segment slopes derived from clones exhibiting biphasic/multiphasic growth curves of strain D: A test of model that pair-wise association of segment slopes occurs at random. Slope Types (GT, hrs) Frequencyc Pair-wise Combinationsa Expectedb Observed a. 14.4 0.10 (a) (c) 3.6 3.0 b. 19.2 0.163 (a) (d) 2.4 1.0 c. 24 0.367 (b) (c) 5.9 4.0 d. 33.6 0.245 (b) (d) 3.9 3.0 e. 48 0.08 (b) (f) 0.6 1.0 f. 96 0.04 (c) (d) 8.8 10.0 (c) (e) 2.9 2.0 (d)(e) 1.9 2.0 (d)(f) 1.0 1.0 aOnly 9 of the 15 possible pair-wise combinations were observed. cexpected number of pair-wise combinations calculated from the frequencies of slope segment types combined in pair-wise combinations and using the binomial expansion where 2 pq is the calculated probability and p and q are the selected frequencies. cfrequency = observed/total observed slope combina- tions out of a total of 49 slope segments. portion of clones having any pair-wise combination of two slope segments is the product of the independent probabilities (2 pq) as given by the expansion of the bi- nomial equation [(p + q2 = 1)], where p and q are the frequencies of the parent slope segments. In every case the observed cases of pair-wise combination of slopes involved deceleration in growth rate, i.e., in 37% (10/27) of pair-wise combinations, a 24 hour slope segment was followed by a 33.6 hour slope. Likewise, in 26% (7/27) of pair-wise slope combinations, either a 14.4 hour or a 19.2 hour slope segment was followed by a 24 hour slope. Finally, in 15% (4/27) of combinations a 14.4 or 19.2 hour slope was followed by a 33.6 hour slope, and in 7% (2/27) a 33.6 hour slope was followed by a 48 hour slope. Statistical analysis of the random pair-wise model of association of slope segments validated the null hypothesis with a P value 0.001. Figure 9 shows the daily clonal division history of the 37 clones of strain E. One day after seeding, 47% (17/37) of the clones divided once. Then, on day two, 46% (18/39) had divided twice, and by day three 46% had completed at least three divi- sions. On the fourth day, 44% (16/36) of clones that di- vided at least four times, and on day five 47% (17/36) had divided at least five time. By day six, there was a slight decrease in percent of clones dividing at least six times to 42% (14/33). This trend continued further on day 7, where only 31% (10/32) of clones completed at least seven divisions. Detailed log N plots for each clone Figure 9. Histogram plot showing the frequency distribution of cells as the Fraction of total cells (Ordinate) that divided ac- cording to a binary geometric doubling (2n, Abscissa) for each of the days of 6 post-isolation culture of cells of 37 clones of strain E. of strain E were constructed. Sixty-eight percent (23/34) of the clones had divided at least three times and were analyzed for their AvGT. Seventy-eight percent (18/23) exhibited either monophasic or biphasic logarithmic growth curves with an overall AvGT of 27.4 ± 5.3 (SD) hours. Fifty percent (9/18) exhibited monophasic growth curves of which 78% (7/9) had a GT of 24 hours over the eight days of culture. Two clones had an AvGT of 21.6 hours over the eight-day period of culture. There- fore, for these two clones on average every cell in this clone must be dividing more than once per day. The other nine clones, that proliferated to form clones greater than 21 cells in eight days, exhibited either biphasic or multiphasic growth curves. The remaining five clones that formed colonies of 21 cells or less in eight days had a AvGT of 52.2 ± 9.4 hours. Further analysis of the slopes commonly observed for the 24 clones that dis- played either bi- or multiphasic growth curves revealed a non-random distribution of segment slopes delimited to the seven observed GTs: 21.6, 24, 33.6, 48, 60, 72 and 96 hours. As observed with clones of strain D, 33.6 hours is the sum of 19.2 and 14.4 hours, and 48, 72 and 96 are whole integer multiples of 24, reinforcing the notion that there are modalities that relate directly to difference in subclonal dendritic growth patterns. Table 3 present an analysis of data of Strain E on the frequency of segment slopes, their pair-wise combina- tions, and a test of the hypothesis that the expected pro- portion of clones having any pair-wise combination of two slope segments is the product of the independent probabilities (2 pq) is given by the expansion of the bi- nomial equation [(p + q2 = 1)], where p and q are the frequencies of the parent slope segments. Only 9 of the 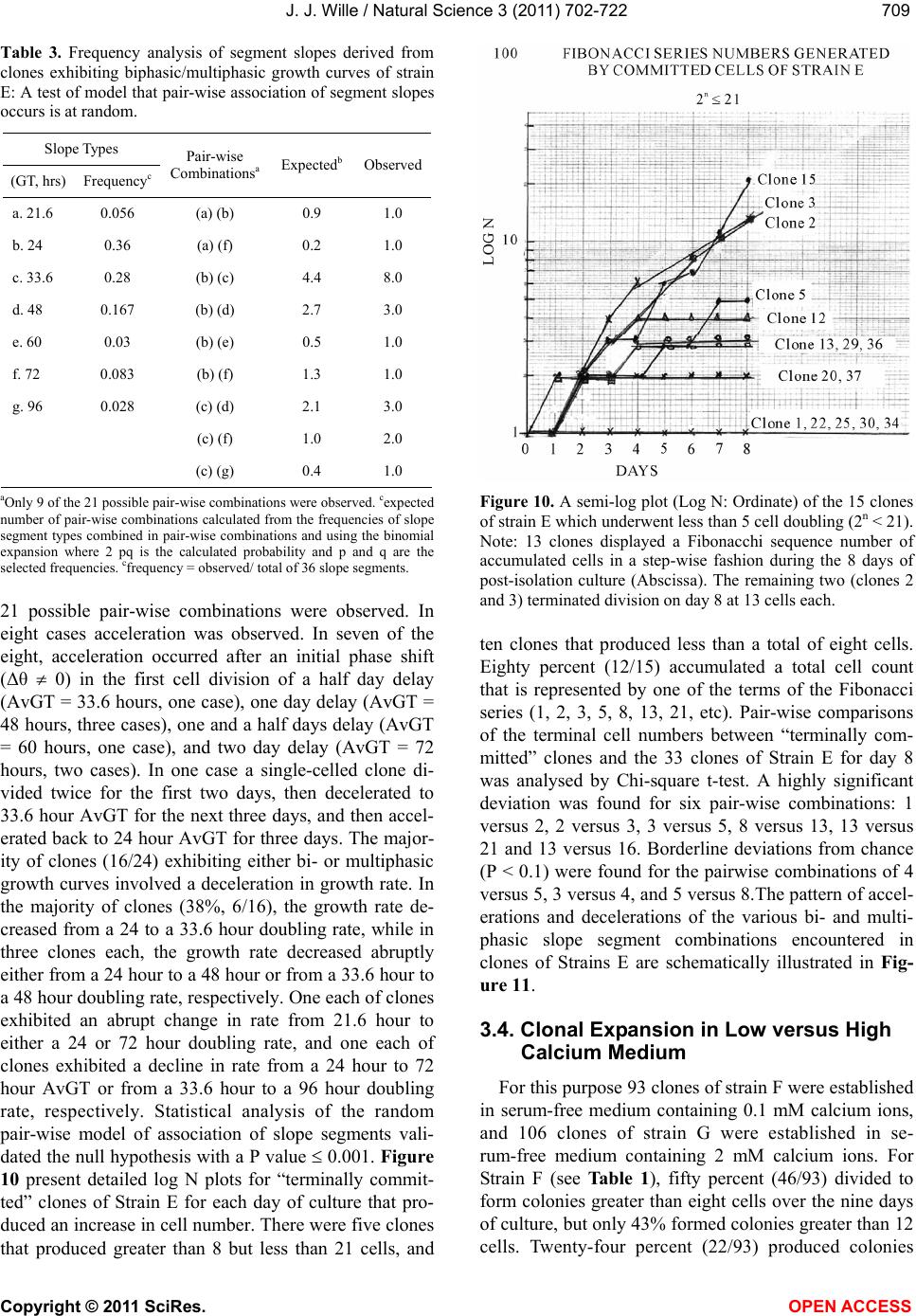 J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 709709 Table 3. Frequency analysis of segment slopes derived from clones exhibiting biphasic/multiphasic growth curves of strain E: A test of model that pair-wise association of segment slopes occurs is at random. Slope Types (GT, hrs) Frequencyc Pair-wise Combinationsa Expectedb Observed a. 21.6 0.056 (a) (b) 0.9 1.0 b. 24 0.36 (a) (f) 0.2 1.0 c. 33.6 0.28 (b) (c) 4.4 8.0 d. 48 0.167 (b) (d) 2.7 3.0 e. 60 0.03 (b) (e) 0.5 1.0 f. 72 0.083 (b) (f) 1.3 1.0 g. 96 0.028 (c) (d) 2.1 3.0 (c) (f) 1.0 2.0 (c) (g) 0.4 1.0 aOnly 9 of the 21 possible pair-wise combinations were observed. cexpected number of pair-wise combinations calculated from the frequencies of slope segment types combined in pair-wise combinations and using the binomial expansion where 2 pq is the calculated probability and p and q are the selected frequencies. cfrequency = observed/ total of 36 slope segments. 21 possible pair-wise combinations were observed. In eight cases acceleration was observed. In seven of the eight, acceleration occurred after an initial phase shift (Δθ 0) in the first cell division of a half day delay (AvGT = 33.6 hours, one case), one day delay (AvGT = 48 hours, three cases), one and a half days delay (AvGT = 60 hours, one case), and two day delay (AvGT = 72 hours, two cases). In one case a single-celled clone di- vided twice for the first two days, then decelerated to 33.6 hour AvGT for the next three days, and then accel- erated back to 24 hour AvGT for three days. The major- ity of clones (16/24) exhibiting either bi- or multiphasic growth curves involved a deceleration in growth rate. In the majority of clones (38%, 6/16), the growth rate de- creased from a 24 to a 33.6 hour doubling rate, while in three clones each, the growth rate decreased abruptly either from a 24 hour to a 48 hour or from a 33.6 hour to a 48 hour doubling rate, respectively. One each of clones exhibited an abrupt change in rate from 21.6 hour to either a 24 or 72 hour doubling rate, and one each of clones exhibited a decline in rate from a 24 hour to 72 hour AvGT or from a 33.6 hour to a 96 hour doubling rate, respectively. Statistical analysis of the random pair-wise model of association of slope segments vali- dated the null hypothesis with a P value 0.001. Figure 10 present detailed log N plots for “terminally commit- ted” clones of Strain E for each day of culture that pro- duced an increase in cell number. There were five clones that produced greater than 8 but less than 21 cells, and Figure 10. A semi-log plot (Log N: Ordinate) of the 15 clones of strain E which underwent less than 5 cell doubling (2n < 21). Note: 13 clones displayed a Fibonacchi sequence number of accumulated cells in a step-wise fashion during the 8 days of post-isolation culture (Abscissa). The remaining two (clones 2 and 3) terminated division on day 8 at 13 cells each. ten clones that produced less than a total of eight cells. Eighty percent (12/15) accumulated a total cell count that is represented by one of the terms of the Fibonacci series (1, 2, 3, 5, 8, 13, 21, etc). Pair-wise comparisons of the terminal cell numbers between “terminally com- mitted” clones and the 33 clones of Strain E for day 8 was analysed by Chi-square t-test. A highly significant deviation was found for six pair-wise combinations: 1 versus 2, 2 versus 3, 3 versus 5, 8 versus 13, 13 versus 21 and 13 versus 16. Borderline deviations from chance (P < 0.1) were found for the pairwise combinations of 4 versus 5, 3 versus 4, and 5 versus 8.The pattern of accel- erations and decelerations of the various bi- and multi- phasic slope segment combinations encountered in clones of Strains E are schematically illustrated in Fig- ure 11. 3.4. Clonal Expansion in Low versus High Calcium Medium For this purpose 93 clones of strain F were established in serum-free medium containing 0.1 mM calcium ions, and 106 clones of strain G were established in se- rum-free medium containing 2 mM calcium ions. For Strain F (see Table 1), fifty percent (46/93) divided to form colonies greater than eight cells over the nine days of culture, but only 43% formed colonies greater than 12 cells. Twenty-four percent (22/93) produced colonies  J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 710 Figure 11. Pattern of Decelerations (→) and accelerations (←) in biphasic growth curves of 17 clones of strain G. Cases (n) of modal decelerations in AvGT occur from 24 hour to 33.6 hours, 33.6 hours to 48 hours, 24 hour to 48 hours, 24 hours to72 hours, 33.6 hours to 96 hours. Cases of model accelerations in AvGT occur from 48 hours to 24 hours, 72 hours to 24 hours, and 33.6 hours to 24 hours. with less than a total of 12 cells and were considered committed to loss of clone forming potential. Thirty- three percent of the single-celled clones never divided. Detailed Log N plots were constructed for forty prolifer- ating clones of Strain F. Only eighteen percent (7/40) of the clones exhibited no phase shifts and continued to divide exponentially for the nine days of culture; they had an AvGT of 30.3 ± 9.5 hours. This, incidentally, was the average population doubling time of the parent cul- ture from which the single-celled clones were seeded. Three of the clones in this group had AvGTs of 28.8, 38.4 and 48 hours, while the rest had a AvGT of 24 hours resulting in a larger variance than the monophasic growth curves witnessed earlier for clones of Strains A through E, whose AvGTs were approximately 24 hours, which was the AvGT of their parent cultures (see Table 1). The majority of clones (33/40) exhibited either bi- phasic or multiphasic growth curves. Seven clones failed to produce more than 13 cells in nine days of culture. Of the remaining twenty-six clones fifty percent (13/26) exhibited biphasic growth kinetics. Seven clones exhib- ited a pattern of deceleration starting from a 24-hour slope segment and changing to a 33.6, a 48-hour or a 72-hour slope segment or starting from a 33.6-hour slope segment to a 96-hour slope segment. By contrast six clones exhibited a pattern of acceleration starting from 33.6-hour slope segment and changing abruptly to a 24-hour slope segment or starting from a 72-hour slope segment and changing to 24-hour slope segment. Among the clones exhibiting multiphasic growth curves, two major patterns emerged. In sixty-two percent (8/13) of clones immediately following a 24 hour slope segment lasting one or two days, there was either a one day phase shift Δθ = 1, five case) or a two day phase shift (Δθ = 2, three cases), followed by resumption to either a 24 hour a 28.8 hour, a 33.6 hour slope or a 48 hour slope, respec- tively, and in four of these cases resumption was to a 48 hour slope segment. In two of these latter cases, the 48-hour slope segment preceded an abrupt transition back to a 24-hour slope segment. In the remaining five of thirteen clones exhibiting a multiphasic growth curve, a 24-hour slope segment was followed by a 33.6 hour, a 48 hour, or a 72 hour slope segment, respectively. In one case, a 24-hour slope segment was preceded by 72-hour slope segment and followed by a 33.6 our slope segment. Decelerating slope segments outnumber accelerating slope segment by 2 to 1, and all accelerating slope seg- ment began from either 48 hour or 72-hour slope seg- ments. Regarding the eight clones of strain F that had a phase shift of one or two days interspersed between slope changes, no increase in cell number can be inter- preted as skipped cell cycles. This interpretation is enlarged upon in the section below detailing the evi- dence linking skipped cycles to an underlying mitotic clock. For clone G (see Table 1), fifty-three percent (56/106) of the clones divided within the first 24 hours and 73% (77/106) divided within the first 48 hours. Correspondingly, 20% only divided once in 48 hours, while 40% (42/106) divided at least twice in 48 hours. Finally, 48% (51/106) of clone G divided more than once in 24 hours. Twenty-nine percent (31/106) of clones failed to achieve a total of greater than eight cells in seven day; they were considered to be committed cells. Fifteen of these failed to divide at all; eleven divided once, four formed a total of three cells, and one divided to form a total of eight cells. As observed with commit- ted cells from clones reared in low calcium medium, the committed cells formed clones with the Fibonacci series terms (1, 2, 3 and 8). Detailed Log N plots of the seventy-five proliferating clones revealed that 46% (42/91) exhibited rapid logarithmic growth curves with no phase shifts. The various monophasic growth rates observed, expressed as AvGT, were 19.2 hours (13%), 21 hours (7%), 24 hours (23%), 48 hours (2%) and 72 hours (1%). The overall mean AvGT for all 42 clones is 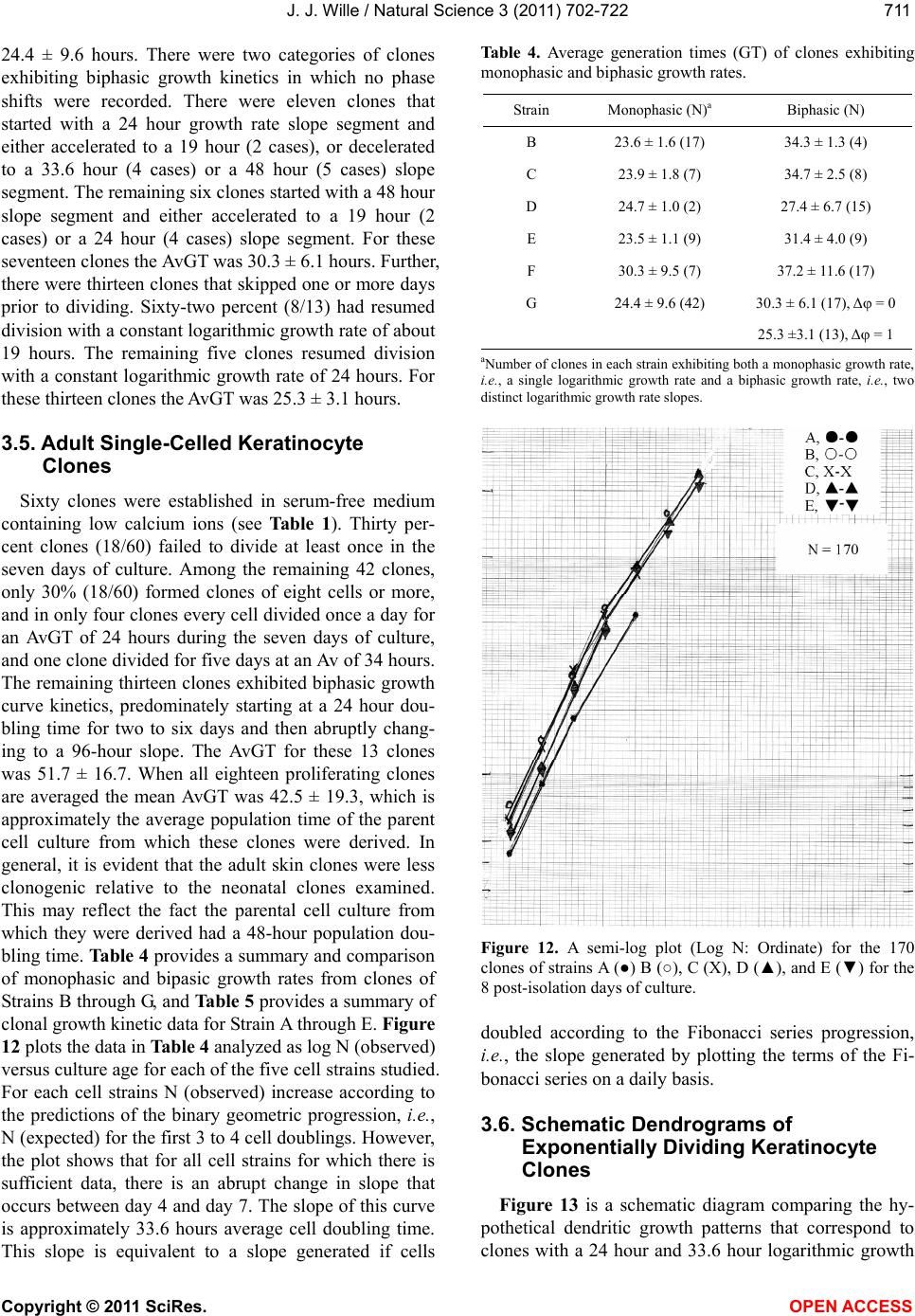 J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 711711 24.4 ± 9.6 hours. There were two categories of clones exhibiting biphasic growth kinetics in which no phase shifts were recorded. There were eleven clones that started with a 24 hour growth rate slope segment and either accelerated to a 19 hour (2 cases), or decelerated to a 33.6 hour (4 cases) or a 48 hour (5 cases) slope segment. The remaining six clones started with a 48 hour slope segment and either accelerated to a 19 hour (2 cases) or a 24 hour (4 cases) slope segment. For these seventeen clones the AvGT was 30.3 ± 6.1 hours. Further, there were thirteen clones that skipped one or more days prior to dividing. Sixty-two percent (8/13) had resumed division with a constant logarithmic growth rate of about 19 hours. The remaining five clones resumed division with a constant logarithmic growth rate of 24 hours. For these thirteen clones the AvGT was 25.3 ± 3.1 hours. 3.5. Adult Single-Celled Keratinocyte Clones Sixty clones were established in serum-free medium containing low calcium ions (see Table 1). Thirty per- cent clones (18/60) failed to divide at least once in the seven days of culture. Among the remaining 42 clones, only 30% (18/60) formed clones of eight cells or more, and in only four clones every cell divided once a day for an AvGT of 24 hours during the seven days of culture, and one clone divided for five days at an Av of 34 hours. The remaining thirteen clones exhibited biphasic growth curve kinetics, predominately starting at a 24 hour dou- bling time for two to six days and then abruptly chang- ing to a 96-hour slope. The AvGT for these 13 clones was 51.7 ± 16.7. When all eighteen proliferating clones are averaged the mean AvGT was 42.5 ± 19.3, which is approximately the average population time of the parent cell culture from which these clones were derived. In general, it is evident that the adult skin clones were less clonogenic relative to the neonatal clones examined. This may reflect the fact the parental cell culture from which they were derived had a 48-hour population dou- bling time. Table 4 provides a summary and comparison of monophasic and bipasic growth rates from clones of Strains B through G, and Table 5 provides a summary of clonal growth kinetic data for Strain A through E. Figure 12 plots the data in Table 4 analyzed as log N (observed) versus culture age for each of the five cell strains studied. For each cell strains N (observed) increase according to the predictions of the binary geometric progression, i.e., N (expected) for the first 3 to 4 cell doublings. However, the plot shows that for all cell strains for which there is sufficient data, there is an abrupt change in slope that occurs between day 4 and day 7. The slope of this curve is approximately 33.6 hours average cell doubling time. This slope is equivalent to a slope generated if cells Table 4. Average generation times (GT) of clones exhibiting monophasic and biphasic growth rates. Strain Monophasic (N)a Biphasic (N) B 23.6 ± 1.6 (17) 34.3 ± 1.3 (4) C 23.9 ± 1.8 (7) 34.7 ± 2.5 (8) D 24.7 ± 1.0 (2) 27.4 ± 6.7 (15) E 23.5 ± 1.1 (9) 31.4 ± 4.0 (9) F 30.3 ± 9.5 (7) 37.2 ± 11.6 (17) G 24.4 ± 9.6 (42) 30.3 ± 6.1 (17), Δ = 0 25.3 ±3.1 (13), Δ = 1 aNumber of clones in each strain exhibiting both a monophasic growth rate, i.e., a single logarithmic growth rate and a biphasic growth rate, i.e., two distinct logarithmic growth rate slopes. Figure 12. A semi-log plot (Log N: Ordinate) for the 170 clones of strains A (●) B (○), C (X), D (▲), and E (▼) for the 8 post-isolation days of culture. doubled according to the Fibonacci series progression, i.e., the slope generated by plotting the terms of the Fi- bonacci series on a daily basis. 3.6. Schematic Dendrograms of Exponentially Dividing Keratinocyte Clones Figure 13 is a schematic diagram comparing the hy- pothetical dendritic growth patterns that correspond to clones with a 24 hour and 33.6 hour logarithmic growth 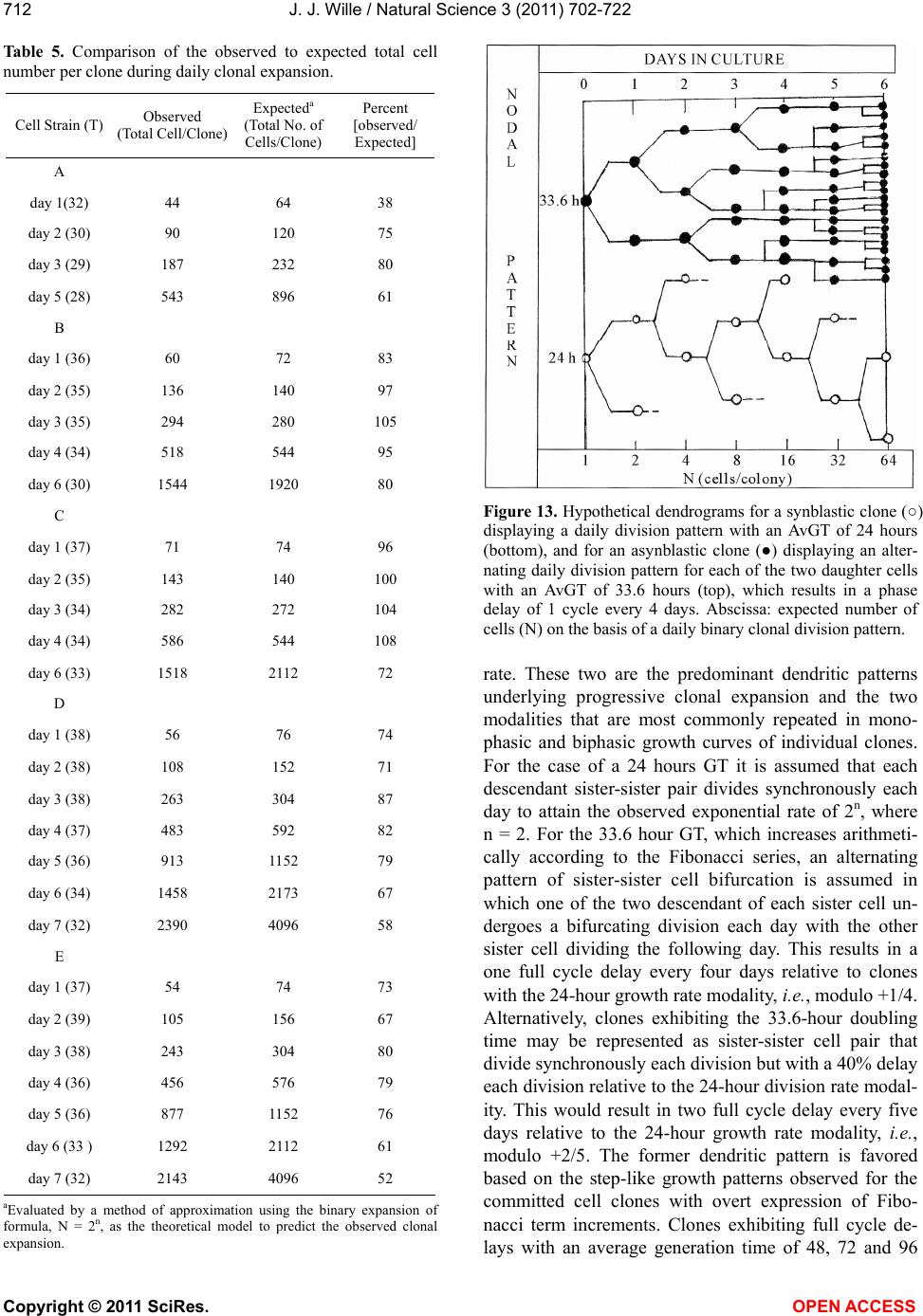 J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 712 Table 5. Comparison of the observed to expected total cell number per clone during daily clonal expansion. Cell Strain (T) Observed (Total Cell/Clone) Expecteda (Total No. of Cells/Clone) Percent [observed/ Expected] A day 1(32) 44 64 38 day 2 (30) 90 120 75 day 3 (29) 187 232 80 day 5 (28) 543 896 61 B day 1 (36) 60 72 83 day 2 (35) 136 140 97 day 3 (35) 294 280 105 day 4 (34) 518 544 95 day 6 (30) 1544 1920 80 C day 1 (37) 71 74 96 day 2 (35) 143 140 100 day 3 (34) 282 272 104 day 4 (34) 586 544 108 day 6 (33) 1518 2112 72 D day 1 (38) 56 76 74 day 2 (38) 108 152 71 day 3 (38) 263 304 87 day 4 (37) 483 592 82 day 5 (36) 913 1152 79 day 6 (34) 1458 2173 67 day 7 (32) 2390 4096 58 E day 1 (37) 54 74 73 day 2 (39) 105 156 67 day 3 (38) 243 304 80 day 4 (36) 456 576 79 day 5 (36) 877 1152 76 day 6 (33 ) 1292 2112 61 day 7 (32) 2143 4096 52 aEvaluated by a method of approximation using the binary expansion of formula, N = 2n, as the theoretical model to predict the observed clonal expansion. Figure 13. Hypothetical dendrograms for a synblastic clone (○) displaying a daily division pattern with an AvGT of 24 hours (bottom), and for an asynblastic clone (●) displaying an alter- nating daily division pattern for each of the two daughter cells with an AvGT of 33.6 hours (top), which results in a phase delay of 1 cycle every 4 days. Abscissa: expected number of cells (N) on the basis of a daily binary clonal division pattern. rate. These two are the predominant dendritic patterns underlying progressive clonal expansion and the two modalities that are most commonly repeated in mono- phasic and biphasic growth curves of individual clones. For the case of a 24 hours GT it is assumed that each descendant sister-sister pair divides synchronously each day to attain the observed exponential rate of 2n, where n = 2. For the 33.6 hour GT, which increases arithmeti- cally according to the Fibonacci series, an alternating pattern of sister-sister cell bifurcation is assumed in which one of the two descendant of each sister cell un- dergoes a bifurcating division each day with the other sister cell dividing the following day. This results in a one full cycle delay every four days relative to clones with the 24-hour growth rate modality, i.e., modulo +1/4. Alternatively, clones exhibiting the 33.6-hour doubling time may be represented as sister-sister cell pair that divide synchronously each division but with a 40% delay each division relative to the 24-hour division rate modal- ity. This would result in two full cycle delay every five days relative to the 24-hour growth rate modality, i.e., modulo +2/5. The former dendritic pattern is favored based on the step-like growth patterns observed for the committed cell clones with overt expression of Fibo- nacci term increments. Clones exhibiting full cycle de- lays with an average generation time of 48, 72 and 96  J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 713713 hours can be represented in dendrograms as sister-sister cell pairs that divide synchronous but only after the lapse of one, two or three full daily cycles of logarithmic growth relative to the 24 hour growth rate modality. They are exactly modulo 1, 2 and 3 cycles out of phase with single-celled clones that divide once a day. Clones were observed that displayed a portion of their biphasic growth with a 28.8-hour logarithmic rate of growth. This can be diagramed as a dendrogram with bifurcating sis- ter-sister cell pairs that are delayed fractionally 20% longer than logarithmically dividing clones that exhibit an average GT of 24 hours. This results in a one full cy- cle delay every 6 days of logarithmic growth relative to the 24-hour modality, i.e., modulo +1/6, this is depicted with reference to the dendrogram for a synblastic clone in Figure 14. Likewise clones were observed that dis- played a 38.4-hour logarithmic rate of growth over some portion of their biphasic growth. This can be diagramed as a dendrogram with bifurcating sister-sister cell pairs that are fractionally delayed 60% longer than logarith- mically dividing clones with an average GT of 24 hours. This results in a three full cycle delay every eight days, i.e., modulo +3/8, relative to the 24-hour growth rate modality. A number of clones were observed that dis- played either a 19.2 or a 21.6-hour logarithmic growth rate either as monophasic or biphasic clonal expansion growth rates. For the case of 19.2-hour logarithmic dou- bling time, sister-sister cell pairs divide 40% faster each day than the 24 hour modality. This results in a one full cycle advance every four days relative to the 24-hour cycle of division, i.e., modulo –1/4. Figure 15 is a hy- pothetical dendrogram for the case of 21.6-hour loga- rithmic growth rate, sister-sister cell pairs divide 20% faster each day relative to the 24-hour cycle (shown at the bottom of the figure). This results in a one full cycle advance every 10 days relative to the 24-hour cycle mo- dality, i.e., modulo –1/10. Finally, some clones were observed which displayed a 14.4-hour growth rate over some portion of their biphasic or multiphasic logarithmic growth rate. These clones can be diagramed as a den- drogram in which sister-sister cell pairs divide synchro- nous 60% faster each day than the corresponding clones with a 24 hour-cycle modality; this results in a phase advance of three cycles every eight days relative to the 24-hour cycle modality, i.e., modulo –3/8. In addition, we propose the “Rule of Deceleration,” in which sis- ter-sister cell pairs abruptly change their bifurcation pat- tern from a 24, 33.6 or 48 hours exponential doubling rate to a progressively restricted subset of sister-cell branches, that continue to divide logarithmically but at decelerated growth rate, resulting in either a switch a lower full cycle modality (2, 3, or 4) or to a decelerating dendritic pattern in which one sister cell branch alter- Figure 14. Hypothetical dendrograms comparing the synblastic clone (○) with a daily division pattern with an AvGT of 24 hours (bottom), to an asynblastic clone (●) displaying an decelerated division pattern with an AvGT of 28.8 hours (top), which results in a phase delay of 1 cycle every 6 days. Abscissa: expected number of cells (N) on the basis of a daily binary clonal division pattern. Figure 15. Hypothetical dendrogram of clones displaying an AvGT of 21.6 hours with bifurcating sister-sister cell pairs that are fractionally 20% faster than logarithmically dividing clones with an average GT of 24 hours. nates every other division and assumes an intermediate cycle modulus (+1/6, +1/4, etc). All possible combina- tions have been observed in the bi- and multiphasic growth curves presented above. By contrast, a number of clones exhibited acceleration from a lower modulus to a higher modulus, e.g., modulus 1 to modulus –1/4 or –1/10, and from a modulus +1/4 to modulus 1 or –1/4. These abrupt changes in logarithmic growth rate can be  J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 714 depicted as dendrograms in which sister-sister cell pairs resume synchronous daily division once a day or accel- erate to fractional cycle advances. For these, we propose the “Rule of Fractional Phase Advance,” for intraclonal phase advances. The above hypothetical dendrograms can be reinterpreted in terms of a phase resetting curve. Figure 16, plots the amount of phase shift (ΔΦ ≠ 0) as a whole or fractional cell cycle, Modulus equal to 0 (ΔΦ = 0) of the unshifted cell cycle so that Modulus 1 is shifted one circadian period of 24 hours. Phase delays are plot- ted as positive whole or fractional cycle delays (+1, +2 n), and phase advances are plotted as negative whole or fractional cell cycle. The data are derived from bi- phasic and multiphasic segment slope data. Inspection of the curve shows both phase adavances and phase delays occur. Phase delays appear to reach an asymptote at Modulus 3, which concurs with the greatest 72 hour segment slope observed. Phase advances extrapolate to the ordinate to yield a cell cycle with an average GT equal to 12 hours. No average generation time (GT) of 12 hours was observed but such a cell cycle would be equivalent to a G1-less cell cycle which are often en- countered in embryonic cell cycles. 4. DISCUSSION 4.1. Interdivision Interval On first inspection, the kinetic of the first inter-divi- sion observed for these two different cell strains grown under two different calcium conditions are remarkably similar. However, any analysis of these results must take into account the effects imposed by trypsin treatment on steady-state conditions prevailing before seeding on to cloning chips. Dissociation of cellular aggregates into individual cells by “trypsin treatment” surely must dis- rupt many on-going cellular processes the consequences of which are too numerous and indeterminate to either enumerate or understand. Yet, they are likely to account to a lesser or greater extent for the observed division delays. Earlier studies [7] attempting to assess mitotic delay following deliberate temperature shock treatments and other so-called perturbations when administered at a given phase of the mitotic cycle in mitotically synchro- nized cells subtracted out the time lags due to the dura- tion of the treatment per se and defined the remainder of the delay as “excess mitotic delay.” This phase-resetting map is called a type 1 phase response curve [8]. Unfor- tunately, this stratagem only works when the delays are referenced comparatively to specific phases of the mi- totic cycle. Here, we have adopted the assumption that prior to trypsin treatment the cells were randomly dis- tributed with respect to phase according to a “log normal age distribution.” If the perturbation preceding cell Figure 16. A theoretical phase resetting curve of NHK cells. The prior unshifted phase (ΔΦ = 0) is plotted on the ordinate as AvGT (hours) versus the reset phase (ΔΦ ≠ 0) on the abscissa (Cell Cycle). The reset phase is plotted as fractional or whole moduli of 24 hour cell cycle, and shows phase advances (– modulus) and phase delays (+ modulus) of modular cell cycles. seeding synchronized all cells to a common phase, the interdivision interval, i.e., excess mitotic delay would have led to a single model time of delay. This was not observed. Therefore, the dissociation treatment did not interfere with the set of contingent phases present prior to dissociation. The simplest outcome expected would be a monophasic log exit from the division cycle. This did not occur either. Under two different conditions of re- covery the result was a biphasic log exit from the divi- sion cycle. Possible reasons for this are discussed below in conjunction with the other clonal division results. 4.2. Clonal Division Rates The clonal data presented show that 60% of single- cell derived clones isolated at random from asynchro- nously dividing logarithmic cultures can form colonies of greater than eight cells, and about 50% will form an exponentially dividing clone in which each cell divides on average once a day for up to six to eight days of cul- ture in a serum-free medium. The study also provided new data showing that individual cells have different propensities to divide at different rates. The majority of clones that retain proliferative potential have an average generation time of approximately 24 hours (circadian division rate). Nevertheless, a significant number of clones display exponential rates of growth that are greater or less than 24 hours. Of interest was the finding that these clones are restricted to only a few of the many possible logarithmic growth rates. Clones exhibiting exponential growth rates greater than 24 hours (infradian  J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 715715 rates) are restricted to modes at 33.6, 38.4, 48, 72 and 96 hours, while clones exhibiting exponential growth rates less than 24 hours (ultradian rates) are restricted to modes of 21.6, 19.2 and 14.4 hours. In addition, indi- vidual clones may switch abruptly from a circadian growth rate to either an infradian or ultradian growth rate. This was amply demonstrated in the pattern and frequencies of growth rate switches seen in biphasic and multiphasic growth curves. Another phenomenon en- countered in single-celled clones was the pattern of phase delays exhibited by isolated single cells prior to their first cell division. These delays were restricted to 24 hour multiples of the circadian inter-division periods, i.e., 48, 72, and 96 hours. Such delays were also en- countered during exponential growth of clones indicat- ing that cycle skips can occur after single cells establish a colony of cells. In particular, the biphasic exponential rate of entry into the first cell division suggested a cell cycle-dependent process was perturbed by the serial passage of cells from steady-state growth to the sin- gle-celled state. We reasoned that cells in S and G2/M were less affected by this perturbation as represented by 30% - 40% of cells (Fraction-A see Figures 2 and 3) that advanced through to cell division in the expected 16 hour interval for cells randomly arrayed between the start of S phase and the end of the G2 phase. Further, the remaining undivided cells after a 16-hour delay exited from the cell cycle at the circadian rate as if they were randomly arrayed at all possible phase of the circadian cell cycle (Fraction-B, see Figures 2 and 3). Again, we reasoned that these must represent cells that were in the G1 phase of the cell cycle at the time of perturbation. If so, why did it take 24 hours rather than the expected 12 hours for this cohort to advance through the cell cycle? In an attempt to reconcile these observations with pre- vious reports, we examined their cinemicrographic data [2]. Not unlike our findings, they reported 70% (7/10) of clones and 67% (14/21) of clones completed cell divi- sion the first day of observation (Cultures 2 and Culture 3, respectively of Figures 2 and 3). In addition, they supply dendrograms showing sister cells, in which one member divides 24 hours after the first division, while the other sister cells is delayed approximately 72 hours before its next division (Culture 3, Clone A, of Figure 2). Equally, they show sister cells that divide synchronously on the second division cell cycle 24 hours after the first cell division (Culture 3, Clone C and J of Figure 3). Also, they show non-synchronous patterns of sister cell division following the first cell division that are not mul- tiples of 24 hour cell cycle (Culture 2, Clones A, B, C and D and Culture 3, Clones K, Q, S, and T). Most in- terestingly, they observed cases in which the interdivi- sion time accelerated from 28.7 hours to 21.1 hours, and cases in which the inter-division time of sister cells ac- celerated from 33.7 hours to 28.8 hours (Culture 2 of Figure 2). Finally, they provided a histogram showing the frequency distribution of inter-divisions times (Fig- ure 6) in which the modal inter-divisions times were 14.4, 19.2, 21.6, 24, 28.8, where clusters of data points center around these modal times in their plot of the cor- relation between filial and parental inter-division times (Figure 7). 4.3. Significance of Fibonacci Series in Keratinocyte Clonal Growth Our results reveal yet another phenomenon: the fre- quency with which non-proliferating clones exhibit the Fibonacci series of terms as end-points in terminal number of cells per clone. A standard explanation for the appearance of the Fibonacci sequence in biological phe- nomena is “edge-effects” consequences. In this case, those clones comprised of daughter cells with unequal probabilities of dividing in a 24-hour period arise from unequal or unbalanced steady state conditions. This pos- sibility is discussed in relation to a more general descrip- tion of Fibonacci numbers. The Fibonacci series is one example of a periodic continued fraction that converges on the numerical value of 1.618, a ratio often called the” golden mean of unity.” It is deep-set in the regular pen- tagon, and dodecahedron, where it enters (as the chord of an angle of 36˚ into the three-isosceles triangles, and by continued bisection generates apices that have their locus upon an equiangular spiral. In other biological contexts, the Fibonacci series has been encountered in plant morphogenesis as the phenomenon of Phyllotaxis seen in the spiraled seeds of the sunflower, and fir cones, and in animal morphology such as the spiraled shells in foraminiferan and in molluscan shells [9]. There does not appear to be any references to it in animal tissue or- ganization and growth. He believed that, it simply re- flects continuation of constant conditions of growth where there is a steady production of similar growth units (i.e. , cells) that are similarly situated at similar successive intervals of time, This is where the time- element enters in the development of equiangular spirals for according the theory of gnomons “one never expects to find the logarithmic spiral manifested in a structure whose parts are simultaneously produced [9], see p. 766).” This idea has a parallel in the con text of the gen- eration of recursive structures by computer algorithms [10], where the Fibonacci series can be simulated as a recursive transition network (RTN) This was diagramed as bifurcating tree of nodes that are recalled successively in time (see Diagram G, Figure 30). By analogy, we pro- pose a similar recurrent branching pattern (RTN) under- lies the clonal expansion of non-synblastic proliferating  J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 716 clones (see Figure 12) as well as the remarkable finding of a 33.6-hour doubling time in clonal subsets of clones exhibiting biphasic growth rates. The 33.6-hour doubling time is simply obtained when the Fibonacci series terms are plotted on semi-log paper as a function of 24-hour intervals (days) of culture. 5. HYPOTHESES 5.1. Circadian Cell Cycling Hypothesis Various hypotheses have been proposed to account for circadian clock control of cell division including gene transcription of clock genes that have kinetics of synthe- sis, and negative feedback regulation on gene transcrip- tion to account for the oscillation in timing of mitosis [11-13]. The circadian transcription clock is a determi- nistic sequence; it predicts phase advances and phase delays occur by restarting the transcription clock at ear- lier or later entry points in the transcription loop of genes. Uncoupling the transcription clock from the mitotic clock probably is limited to restriction points in the G1 phase of the cell cycle and in this respect could account for the multiple cell cycle “on-phase” coupling and un- coupling of the transcription clock from the mitotic clock. Finally, it would be hard to construct an interpre- tation of the transcription clock that can account for “cross-talk” between cells, leading to phase synchroni- zation between sister cells or phase dispersion among a cohort of cells in a clone based solely on the order of appearance of mRNA transcripts or how this could ac- count for the observed Fibonacci series in the total cell numbers arising in committed clones. Although, mo- lecular models provide a mechanism for sequential pro- gression through the cell cycle, and provide a molecular escapement component for the circadian control system [8]. They all fail to account for the recurrent 24-hour periods per se, nor deal with the effect of perturbations to clock functioning, i.e., phase shifting and period al- terations, and multiple cycle “on-phase” recurrent rhythms. 5.2. Evidence for Biochemcial Oscillator: Retrodictions and Predictions Many results observed in these studies can be ex- plained by the hypothesis of a biochemical oscillator with underlying stable limit cycle dynamics [14] as the temporal control system for human epidermal cell divi- sion [15,16]. Earlier, we proposed a two-dimensional (X, Y) biochemical oscillator of the sort previously de- scribed as a model for explaining the behavior of the mitotic division rhythm in the acellular slime mold, Physarum polycephalum [11], and later extended to mammalian cell cycles [12]. In particular, the proposed mitotic oscillator model further associates this clonal division pattern with events leading to the loss of cycling among proliferating cells during their transition to G1/G0 arrest, and is ultimately involved in the mechanism of commitment to terminal cell differentiation (see Figure 17). In addition, the puzzling modal frequencies of divi- sion rates led us to consider that epidermal keratinocytes may find an explanation in a mitotic oscillator-based clock that controls cell division timing. Figure 17 arbi- trarily maps the different cell cycle phases as sets of neighboring states along the limiting trajectory. The lim- iting trajectory moves smoothly through each of the states in a counter-clockwise motion. In addition, it means something dynamically to be at states of the sys- tem that lie off the limiting trajectory. These are real biochemical states that lie in the state space of the limit cycle. Perturbations may carry the system to point off the limit cycle through a destruction vector that drives the system toward the origin (Xo, Yo), or by changing the parameters of the constant kinetic scheme. In either case, once transients die down, return from these states occurs along trajectories that wind back on to the limit- ing trajectory arrive at their ultimate phase according to the isochron structure of the oscillator. The model ret- rodicts that the temporal control of cyclins during the cell cycle occurs as the closed path trajectory of the limit cycle mitotic oscillator passes through biochemical states that could trigger transcription of specific cyclin genes and their gene products [19]. Figure 18 simulates the results of a perturbation given at a critical phase and critical intensity along the limiting trajectory that map to sets of neighboring states in the G2 phase of the cell cy- cle. As shown, the perturbation drives the system “in- side” the limiting trajectory on a destruction vector that points toward the origin. Upon release, the system re- turns along a winding trajectory that crosses the Yc trig- gering mitosis from the inside the limit cycle. In this scenario, there is little or no delay to mitosis, and the next cycle will occur in phase with identical copies of an unperturbed oscillator. However, if the system is driven further inside the limit cycle along the same isochron in the G2 phase, upon release it will return on a “grazing” or sub-threshold trajectory that does not cross the Y-axis at the Yc value as required to trigger mitosis. These lat- ter copies of the oscillator will not undergo mitosis/cell division. Nevertheless, after a slightly less than one cy- cle delay the next cell cycle will recur in phase with an unperturbed copy of the oscillator, since the” grazing” trajectory crosses the limiting trajectory in phase. A still more intense perturbation given at the same critical phase in G2 will drive the system toward the singularity. Since this is a phase-less (timeless) locus where all  J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 717717 Figure 17. Diagram showing the effect of a perturbation given at a state in the G2 phase (open shaded ellipse) that instantane- ously drives the limiting trajectory “inside” the closed path trajectory where it continues to cycle in phase (θ1) but follows a new trajectory (dotted arrow) that fails to achieve sufficient concentration of Y, Yc (shaded circle) to trigger mitosis result- ing in a one or more full cycle delays. Figure 18. Oscillator clock control of recurrent events of the cell cycle. Different events of the cell cycle are driven when the limiting trajectory of the oscillator pass through the arbi- trarily drawn regions of state space (circles and boxes). Mitosis is triggered at Yc, a critical concentration of active mitogen (Y). Cytokinesis occurs when the trajectory reaches a near maxi- mum of Y. The G1 phase of the cell cycle is associated with a region of state space near a secondary steady state, SS1 that is surrounded by a low amplitude oscillation. Cells cycle there until reentering the high amplitude limit cycle oscillation, de- pendent on growth contingencies (variable kinetics). Alterna- tively cells in the SS1 region can drift into a tertiary steady state labeled SS2 in the region of state space surrounded by an anti-limit cycle (curved line) and become extremely slow cy- cling in a resting state, G0. The S phase is denoted by the boxed region with minimal Y and increasing concentration of X, the inactive mitogen. Finally, the boxed region of state space de- noted by G2 occurs when the limiting trajectory passes through set of states with high concentration of X and increasing con- centration of Y. isochrons converge at or near the singularity (indetermi- nate point), copies of the system that are at or near the singularity can take up as many different ultimate phases as the isochrons upon which they lie. Upon release, the various copies of the system will appear to have random phases with respect to one another, i.e., high phase dis- persion. The account given above is in accord with the observed phase resetting behavior of cells presumed to be in the mid-G2 phase of the cell cycle. These cells ex- hibit little inter-divisional delay, and their succeeding cell divisions recur in phase. Still other cells behave as if they were driven to a “grazing” or sub-threshold trajec- tory and skip mitosis/cell division, but since they are on a G2 isochron, their succeeding cell divisions recur in phase with an unperturbed cell another, i.e., high phase dispersion. This brief account has been provided to the reader to facilitate understanding further predictions of the model. 5.2.1. Model Confirmation: G2 Phase Perturbation The following are predictions if the perturbation drives the initial state of the system to a new set of states that return to the limiting trajectory in phase after one cycle delay. By inference, cells exhibiting multiple cycle delays prior to resuming in phase cell cycling are copies of the oscillator that are driven yet deeper inside the state space of the oscillator but are still on a G2 isochron, and upon release follow a sub-threshold trajectory that re- quires multiple cycles before winding onto the limiting trajectory. The biochemical oscillator system used to model these G2 perturbations had a “winding number of three, which easily could explain multiple cycle delays and return in phase for clones exhibiting 48, 72 and 96 hour delays before cycling, which, incidentally, was the longest delays observed. In our model oscillator the an- gular velocity is the same off the limiting trajectory as drawn by straight-line isochrons. Otherwise, events would get out of phase if the angular velocity is faster or slower on isochrons that lie inside the state space of the oscilla- tion. This does not appear to be the case for the underly- ing oscillation preserving the cell division rhythm in normal human epidermal keratinocytes. 5.2.2. Model Confirmation: G1 Perturbation Cells presumed to be in the G1 phase at time of per- turbation, were delayed 16 - 24 hours (i.e., a skipped one cell cycle) before they started to divide, and then it took them another 24 hours to complete this first inter-divi- sion cycle, as if upon release the cells were randomly arrayed at different phases of the cell cycle. In order to account for the phase behavior cells perturbed during the G1 phase of the cell cycle, it is necessary to explore the  J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 718 state space surrounding the G1 isochrons. A destruction vector at G1 drives the oscillating system off the cycle in the direction of the origin, but on a G1 isochron. There- fore, identical copies of the oscillating system released from these “off cycle” states should resume cell cycling in phase with unperturbed copies of the oscillator that were cycling from phases near a G1 isochron. This did not happen. Why? Previous work on mammalian cell cycles suggested that G1 phase of the cell cycle may not be part of the constant kinetic high amplitude oscillation, i.e., an inde- terminate or phase-less portion of the cell cycle [11]. As cells complete mitosis and cytokinesis, the synthesis of many messages stops while chromosomes undergo a chromatin condensation in prophase, and then decon- densation in telophase. A realistic biochemical oscillator model would posit that cells in early inter-phase enter a G1 checkpoint and depending on the availability of pre- formed precursors and the presence of active synthetic machinery for synthesizing X and Y, temporarily exit off the cell cycle to become G1/G0-arrested cells. It is pro- posed that the high amplitude oscillation under variable parameterization becomes a low amplitude oscillation, existing entirely within the state space of G1 isochrons (see Figure 18). Since constant kinetics no longer pre- vail, the limit cycle amplitude would decrease and its singularity drift toward the X, Y origin, at which point the systems dynamics can undergo a bifurcation. One parameterization of the coefficients of the state variables, for example, is a decrease in the rate of synthesis of X. Bifurcation can occur in one of two ways. The first is by a “soft” bifurcation, occurring at the G1 checkpoint, which would carry the oscillating system near to but not within a neighboring basin of attraction. Simply by in- creasing the rate of synthesis of X to the original rate would return the system to the high amplitude oscillation. Cells would then be back on the G1 isochrons, and stay in phase. It is posited that unperturbed cells, having a minimum 12 hour G1 phase, routinely undergo a soft bifurcation in early inter-phase, and cycle with a 12 hour period in the low amplitude oscillation before they gain enough rate of synthesis of X to return to the high am- plitude oscillation. The high amplitude oscillation is also a 12-hour period. Together the small and large amplitude oscillations constitute a 24-hour or circadian cell cycle [11]. The second mode of exit from the cell cycle, also occurring in early inter-phase, is by way of a “hard” bi- furcation, which would carry the oscillating system into an anti-limit cycle trapped within a basin of attraction (see Figure 17). A “hard” bifurcation creates a “hystere- sis effect” whereby a higher rate of synthesis of X is required to restore the low amplitude oscillation than that prevailing under constant kinetics of the high am- plitude oscillation. Cells trapped in an anti-limit cycle are non-cycling, i.e., phase-less. Hence, they exit through a stochastic process in achieving sufficient syn- thetic rate to restore the low amplitude oscillation. Events beyond the restoration of the low amplitude limit cycle oscillation are the same as in the soft bifurcation. In summary soft to hard bifurcation switch accounts for the variable dwell time of G1/G0 cells, while a hard bi- furcation accounts for random process whereby G1 phase cells return to the cell cycle. This modeling predicts that there will be large phase dispersions in perturbed G1 cells. 5.3. Limit Cycle Dynamics Model: Diffusively Coupled G1 and G2 Phase Cells We now attempt to account for the fact that many clones experienced a 33.6-hour exponential growth rate upon resumption of clonal growth from a perturbed cell cycle. This can be accounted for by considering the fact that epidermal keratinocytes grow in a colony with each cell in contact with one or more sister cells. The possi- bility exists for neighboring sister cells within a colony to interact diffusively with each other. Consider the case where two independent copies of the oscillating system are located in adjacent boxes connected by a semi-per- meable membrane. If we let the variables of the dy- namical system be the diffusible components, and de- pending the allowed rate of diffusion, one can obtain either synchrony (homogeneous solution) or persistent asynchrony (inhomogeneous solution), i.e., phase cou- pling results in synchrony if the two copies of the oscil- lator are close in phase, while phase coupling between far apart phases may fail to synchronize. Computer simulations (data not given) of phase coupled limit cycle oscillators that are diffusively coupled demonstrates that a line connecting phases between an oscillator in G1 phase with a second oscillator in G2 phase may result in the persistent sub-threshold amplitude oscillation in the box containing the G2 oscillator while the oscillator in the G1 box continues to cycle with a high amplitude os- cillation. In this situation, the G2 cell oscillates below the threshold (YC) required to trigger mitosis/cell divi- sion. At greater diffusion rates, both oscillations may be dampened to have sub-threshold oscillations, in which case cell division would cease in both cells even though the oscillation preserving rhythmicity of cell division persists. Uncoupling of the oscillator pair would restore the cell division rhythm. It is precisely, this phase cou- pling by diffusion (electrical, chemical, etc) that is in- voked to occur in epidermal cell colonies that account for an alternating sequence of threshold and sub-thresh- old oscillations between pairs of sister cells. Such an  J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 719719 mechanism would produce a pattern of alternating cycles of division such that one member of the pair divides while the other does not, but then on the succeeding cy- cle the sister cell that failed to divide now divides, while the other sister cell fails to divide. This situation is de- picted in the dendrogram representing the case of clones exhibiting a 33.6-hour AvGT (see Figure 13). Obviously, uncoupling of sister cells results in a return to synchro- nous 24-hour cell cycles. 5.3.1. Predictions (“p”) p1: Phase compromise occurs at a phase off the limit- ing trajectory of the oscillating system (θi, xi, yi) that results in a one-cycle delay for the G2 cycling cell, and to a phase advance for the G1 cycling sister cell. p2: The G2 cycling sister cell misses one cell division cycle (cycle 2), but it eventually winds back onto the limiting trajectory of the large amplitude oscillation and then re-enter G2 phase in phase with Cycle 3. Here it again comes into a relationship to phase-couple with a G1 cycling cell that has completed its second cycle and is now in cycle 3. This reinitiates the phase compromise situation and results in a perpetuated pattern of alternat- ing sister cell divisions as witnessed by Fibonacci series terms in total cell number per clone per day, i.e., GT = 33.6 hours (this model is further discussed below in dual oscillator-probabilistic mitotic control model. 5.3.2. Confirmation 1) These predictions are confirmed in data presented in Figures 2 and 3 (biphasic inter-division kinetics) based on the expected proportion of G1 cells present at the time of cloning from an asynchronous logarithmi- cally dividing parent culture. 2) In asynchronous culture with a 24 hour cell cycle time (data from clones of strains F and G). The predictions are also fulfilled in data presented in Table 2 (biphasic growth curves alternating from 24 hour to 33.6-hour GT slopes. 3) The prediction is also verified for clones exhibiting the Fibonacci series of limited or “committed” prolifera- tive potential (e.g., see data in Figure 6 and Figure 12 of “committed cell” clones of strain D. 5.4. Stochastic Cell Cycle Models: Evidence for Probabilistic Control of G1 States of the Cell Cycle Although the stable limit cycle oscillator model does a good job in accounting for “multiple” skipped cell cycles, phase resetting of G2 perturbed cells, and phase com- promise behavior in diffusively coupled G1 and G2 cells, it requires an ad hoc hypothesis to explain G1 phase re- setting following a perturbation, i.e., a stochastic com- ponent as part of the cell cycle and collapse of the limit cycle following a “hard bifurcation” in the G1 phase. In addition, the mitotic oscillator hypothesis does not easily explain period shortening without resort to an ad hoc provision that would increase the size of the high ampli- tude oscillation, e.g., by increasing the synthesis rates of the one or more variables of the system. In the discus- sion, a clone dividing exponentially at AvGT of 21.6 hours was explained by introducing a 10% increase in the rate of cell division. Likewise, for clones that showed a consistent delay that is an integral fraction of the 24-hour cycle, e.g., 28.8 hours, a 10% bias in lengthen- ing the cell cycle relative to a standard 24 cell cycle was introduced. We now attempt to show that this fits a probabilistic mode for temporal control of the G1 phase of the cell cycle in which there is a sensitive dependence on initial conditions with resultant chaos in the oscillator dynamics. Earlier, a theoretical model was examined for the distribution of cell size and generation time that in- corporated size control and random transition [17,21]. They found that a model with a random-exiting phase of the cell cycle and a minimum size requirement for entry into the random-exiting phase predicted exponential beta-curves that are characteristic of sister cell genera- tion times. They argue against unequal mother cell divi- sion as the source of random fluctuations in determinis- tic cell size models, and that transition-prob- ability models with no feedback from cell size are unable to account for the rapidity with which new, stable size dis- tributions are reestablished. Indeed, deterministic cell cycle models with transition probability like properties have been proposed [19]. 5.5. Daughter Cell Size/Cell Mass Variance Can Perpetuate Lineage Differences We begin this discussion with observations drawn from two well-documented An example of cell lineage differences due to a consistent bias in the division rate of cell division products comes from prokaryotes in which careful measurements of inter-division period indicated that there was as much as a 20% variation in inter-divi- sion times between sister cells, and further that there is a correlation between filial and grand-parental generations in inter-division timing and size (Kubitchek, per. com- mun). These results have been interpreted as due to bio- logical variation in the cell division process with smaller daughter cells requiring longer inter-division period and larger daughters requiring less time to reach the doubled cell mass before dividing to form smaller cells. The lar- ger notion is that there is inherent variation of the cell division process that leads to cell size/mass variability, which in turn is compensated for by a mass to DNA ratio threshold mechanism. In this regard, unequal division in  J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 720 “conical,” a mutant of the ciliated protozoan Tetrahy- mena, leads to sublines of different sizes. Regulation to normal size occurs over several generations by slowing the rate of cell division of the smaller cell with all phases of the cell cycle being prolonged [22]. In the ciliated protozoan Paramecium the two products of division are readily recognized, separately cloned separately. The clonal lineage of the anterior product, called the proter, can be compared with that of the posterior product of division, the opisthe. In essence each half-cell must build a new half, the proter builds a new opisthe and the 0pithe builds a new proter. Other asymmetries may also exist with regard to the distribution of mass and organelles. Proter lineages display shorter inter-division times rela- tive to opisthe lineages (T. M. Sonneborn, per. commun). These results indicate that consistent biases in cell divi- sion can generate proportional cell cycle advances and delays. Finally, new genetic and molecular studies in the yeast Saccharomyces cerevisiae have provided evidence in favor of the “critical cell size model” for coordination of cell growth with division [21]. They reported that both the expression and the activity of G1-phase cyclins is modulated by growth rate and cell size in yeast. Other data obtained in these studies suggested that the prolif- erative capacity also correlates with cell size. 5.6. Probabilistic Model for G1 Phase Duration Variability in the duration of the G1 phase has been well established [22] and G1-less cells occur during em- bryogenesis, in mutant mammalian cells [23]. Here, we propose that each cell division creates a proter-like and opisthe-like cell line, and further that the proter-like cell (arbitrarily) has an inherent bias in its expected in- ter-division time while the opisthe-like cell line does not. The reasoning for this has been given above, e.g., bias in cell mass for anterior cell products. The bias is con- strained to values that lie within the standard deviation for the process of cell division as a whole. We then as- sign probability values, p and q to the proter and opisthe cell division rates, respectively, to reflect a fractional deviation of the proter-like cells from the standard rate (mean inter-division time). If there were no biases, the value of p and q are both equal to one-half (0.5), then according to the expansion of the binomial equation an equilibrium distribution of proter and opisthe cell lines are predicted to remain stable over succeeding genera- tions. In the following example, we let the proter cell line have a 10% decrease in mean inter-division time. Table 6 compares the predicted number of proter and opisthe cells having ± 10% bias in mean inter-division rate with the combined proter or opisthe cell increase without a bias in mean inter-division time, assuming the 24 hours cell cycle for unbiased rate. When the com- bined number of proter and opisthe cells are plotted against each successive mean inter-division interval, it yields a logarithmic rate of increase at a GT = 21.6 hours. Likewise, if we had let p remain constant at 0.5 and in- stead let the value of q be 10% greater than the mean inter-division time, the combined number of proter and opisthe cells plotted at each successive mean inter-divi- sion interval yield a logarithmic rate of increase clone expanding at a GT = 28.8 hours. Another consequence of a continuing bias in mass of the anterior versus the pos- terior cell products is the alternating reappearance of the anterior product in the opisthe of the preceding genera- tion, perpetuating an alternating pattern of delays and advances in each succeeding clonal line as detailed be- low. 5.7. Dual Oscillator-Probabilistic Control of Cell Cycle Diffusive Coupling between Proter and Opisthe Cells that are in the G1 and G2 Phases in Epithelial Cell Colonies Here we propose that soon after division, daughter cells that have a cell mass 10% than the mean cell mass at division, termed the Proter, leave the limit cycle by a soft bifurcation undergo a 12 hour small amplitude os- cillation and return to the limiting trajectory of the large amplitude oscillation in the neighborhood of a G1 isoch- ron. In this second cycle, it traverses the S phase and then now becomes diffusively coupled to a cell in the G1 phase. As we indicated before, this coupling results in phase compromise between the G1 - G2 pair, in which the G2 phase cell is driven inside the oscillation to a sub-threshold G2 trajectory where it fails to trigger mito- sis/cell division. It now exits the limit cycle in the neighborhood of G1 isochrons that cause it to undergo a hard bifurcation. This drives it to an anti-limit cycle where it must cycle at least twice before gaining enough amplitude to renter the large amplitude oscillation. In this way a proter cells in cycle 1 is converted to an opis- the cell in cycle 3, and an opisthe cell in cycle 1 is con- verted to a proter cell in cycle 3. This pattern of clonal proliferation is illustrated by the dendrogram depicted in Figure 19. 5.8. Deterministic versus Stochastic Control of Cell Cycling or is it both? As we showed, certain aspects of the cell cycle such as G2 phase perturbation were well handled by the limit cycle dynamics while other aspects such as G1-phase duration seemed to be better accounted for by a probab- listic model. This subject has recently been modeled as a  J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 721721 Table 6. Probabilistic model prediction of clone size of cell lineages with examples of biased versus unbiased mean inter- division times. Successive Generationsa (n)Clone Sizea GTb Condition: 0 1 24 1 2 2 4 1. p = q = 0.5 3 8 0 1 21.6 1 2.1 2 4.4 2. p = 0.55, q = 0.5 3 10.2 0 1 19.2 1 2.2 2 4.8 3. p = 0.6, q = 0.5 3 12.3 aSuccessive generations are calculated from the terms of the binomial ex- pansion, (p + q)n 1. bGT, average generation time derived from semi-log plot of clone size versus mean inter-division time of unbiased cell lineages. d d d d d d d Figure 19. Hypothetical dendrogram illustrating diffusive cou- pling (d) between the proter and opisthe products of cell divi- sion which occur on a daily basis (days 1, 2 , 3, and 4) for each proter descendant, but only on days 3 and 4 for each of the opisthe descendants, resulting in a Fibonacci sequence of ac- cumulated cells (N) and an AvGt = 33.6 hours. complex network of regulatory protein (cyclin-depend- ent kinases and their activator and inhibitors [26], where by the stochastic fluctuations arise both from intrinsic molecular noise and extrinsic noise from unequal divi- sion. In particular, t the eukaryotic cell cycle is actually evolved from a prokaryotic deterministic G1-less cell cycle through acquisition of an environmentally-sen- sitive stochastic transitional phase inserted between cy- tokinesis and chromosome decondensation, i.e., a time- less phase in the cell cycle. Here, we propose that the epidermal keratinocyte cell cycle has both a determinis- tic portion and a stochastic portion. The former appears to be a fixed sequence of events that begins at the G1/S checkpoint in S and to terminate with mitosis and cyto- kinesis. We suggested earlier that this fixed sequence is underlain by a biochemical oscillator that triggers events during the S-G2/M phases in the acellular syncytial slime mold [10]. In mammalian cells, however, cytokinesis not only partitions the mother cell into two compartments, but results in a sharp phase discontinuity between sister cells, as well as halving the mass to DNA ratio, thus, severing the steady-state conditions upon which the ex- istence of limit cycle deterministic control system was predicated. The void in mitotic control may be just that. In its place we propose a stochastic system of control of G1 phase control in which a critical threshold of cell size/cell mass is required before reentry into the deter- ministic portion of the cell cycle. In effect upon cell di- vision, cells entering early G1 are faced with whims of environmental vicissitudes. Mammalian cells have “learned” to cope with this evolutionarily by inventing a series of environmental checkpoints that limit entry into the deterministic portion of the cell cycle. The plethora of decision points converge on that portion of the deter- ministic control system in the limit cycle dynamics where the trajectory naturally collapses in the G1 isochron re- gion of the limit cycle and where it may undergo transi- tion to an unstable singularity with a much diminished oscillation, or it but may rapidly return to the large am- plitude limit cycle upon stochastically attaining thresh- old cell size/cell mass. This situation appears to occur routinely for cells meeting all of the environmental con- tingencies, nutrients, growth factors, matrix support, but to be lacking in cells that sink deeper into G1/Go arrest, and cell death. Ultimately only continued cell cycling or initiation of a terminal cell differentiation program can assure cell survival. 6. DEDICATION AND ACKNOWLEDGEMENTS This paper is dedicated to Dr. Charles F. Ehret, who died February 24, 2007 at aged 83. He introduced me to the exotic yet ubiquitous world of chronobiology. Theoretical and experimental work was con- ducted on a biochemical oscillator model for the synchronous mitosis in Physarum polycephalum at the University of Chicago in collabora-  J. J. Wille / Natural Science 3 (2011) 702-722 Copyright © 2011 SciRes. OPEN ACCESS 722 tion with Dr. Stuart A. Kaufman and Dr. John J. Tyson. The initial laboratory work was undertaken at the Mayo Medical School in the laboratory of Dr. Robert E. Scott. I wish to thank Ms. Nell Swanson for her diligent technical assistance and my gratitude to Dr. Mark Pit- telkow of the Mayo Medical School for his invaluable scholarship. REFERENCES [1] Riley, P.A. and Hola M. (1980) Clonal differences in generation times of GPK epithelial cells in monolayer culture. Experimental Cell Biology, 48, 310-320. [2] Kitano, Y., Nagase, N., Okada, N. and Okano, M. (1982) Cinemicrograhic study of cell proliferation patterns and interdivision times of human keratinocytes in primary culture. Normal and Abnormal Epidermal Differentiation, University of Tokyo Press, Tokyo, 97-108. [3] Barrandon, Y. and Green, H. (1985) Cell size as a deter- minant of the clone-forming ability of human keratino- cytes. Proceedings of the National Academy of Sciences of the United States of America, 82, 5390-5394. doi:10.1073/pnas.82.16.5390 [4] Barrandon, Y. and Green H. (1987) Three clonal types of keratinocytes with different capacities for multiplication. Proceedings of the National Academy of Sciences of the United States of Amrica, 84, 2302-2306. doi:10.1073/pnas.84.8.2302 [5] Wille, J.J. (1999) Heterogeneity in clonal potential and life histories in individual keratinocytes propagated in serum-free medium. Journal of Inventigative Dermatol- ogy, 112, 573. [6] Wille, J.J., Pittelkow, M.R., Shipley, G.R. and Scott, R.E. (1984) Integrated control of growth and differentiation of normal human prokeratinocytes cultured in serum-free medium: clonal analyses, growth kinetics, and cell cycle studies. Journal of Celluar Physiology, 121, 31-44. doi:10.1002/jcp.1041210106 [7] Sheffey, C. and Wille, J.J. (1978) Cycloheximide-in- duced mitotic delay in Physarum polycephalum. Experi- mental Cell Research, 11 3, 259-262. doi:10.1016/0014-4827(78)90365-8 [8] Bass, J. and Takahashi, J.S. (2010) Circadian integration of metabolism and energetic. Science, 330, 1349-1354. doi:10.1126/science.1195027 [9] Thompson, D.W. (1943) On growth and form. Cam- bridge University press, New York. [10] Hofstadter, D.R. (1980) Godel, esher, bach: An eternal golden braid. Chapter V: Recursive Structures and Proc- esses, Vintage Books. A Division of Random House, New York, 131-137. [11] Kauffman, S.A. and Wille, J.J. (1975) The mitotic oscil- lator in Physarum Polycephalum. Journal Theory Biol- ogy, 55, 47-93. doi:10.1016/S0022-5193(75)80108-1. [12] Barnett, A. Ehret, C. and Wille, J.J. (1969) Testing the chronic theory of circadian timekeeping, Biochronometry, Proceeding of the Symposium on Biochronometry. Fri- day Harbor, Washington, D.C. [13] C.F. Ehret and E.Trucco E, (1967) “Molecular models for the circadian clock. I. the chronic concept”. Journal The- ory Biology, 15, 240-262. doi:10.1016/0022-5193(67)90206-8 [14] Glass, L. and Mackey, M. (1988) From Clocks to Chaos. The Rhythms of Life, Chapter 2. Princeton University Press, Princeton. [15] Tyson, J.J. and Novak, B. (2008) Temporal organization of the cell cycle. Current Biology, 18, R759-R768. doi:10.1016/j.cub.2008.07.001 [16] Novak, B. and Tyson, J.J. (2008) Design principles of biochemical oscillators. Nature Reviews, 9, 981-991. doi:10.1038/nrm2530 [17] Brooks, R.F. Bennett, D.C. and Smith, J.A. (1980) Mam- malian cell cycles need two random transitions. Cell, 19, 493-504. doi:10.1016/0092-8674(80)90524-3 [18] Wille, J.J. and Scott, R.E. (1984) Cell cycle-dependent integrated control of cell proliferation and differentiation in normal and neoplastic mammalian cells. Cell Cycle Clocks, New York, 433-453. [19] Tyson, J.J. and Hannsgen, K.B. (1985) The distributions of cell size and generation time in a model of the cell cy- cle incorporating size control and random transitions. Journal Theory Biology, 11 3, 29-62. doi:10.1016/S0022-5193(85)80074-6 [20] Kar, S. Baumann, W.T. Paul, M.R. and Tyson, J.J. (2009) Exploring the roles of noise in the eukaryotic cell cycle, http://www.pnas.org/cgi/doi/10.1073/pnas.0819934106 [21] Mackey, M.C. Santavy, M. and Selepova, P. (1986) A mitotic oscillator model for cell cycle with a strange at- tractor, Non-linear Oscillations in Biology and Chemistry. Springer-Verlag, Berlin, 354-454. [22] Schafer, E. and Cleffmann, G. (1982) Division and growth kinetics of the division mutant “conical” of Tetra- hymena, Experimental Cell Research, 137, 277-284. [23] Zhang, R. Aguila, D. Schneider, C. and Schneider, B.L. (2005) The importance of being big. Journal Invest Der- matol Symp Products, 10, 131-140. doi:10.1111/j.1087-0024.2005.200414.x [24] Mitchison, M. (1971) The Biology of the Cell Cycle. Cambridge University press, London. [25] Kubitschek, H.W.E. (1970) Evidence for the generality of linear cell growth. Journal Theoretic Biology, 28, 15-29. doi:10.1016/0022-5193(70)90061-5. [26] Baserga R., (1976) Multiplication and Division in Mam- malian cells. Marcel Dekker, New York.
|