 Circuits and Systems, 2016, 7, 701-708 Published Onlin e May 2016 in Sci Res. http://www.sc irp.org/journal/cs http://dx.doi.org/10.4236/cs.2016.76059 How to cite this paper: Prema, S. and Umamaheswari, P. (2016) Multitude Classifier Using Rough Set Jelinek Mercer Naïve Bayes for Disease Diagnosis. Circuit s and Syste ms, 7, 701-708. http://dx.doi.org/10.4236/cs.2016.76059 Multitude Classifier Using Rough Set Jelinek Mercer Naïve Bayes for Disease Diagnosis S. Prema1, P. Umamah eswari2 1Information and Communication Engineering, Anna Univer sit y, Chennai, India 2Comput er Science and Technology, MIT Campus, Anna Uni ver sity , Chennai, India Received 29 February 2016; accepted 9 May 2016; published 12 May 2016 Copyright © 2016 by authors and Scientific Resea rch Publishing Inc. This work is lic ensed under the Creati ve Commons Attribution International License (CC B Y). http://creativecommons.org/licenses/by/4.0/ Abstract Classificati on m odel has r eceived grea t atte nti on in any d om ain of rese a rch an d al so a reliable to ol for medic al dise ase diag nosis. The d om ai n of clas sific ation m odel is us ed in disease d iagnosis, disease predic tio n, bio i nform atics, c rime p redictio n and so on. H owever , an efficie nt diseas e di- agnosis model w as comp rom is ed the disease predictio n. In this paper, a Rough Set Rule-based Multitude Classifier (RS -RMC ) is dev eloped to improve the disease p redict ion r ate and enhance the cla ss accur acy of dise ase bei ng d iagnosed . Th e RS-RMC involve s two steps. I nitiall y, a Roug h Set mod el is used fo r F eat u re Sel ectio n ai ming at mi nimizin g th e execu tion tim e f or obt aining the disease fe atu re s et. A Multitude Classifier mod el is presented in second step fo r detecti on of he ar t disease and for efficient cl assification . The Naï ve Bayes Classifie r algo rith m is designed for effi- cient identification of clas ses to meas u re th e rel atio nship bet ween d isease fea tu res and i mproving disease prediction ra te. Ex p erimental an alysis sh ow s that RS -RMC is u sed to reduce the ex ecution time for ext rac tin g the d is ease fe ature w ith minimu m false posi tive r ate compared to th e sta te-of- the-art w orks . Keywords Classificati on M odel , Disea se Diagnosis, Rou gh Set M odel , Fe ature Sel ection , Multitude Classifier, Mercer Naïve 1. Introduction In a conventional classification model, the classification strategy identifies and selects the best classifier on the basis of experimental assessment with various individual classifiers. In a diversion from the conventional ap- proach, the use of Multitude Classifier System (MCS) has been presented as an alternative approach to improve classification accuracy of the disease being detected. 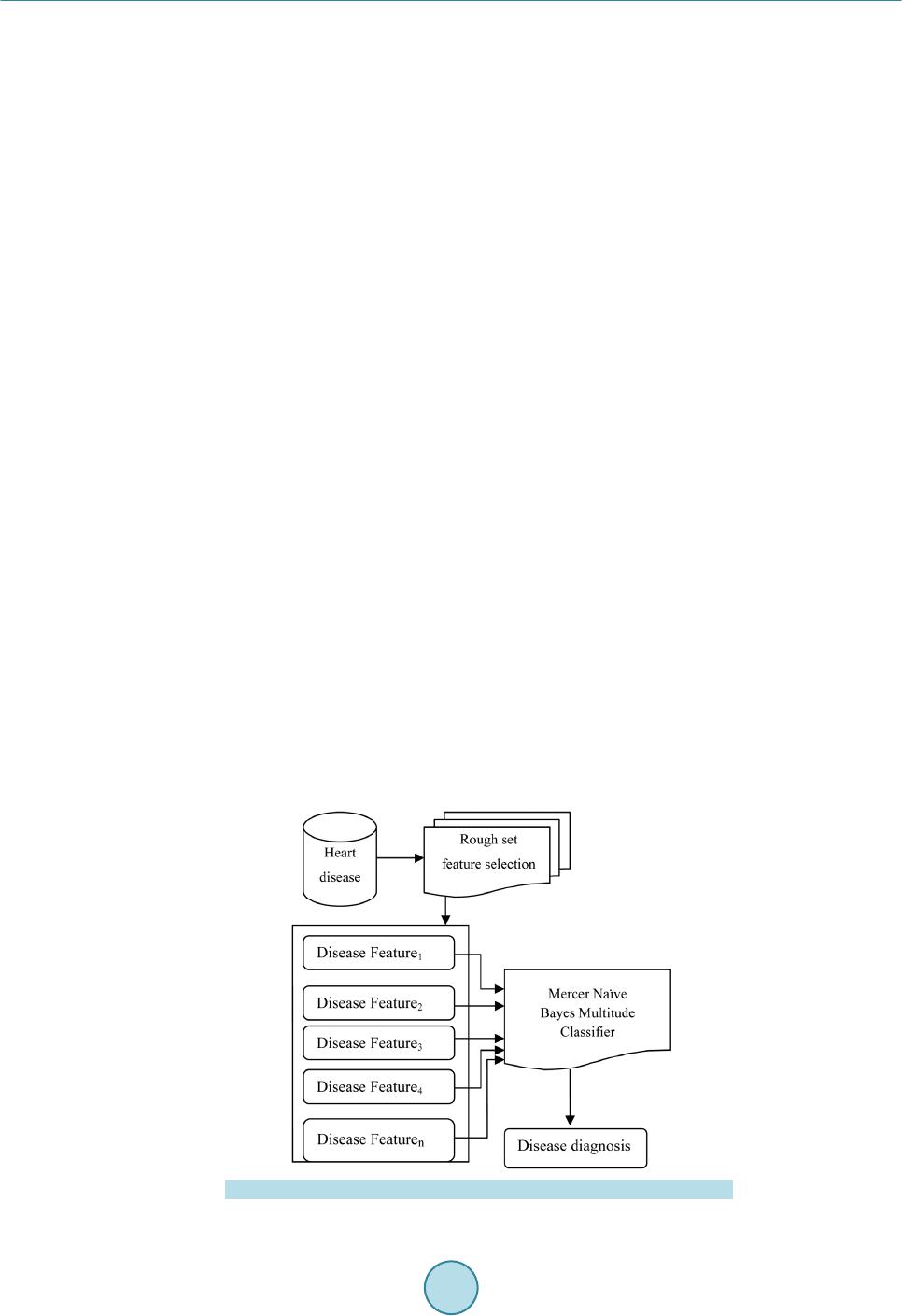 S. Prema, P. Umamaheswari Potential Management of Ventricular Arrhythmias (PPM-VA) [1] identified the stroke pattern and manage- ment of ventricular Arrhythmias that wer e largely related to stroke for the effective diagnosis of disease at an early stage. Prediction of Events using Spatio Spectro Temporal Data (PE-SST D) [2] provided with a case st udy on stroke resulting in the accuracy of the disease being diagnosed. However, both the above methods lack the class accuracy of disease diagnosis with the increase in the feature. To improve the detection rate of disease for Interstitial Lung Disease (ILD) in [3], a method was presented improving the detection of disease at an early stage. However, the classification accuracy remained unsolved. To address the issues related to class accuracy, Striatial Binding Ratio (SBR) was used in [4] to improve the class accuracy rate. A functional classification model [5] for early detection of heart failure using classification sche- ma was presented for improving class accuracy rate. Classification methods for bipolar disorders [6] were de- signed to improve diagnostic reliability. Another method using neural network and decision based support sys- tem was designed [7] to improve the classification accuracy rate. However, accuracy with respect to scalability remained unaddressed. An efficient classification approach using ANN and Feature Subset Selection [8] was presented to improve the accuracy of disease being detected. In [9], with the objective of i mproving classification rate, Diagnostic and Statistical Manual of Mental Disorders was presented. In accordance with the above-mentioned advantages of both disease classification and disease diagnosis, in this paper, a new framework called Rough Set Rule-based Multitude Classifier (RS-RMC) is proposed to in- crease the disease prediction rate and efficiency of the classification accuracy of disease being diagnosed. 2. Design of Rough Set Rule-Based Multitude Classifier To address the problem of disease diagnosis at an early stage, a framework is proposed based on Rough Set Rule -based Multitude Classifier. The Rough Set Rule-based Multitude Classifier uses disease features based on similar type of medical diseased data to identify the relationship between the disease features for efficient classi- fication. Figure 1 shows the b lock diagram of Rough Set Rule-based Multitude Classifier. As sho wn in Figure 1, Cleveland Heart Disease dataset extracted from UCI reposito ry is given as input. T he blo ck di agra m shows a two -stage process. In the first stage, Rough Set Feature Selection model is applied to the input dataset to extract the disease features. Feature Reduct applied in Rough Set model reduce the disease fea- ture without losing significant information. This is performed through lower and upper approximation without chan ging the values of d i s eas e features. The second stage goes through the Multitude Classifier model called as the Mercer Naïve Bayes Classifica- tion model. This is performed using Naïve Bayes Disease Classifier algorithm aiming to improving the class accuracy. Finally, the Jelinik Mercer Multitude Classifier is used as a smoothing technique for obtaining ap- pro xi mati on f unc t ion increasing the disease prediction rate. Figure 1 . Block diagram of rough set rule-based multitude classifier.  S. Prema, P. Umamaheswari 2.1. Construction of Rough Set (Model) Feature Selection (Minimize Execution Time) The first step in Roug h Set Rule-based Multit ude Classifier ( RS-RMC) fr amewor k is the const ructi on of Ro ugh Set Feature Selection (RSFC) to reduce the complexity by minimizing the redundant disease features contained within the set of feature patterns. The feature selection using Rough Set model minimizes the redundant disease features by selecting those significant disease features that are most essential for Multitude Classifier repre- sented in the pattern set. Let us consider “DFP” the set of all disease feature patterns, “F” the set of all disease features, then the value of disease feature “a” in disease feature pattern “P” is given as below (1) From (1), all disease features with disease feature patterns are identified. In order to reduce computational complexity and memory requirements, the disease feature patterns with the features identified is herein treated as a Rough Set model. The objective of using Rough Set model is to reduce the redundant features and therefore minimizes the execution time to obtain relevant features. With the reduced features, the indiscernible relationship for identifying the features is obtained. For any two disease patterns, in “DFP” indisce r nib le relatio n for disease features is given as below () ( ) ,,,for all ii faPfbPa P= ∈ (2) For example, given the disease features (Chest Pain, Blood Pressure, Heart rate), the indiscernible set are, “ () ()() ( ) { } 1326354 ,,,,,,PP PP PP P ”. The Rough Set Feature Selection in RS-RMC framework performs two op- erations called, lower and upper approximation to measure the significance of the disease feature from the dis- ease feature pattern set. Then, the lower and upper approximation is obtained as given below (3) ( ){} [ ] | R DFP AxDFPxA ′=∈⊆ ≠∅ (4) From (3) and (4), the disease features using lower approximation is identified, where “A” is t he set of dise ase feature patterns in “DFP” tha t are surely in “A”. On the o the r hand the upp er ap proxi mat ion with “A” a s the set of disease feature patterns in “DFP” i s probably in “A”. Finally, with the obj ective of reducing the disease fea- ture set without losing significant information, disease feature Reduct Set is applied to the resultant set. A dis- ease feature Reduct Set is then defined as a subset “P” of disease features (i.e., reduct disease features) “f” is given as be lo w, (5) By applying (5), the RS-RMC framework searches for disease feature reduct set of least cardinality. As a re- sult, RSFC preprocesses disease feature patterns without changing the values of the disease features, aiming at minimizing the execution time for obtaining the disease features. Using this resultant disease features relation- ship between features helps in the easy and early diagnosis of disease. 2.2. Jelinik Mercer Naïve Bayes Classifier The second step in Ro ugh Set Rule-based Multitude Classifier ( RS-RMC) fra me wor k is the effective identifica- tion of disease features relationship using Jelinik Mercer Naïve Bayes Classifier. With the objective of increas- ing the disease prediction rate, Jelinik Mercer Naïve Bayes Classifier is applied for disease diagnosis after mi- nimi zi ng the disease features. Jelinik Mercer Naïve Bayes Classifier for RS-RMC framework is based on the Bayes rules. The Bayes rules for Classifier a pplies the cond itional probability rule that measures the maximum likelihood of a property for the given d isease features. Let us consider a scenario with a patient observed to have disease with certain symptoms (i.e., features). In order to perform Multitude Classifier, Jelinik Mercer Naïve Bayes is applied to measure the relationship between the disease features and identify whether the disease being diagnosed is correct or not. The Naïve Bayes Classifier assumes that the presence of a specific disease feature is unrelated to the presence of any other disease feature. The Naïve Bayes Classifier then efficiently predicts that given the reduct disease  S. Prema, P. Umamaheswari features “f”, belongs to the class “ ” then there exists disease feature relationship between “ ” and “f” as given as be lo w ( ) ( ) i i i f PPcf cf cf Pf Pf ∗ = (6) where denotes the maximum posterior hypothesis for class “ ”. By applying the above formula, conditional probability of a disease pattern belonging to each disease is efficiently identifie d improvi ng the cla ss accuracy. Based on the conditional probability of disease pattern, the instance (i.e., feature) is classified as the class with the hi ghest conditional prob a bility. Figure 2 shows t he Naïve Ba yes Disease Classi fier algorit hm for efficie nt identificatio n of classes that helps in measuring the relationship between disease features. As a result, disease diagnosis is made in an efficient manner improving the disease prediction rate. Figure 2 shows the design of Naïve Bayes Disease Classifier al- gorithm. From the above algorithm, for each dataset, the features in the dataset are identified. Once the features are identified, the list of patients along with their associated classes is obtained. Based on the disease features, reduct disease features are identified to reduce the complexity without losing the values of disease feature. Fi- nally, maximum posterior hypothesis is applied to the reduct disease features for efficient disease diagnosis. 2.3. Jelinik Mercer Multitude Classifier Model The Jelinik Mercer in RS-RMC framework classifies Multitude Classifier for effective disease diagnosis. Jelinik Mercer Multitude Classifier is used as a smoothing technique to obtain an approximation funct ion for multit ude of disease features. It is formulated as given below () 1 ii f ff P PP cfcf cf ββ =−∗ + (7) From (7), “ ” represent the smoothened probability of a test, given the patient medical information with e xisti ng tests a nd “ ” ranges between “0” and “1”, “ ” representing the maximum li kelihood esti- mation in class feature “ ”. This in turn increases the disease prediction rate. Figure 2 . Naïve bayes diseas e classifier algorithm. 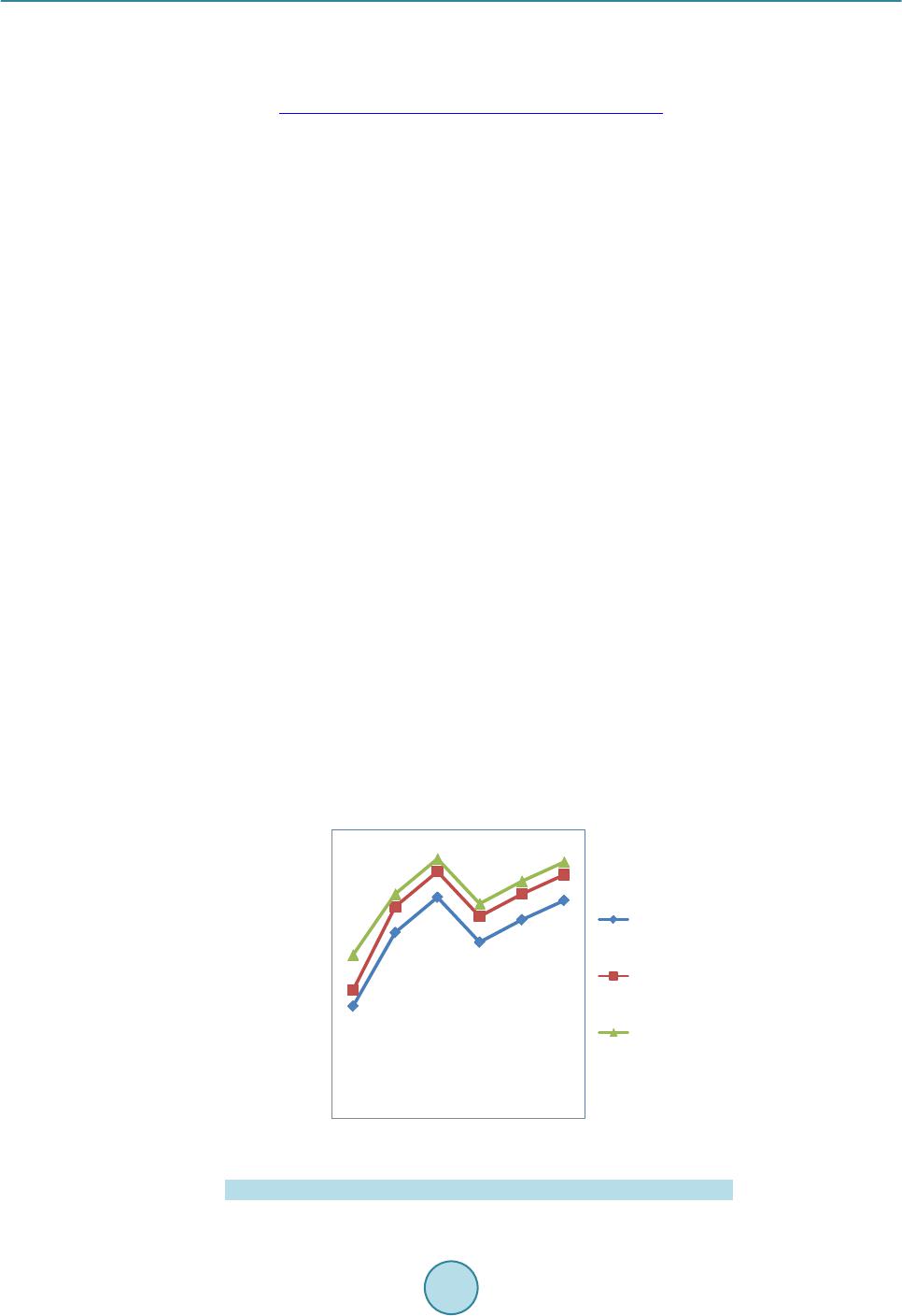 S. Prema, P. Umamaheswari 3. Experimental Settings The performance of the Rough Set Rule-based Multitude Classifier (RS-RMC) framework is experimented us- ing Cleveland Heart Disease Dataset extracted from UCI repository from Cleveland Clinic Foundation. Heart disease data set available at http://arc hive.ics.uci.ed u/ ml/datasets/heart+Disease [10]. The data set has 76 raw attributes. However, all of the published experiments only refer to 11 of them. The RS-RMC framework is si- mulated usin g MATLAB. The experimental work is compared against the existing Prevention and Potential Management of Ventricular Arrhythmias (PPM-VA) [1] and Prediction of Events using Spatio Spectro Temporal Data (PE-SSTD) [2] to identify the effectiveness of RS-RMC framework. The performance of the RS-RMC framework is measured in terms of disease prediction rate, execution time and false positive rate on effective disease diagnosis and class accuracy. 3.1. Execution Time The execution time is the time taken to obtain the disease feature set. It is expressed in terms o f milliseconds and is formulated as given below: (8) From (8), “F” denotes the features in the Cleveland Heart Disease dataset. Lower the time taken to execute, more efficient the method is said to b e In Figure 3, results are reported for various classification methods for Heart Disease Dataset. On classific a- tion using PPM-V A and P E-SSTD, the execution time for obtaining two features were observed to be 0.40 ms and 0.51 ms, whereas using RS-RMC, the execution time reduced to 0.35 ms. Figure 3 shows the time taken to obtain different features with differing sizes where features ranging from 2 to 13 were considered for experimental settings. As it can be seen the execution time steeply increases as the number of features increases, regardless of the method applied. This is because an increasing fraction of the features capacity is employed to obtain the features and diagnose disease at an early stage. For instance wit h fo ur fea tur e s (i. e., Chest Pain, Blood Pressure, Blood Sugar, Heart Rate), the execution time is 0.58 ms using RS-RMC framework, whereas 0.66 ms and 0.70 ms using the existing PPM-VA a nd P E -SSTD respectively. Moreover, the execution time is comparatively minimized using RS-RMC compared to the other methods which are demonstrated in Figure 3. This is because by applying Rough Set model that only selects those features that are highly essential and therefore reduces the redundant disease features. Therefore using RS-RMC the execution time for o btaining the features is reduced by 12.93% compared to PPM-VA. In a similar manner, by applying inter values to the disease features reduces the execution time by 31.45% compared to PE-SSTD. Figure 3 . Measure o f execution time. 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 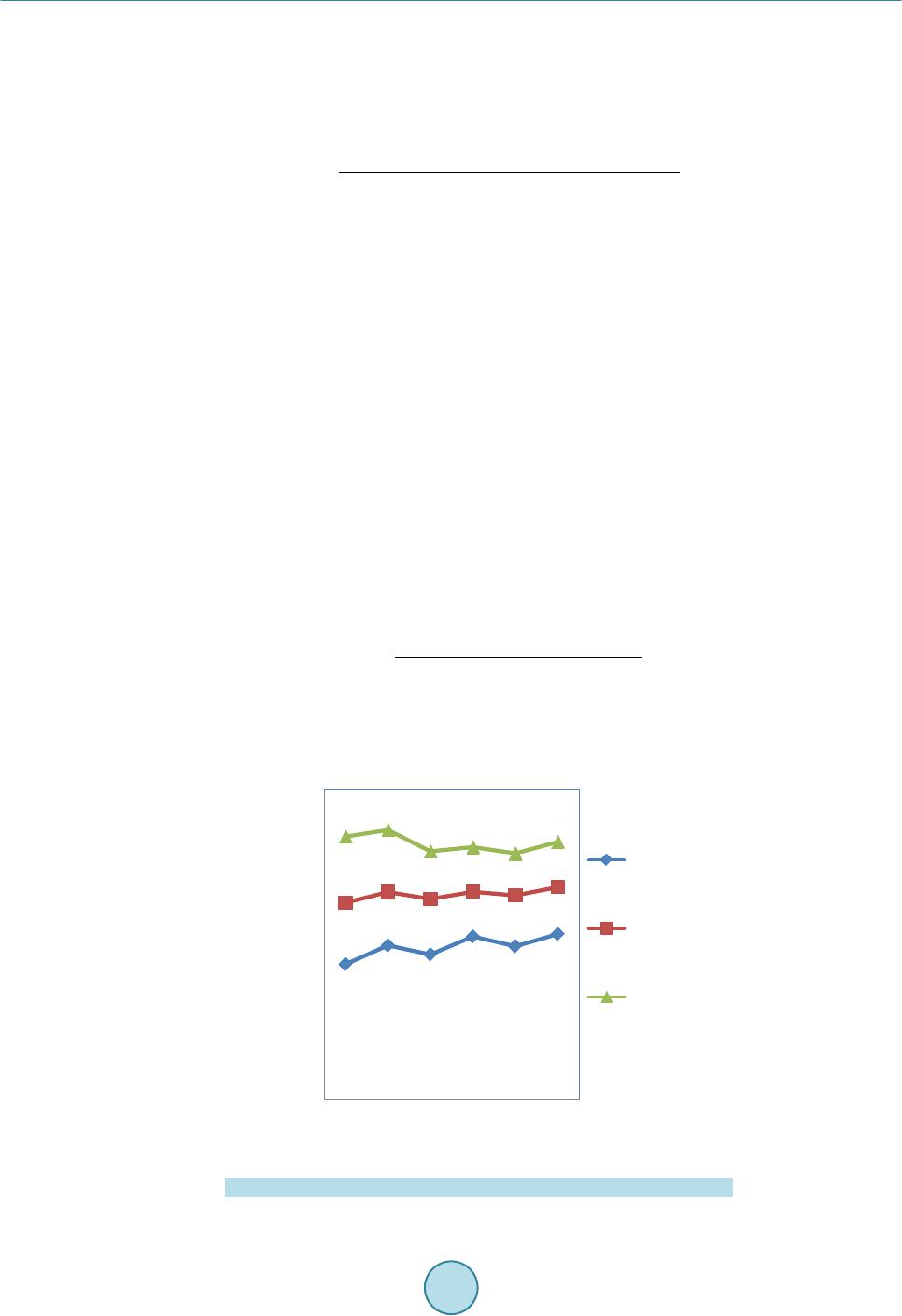 S. Prema, P. Umamaheswari 3.2. False Positive Rate The false posit ive rate on disease diagnosis is the ratio o f absent e vents (i.e. , disea se diagnosis) that yield posi- tive test (i.e. , identified as disease though not) outcomes. Therefore, False Positive Rate (FPR) is the ratio of number of false positives to the total patients for conducting experiments. The mathematical formulation for FPR i s given be l ow ( ) ( ) No of false positivesidentified with disease FPR Total patientsdiagnosed with disease = (9) From (9), denotes the false positive rate. Lower the false positive rate, more efficient the method is said to be and is measured in terms of percentage (%). Figure 4 shows the false positive rate under different simulation setting. The experiments were conducted with different number of patients and the FPR for the correspo nding was measured. From Figure 4 we can see that the value of FPR is comparatively lower in RS-RMC than the other two methods PPM-VA and PE-SST D. Figure 4 illustrates the i mpact of false positi ve rate and compared with t wo state-of-the-art works for 30 pa- tients. Figure 4 compares all the performance improvement based on the false positives provided by the three methods. As the number of patient increases, small rise and small fall off value is recorded irrespective of the methods used. But, RS-RMC recorded low FPR with the application of Feature Reduct in Rough Set model. By applying the Feature Reduct in Rough Set model, false positive rate for disease feature identification is reduced and therefore the disease prediction rate is also minimized. In addition by applying the Feature Reduct disease feat ure set is red uced witho ut losi ng the infor matio n. Thi s sub seque ntly he lps i n reduc ing t he false posit ive ra te by 36.48% compared to PPM-VA and 74.54% compared to PE-SSTD respectively. 3.3. Disease Prediction Rate Disease prediction rate measures the rate of disease being predicted correctly without any assumption. The Dis- ease Prediction Rate (DPR) is the ratio of successful prediction of disease to the total number of patients and is given as be lo w. Successfully predicted as disease DPR Total patients = (10) The performance of the different disease diagnosis method for Cleveland Heart Disease dataset is shown in Figure 5. It is observed that by applying RS-RMC, the disease prediction rate is increased by 6.41% to 30.64% with that of PPM-VA and PE-SST D. Figure 4 . Measure o f false positive rate with respect to patients. 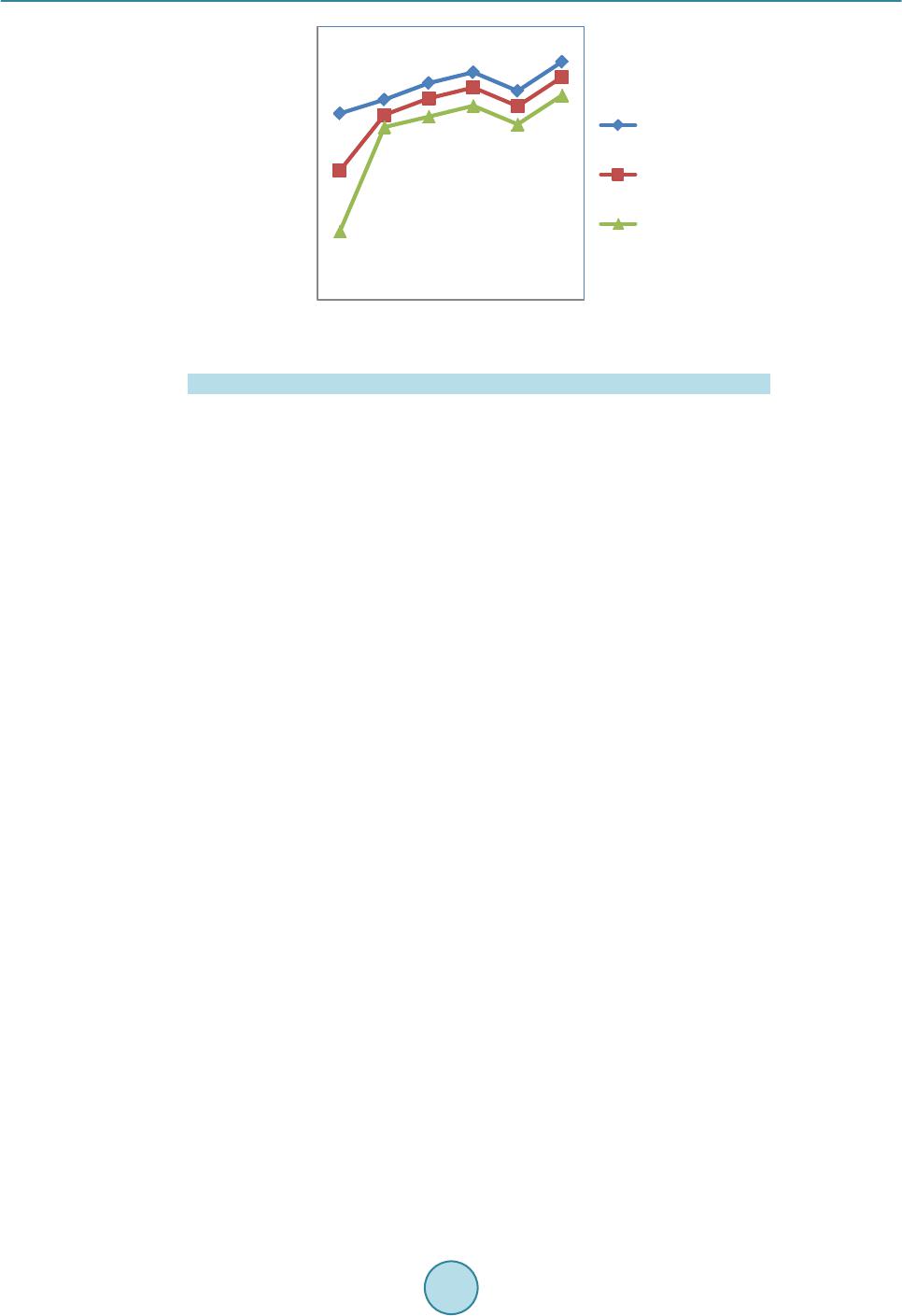 S. Prema, P. Umamaheswari Figure 5 . Measure o f d isease pred iction rate. Figure 5 shows the comparison of the disease prediction rate of traces with number of patients ranging from 5 to 30 and applied in Matlab. Five features were considered for obtaining the disease prediction using the three methods RS-RMC, PPM-VA and PE-SSTD respectively. From the figure, the value of disease prediction rate achieved using the proposed RS-RMC framework is higher when compared to two other existing techniques namely, PPM-VA [1] and P E-SSTD [2]. Besides we can also observe that by increasing the number of patients who provide their disease features, the disease prediction rate is increased using all the methods. But compara- tively, it is higher in RS -RMC framework because the relationship between disease features is efficiently identi- fied using Naïve Bayes Classifier algorithm. By applying Naïve Bayes Classifier algorithm, e fficient identifica- tion of classes through maximum posterior hypothesis is evaluated that helps in measuring the relationship be- tween disease features and significantly improves the disease prediction rate by 18.52% and 38.74% compared to PPM-VA a n d PE-SSTD respectively. 3.4. Classification Accuracy Figure 6 shows the classification accuracy using RS-RM, PPM-VA and PE-SSTD respectively. To extract the classification accuracy, 30 patients with 20 female and 10 male patients in the age group of 40 - 55 years were considered. Figure 6 sh ows the classif ication accuracy recorded using the three methods RS-RMC, PPM-VA and PE-SSTD. From the fig ur e it is illustrative that the classification accuracy is improved in the prop osed RS-RM C fr amework compared to two other methods. RS-RMC offers an improved disease diagnosis model by increasing disease prediction rate and tru e positiv e rate an d decreasin g t he f alse positi v e rate f or disease diag n osis m odel. Unl ike th e existing methods, RS-RMC used Jelinik Mercer where relationship between the features are efficiently identified and obtain approximation function for multitude of disease features. The RS-RMC framework i mprove i t s clas- sification accuracy by reducing the execution time for obtaining the features by 55.51% and by handling over half of its disease feature set in effective disease diagnosis. 4. Conclusion In this paper, we considered the design of a Multitude Classifier Disease Diagnosis framework to improve dis- ease prediction rate and class accuracy in the field of medical domain is presented. A Multitude Classifier framework is introduced, and considered the problem of efficient disease diagnosis in that framework. The RS-RMC fr ame work o ffers le ss fa lse po sitive r ate with lesser execution time using Rough Set Feature Selection and Feature Reduct model. Analysis of disease prediction rate demonstrates that RS-RMC framework provides higher heart disease prediction rate with the aid of Naïve Bayes Classifier algorithm. Finally, Jelinik Mercer in RS-RMC framework significantly classifies Multitude Classifier using approximation function for multitude of disease features. The performance of RS-RMC framework was compared to other disease diagnosis model, Disease Prediction Rate (%)  S. Prema, P. Umamaheswari Figure 6 . Measure of classificat ion accurac y. PPM-VA and PE-SSTD respectively. We compared the performance with many different system parameters, and evaluated the performance in terms of different metrics, such as execution time, disease prediction rate, false positive rate and classification accuracy. The results show that RS-RMC framework offers better performance with an improvement of classification accuracy by 55.51% and disease prediction rate by 28.63% compared to PPM-VA and PE-SSTD respectively. References [1] Koppikar, S., Baranch uk, A., Guzmán , J.C. and Morillo, C.A. (2013) Stro ke and Ventricular Arrhythmias. Internation- al Journ al of Cardiology, Elsevier, 7. [2] Kasab ov, N., Feigin, V., Hou, Z.-G., Chen, Y.X. , Li an g , L., Krishnamurthi, R., Othman, M. and P armar, P. (2014) Evol vin g Spi kin g Neural Ne t wor ks for P erson alised Modelling, Classification and Pred iction of Spatio-Temporal Pat- terns with a Case Study on Stro ke. Neu ro Co mpu ti ng, El sevier, V ol. 134, 269-27 9. [3] Meyer, K.C. (2014) Diagnosis and Man agement of Interstitial Lung Disease. Springer Open Journal. [4] Prashanth, R., Roy, S.D., Mandal, P.K. and Ghosh, S. (2014) Automatic Classification and P red iction M od els for Early Par kin son ’s Disease Diagnosis fro m SPECT Imag ing. Exp ert S ystems with Applications, Elsevier, 41, 33 33 -3342. http://dx.doi.org/10.1016/j.eswa.2013.11.031 [5] Ch awla, L.S., Herzog, C.A., Costanzo, M.R., Tumlin, J., Kellum, J.A., McCullough, P.A. and Ronco, C. (2014) Pro- posal for a Funct ion al Classification System of Heart Failure in Patients with End-Stage Ren al Disease. Journal of the American College of Cardiology, Elsevier, 63, 1246-1252. http://dx.doi.org/10.1016/j.jacc.2014.01.020 [6] de Dios, C., Goikol ea, J.M., Colomb, F., Morenoc, C. and Vietab, E. (20 14) Bipolar Disorders in the New DSM-5 and ICD-11 Classifications. Elsevier, 7, 179-185. [7] Ghwa nme h, S., Mohammad, A. and Al-Ibrahim, A. (2013) Innovati ve Artificial Neural Netwo rks -Based Decisio n Support System for H eart Diseas e s Diagnosi s. Journal of Intelligent Learning Systems and Applications, 5, 176-183. http://dx.doi.org/10.4236/jilsa.2013.53019 [8] Jabb ar, M.A., Deekshatu lu, B.L. an d Chandra, P. (2013) Classification of Heart Disease Using Artificial Neural Net- wor k and Featur e Subset Selection . Global Journal of Computer Science and Technology Neural & Artificial Intelli- gence, 13 , 5-14 . [9] Hecker s, S., Barch , D.M., Bustillo, J., Gaebel, W., Gur, R., Malaspin a, D., Owen , M.J., S chultz, S., Tandon, R., Tsuang, M., Van Os, J. and Carpenter, W. (2013) Structure of the Psychotic Disorders Classification in DSM 5. Sch i- zophrenia Research, Elsevier . [10] Frank, A. and Asuncion, A. (2010) UCI Machine Learning Repository. Universit y of Californ ia, School of Information and Computer Science, Irvine. http://archive.ics.uci.edu/ml
|