 America n Journal of Analy tic al Chemistry, 2011, 2, 392-400 doi:10.4236 /ajac.2011.23048 Published Online July 2011 (http://www.scirp.org/journal/ajac) Copyright © 2011 SciRes. AJAC Comparison Between DNA Immobilization Techniques on a Redox Polym er Mat ri x Vijayalakshmi Velusamy1, Khalil Arshak1*, Catherine F. Yang2, Lei Yu2, Olga Korostynska3, Catherine Adley4 1Electronic and Computer Engineering Department, University of Limerick, Limerick, Irelan d 2Department of Chemistry and Biochemistry, Rowan University, Glassboro, USA 3School of Physi cs , Dublin Institute of Technology, Dublin, Ireland 4Department of Chemical and Environmental Sciences, University of Limerick, Limerick, Ireland E-mail: viji.su b h a@ul.ie Received February16, 2011; revised April 2, 2011; accepted April 16, 2011 Abstract In this paper we report a label-free detection method of unmodified DNA using polypyrrole as a n immobili- zation matrix by impedance measurement. A probe and a target complementary DNA sequence specific for the bacterial pathogen, Bacillus cereus are used. Impedance measurements are performed without using ad- ditional redox probes. The effects of hybridization and non-specific binding are compared when the Probe DNA molecules were immobilized by two different methods: electrochemical adsorption and entrapment. The probe DNA immobilized using electrochemical adsorption yielded better hybridization signals com- pared to that immobilized using the entrapment method. Control experiments were also performed to prove the specificity of the biosensor in the presence of non complementary DNA. Negligible unspecific binding with the immobilized probe was observed with the electrochemically adsorbed probe, whereas the entrapped probe responded to the non complementary target. The performance of the DNA sensor was characterized using both cyclic voltammetry and impedance spectroscopy techniques and proved to be effective in terms of specificity of hybridization events. Keywords: Immobilization, Label Free DNA, Polypyrrole 1. Introduction Biomolecules of interest called bioreceptors/biorecog- nition elements ca n be generally classified into six diffe- rent major categories. These categories include antibody /antigen, e nzymes, nucleic acids/DNA, cellular structures /cells, biomimetic and bacteriophage (phage). Among other biorecognition elements, DNA target has received considerable attention since each organis m ha s its uniq ue DNA sequences and any self-replicating micro-organism can be easily identified. Biosensors based on DNA as bioreco gnition element ar e simple, r apid, and hig hly spe- cific hence widely used in pathogen detection. In co ntra st to enzyme or antibodies bioreceptors, nucleic acid rec- ognition layers can be readily synthesized and regene- rated. However, there are factors which p lay ke y role s in the design of DNA biosensors, 1) The immobilization matrix 2) The method of immobilization and 3) The de- tection technique. The choice of suitable substrate/matrix to immobilize the DNA is a significant factor since the immobilized single stranded ss-DNA has to be stable on the attached surface to facilitate the hybridization event. Various ma- terials being used to modify the electrode (Au, Ag, Pt, Glassy Carbon, and Indium tin oxide (ITO)) surface and include carbon nanotubes, conducting polymers, metal nanoparticles, composites of various electroactive mate- rials. Conducting polymers (CPs) are preferred among those materials not only because they can be synthesized easily but also due to their high stability, excellent elec- trical properties and suitable to immobilize the bio- molecules [1,2]. Among the various CPs polyaniline, polythiophene, and polypyrrole are biocompatible [3-8] —however, polypyrrole (PPy) is used mostly in biosen- sors and immunosensors because of its biocompatibility and the ease of immobilization of various biologically active compounds [9]. To detect bio-analytes at a phy- siological pH, b iosensing mat erials must be electroactive  V. VELUSAMY ET AL. Copyright © 2011 SciRes. AJAC 393 in neutral environments, unlike polyaniline and poly- thiophene. To overcome this problem, PPy is attractive because it can be more easily deposited from neutral pH aqueous solutions of pyrrole monomers [10]. Most prac- tical types of PPy have conductivity in the range of 1 - 100 S/cm [11]. PPy can be prepared by various methods such as chemical polymerization [12-16], electrochemi- cal polymerization [17,18], spreading [19,20], vapour phase polymerization [21], plasma polymerization [22,23]. The electrochemical polymerization of pyrrole has been extensively studied as it is simple and reliable procedure which does not need sophisticated laboratory equipme nts and reagent s . One of the critical factors in biosensor design is the develop ment of im mobiliza tion methodolo gy that strong- ly stabilizes the DNA on the transducer surface. Integra- tion of the DNA with the signal transducer is mostly achieved by immobilizing ss-DNA on the electrode/mo- dified surface. Different mechanisms of immobilization techniques are shown in F igure 1. Mechanisms of immobilization of a label-free umodi- fied DNA probe on the conducting polymer modified surface can be divided into two major categories: i) Ad- sorption and ii) Entrapment. While immobilization matrix and method are impor- tant in DNA sensor design, the choice of a suitable de- tection technique is also of great significance in deter- mination of overall performance of the DNA biosensor, in particular with respect to the immobilization and hy- bridization efficiency of the DNA. The detection tech- niques employed in the DNA biosensors can be optical [24-27], electrochemical [28-30] or mass [31,32] based. Depending on the nature of the target, the detection pa- rameters can be drawn from the detection of the specific sequence to confirm the presence of target micro-organ- isms [33,34], DNA damage [35,36], presence of chemi- cal [37] and biological compounds [38], presence of toxic [39] and genotoxic compound s [40]. In general, compared to the other detection techniques, the electrochemical based detection method is more ad Figure 1 . Mechanism of immobilization. vantageous, not only because it is simple, cost effective and reproducible but also due to its suitability for minia- turized real-time handheld use without sacrificing its sensitivity and specificity. In electrochemical DNA bio- sensors, detection is based on the variation in the electri- cal properties of the DNA- modified electrode before and after hybridization. It is likely that the change may have resulted from the change of double-layer capacitance, heterogeneous electron transfer resistance, impedance or current. DNA sensors based on electrochemical imped- ance spectroscopy (EIS) detection is a device that tran- scribe the changes in interfacial properties between the electrode and the electrolyte induced by DNA hybridiza- tion, DNA conformational changes, or DNA damages to an electrical signal [41]. EI S is becoming more and more popular for electrochemical measurements of molecular interactions and is also widely used in a variety of bio- sensing applications. The principle and applications of EIS to biosensing have been recently summarized in the literature [42]. The method provides unique advantages compared to other electrochemica l me t ho ds, such a s hi g h sensitivity, ease of signal quantification, and ability to separate the surface binding events from the solution impedance [43]. Impedance data are normally recorded in a range of frequencies, using alternating current of small amplitude, thus the EIS is often referred to as AC Impedance. Compared to other electrochemical methods, such as cyclic voltammetry (CV) or differential pulse voltammetry (DPV), known to characterize molecular interactions on the surface of electrodes, AC Impedance is less destructive to the measured biological interac tions because it is performed in a very narrow range of small potentials [43]. To cha racterize the DNA immobilized on the modified electrode surface using EIS, the change in impedimetric response upon hybridization can be meas- ured either in terms of kinetics of electron transfer proc- ess by Faradic impedance measurements or in terms of alterations of capacitance and molecular layer organiza- tion, originating from biorecognition events, by non- Faradic approach [44]. In this paper, we report the label-free detection of DNA hybridization using Faradic impedance measure- ment s . The PPy modified gold electrode surface was employed to optimize the immobilization of probe DNA molecules and to resist the non spe cific bindi ng of target DNA molecules. Further, electrochemical adsorption and entrapment methods of immobilization are compared with respect to hybridization with target complementary and non-c omple menta ry DNA. Here, the present applica- tion is specifically applied to food quality monitoring emphasizing on the detection of the bacterial pathogen, Bacillus cereus.  V. VELUSAMY ET AL. Copyright © 2011 SciRes. AJAC 394 Table 1 . DNA sequences. Functio n Seque nce Probe (5’-3’) A TC GCC TCG TTG GAT GAC GA Complementary (5’-3’) TCG TCA TCC AAC GAG GC G AT Non-Complementary (5’-3’) AAA ATC GAT GGT AAA GGT TGG 2. Experimental 2.1. Material s Pyrro le a nd M gCl2 solution were purchased from Sig ma- Aldrich, USA. All the reagents were analytical grade and used without further purification. Specific sequences for the Bacillus cereus were designed in our laboratory [45] and its synthetic form was purchased from Integrated DNA technologies, USA. All stock DNA solutions were prepared using distilled water and a concentration of 1 µg/µl from the stock solution was made using 10mM Tris-HCl buffer, pH 7.2. The capture probe and the target probe are 20-mer oligonucleotides and the non comple- mentary probe 21-mer in length. Table 1 shows the se- quences of the oligonucleotides used in this work. 2.2. Instrumentation A three-electrode cell comprising—a gold (Au) working electrode (2-mm diameter), a platinum wire counter electrode and a Ag/AgCl reference electrode were used. All electrochemical measurements were carried out using Autolab PGSTAT302N (Eco Chemie, The Netherlands) with a Frequency Response Analyser (FRA) module in- terfaced for impedance measure ments. 2.3. Gold Electrode Surface Modification and Electrochemical Synthesis of Polypyrrole The Au electrode surface was polished to mirror finish prior to use sequentially with 1, 0.3, and 0.05 μm α- Al2O3 paste, and rigorously rinsed with distilled water following each polish. Prior to surface modification, the bare electrode was scanned in 0.1M MgCl2 in 10 mM Tris-HCl buffer (pH 7.2) between –0.3 and 0.8 V until a reproducible cyclic voltammogram was obtained. Two different types of methods were employed for immobili- zation of probe DNA. The first approach involves incorporation of probe DNA into the polymer matrix during the growth of the PPy or during the co-deposition of the pyrrole monomer which is known as the entrapment method. The second approach involves the polymerization of the polypyrrole without the presence of DNA and then i mmobilizing the probe DNA by electro chemical adsorption. 2.4. Immobilization 2.4.1. Immobilization by Entrapment Immobilization by co-deposition was achieved using cyclic volta mmetry (CV) for 20 cycles over –0.3 to +0.8 V at a scanning rate of 50 mVs−1. The 2ml solution for immobilization by co-deposition consists of 0.1M Pyr- role/0. 1 M MgCl2 so lution and 1 µg of probe DNA. 2.4.2. Immobilization by Electrochemical Adsorption First, the electrochemical polymerization of PPy was performed by CV, involved the immersion of an Au disk electrode into a solution of 0.1 M pyrrole containing 0.1 M MgCl2. A cyclic potential from –0.3 to 0.80 V (versus Ag/AgCl) for 20 cycles at a scan rate of 50 mVs−1 was applied. The Au electrode coated with PPy film was then washed with di stilled water a nd d ried under nitroge n gas . The so-prepared PPy-coated electrode was characterized by impedance spectroscopy and CV. Prior to immobilization, the PPy coated electrode was transferred to a 20mM Mg Cl2 and 10 mM Tris-HCl buffer (pH 7.2) for electrochemical oxidation of the PPy layer at 0.5 V (versus Ag/AgCl) for 600 s. Immobiliza- tion was thereafter achieved by applying a constant po- tential of 0.8 V for 600 s and the 2 ml immobilization solution consisted of 20 mM MgCl2 in10 mM Tris-HCl buffer (pH 7.2) and 1 µg of probe DNA. The ss-DNA modified electrode was washed with distilled water to remove loosely adsorbed DNA and dried under nitrogen gas. T he ss-DNA/PP y film was later analysed by imped- ance measurements. 2.5. Hybridization of the Target DNA The hybridization was completed by applying a constant potential of 0.5 V for 600 s and the 2 ml hybridization solution consists of 20 mM MgCl2 in 10 mM Tris-HCl buffer (pH 7.2) and 1 µg of complementary DNA strand. The same procedure was repeated with non-complementary DNA as a co ntrol experiment. 2.6. Characterization of the DNA Bio sensor Both the EIS and CV techniques are used to characterize the performance of the DNA biosensor. All impedance measurements were performed at 0.3 V bias potential in an analysis buffer consisting of 0.1 M MgCl2 in10 mM Tris-HCl buffer, with a pH of 7.2. AC amplitude of 5 mV was used and the data collected in the frequency 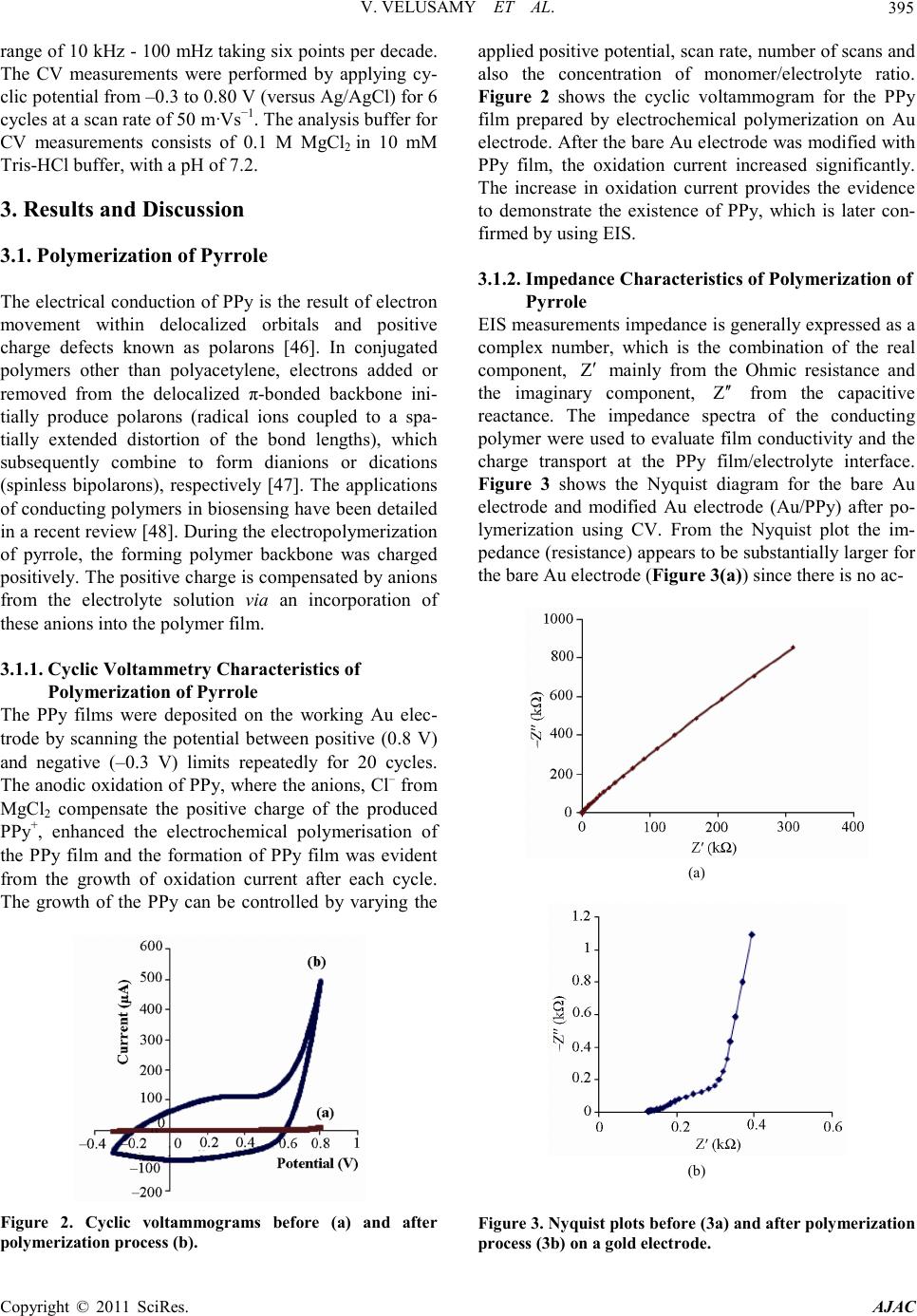 V. VELUSAMY ET AL. Copyright © 2011 SciRes. AJAC 395 range of 10 kH z - 100 mHz taking six points per decade. The CV measurements were performed by applying cy- clic potential fro m –0.3 to 0.80 V (versus Ag/AgCl) for 6 cycles at a scan rate of 50 m·Vs−1. The analysis bu ffer for CV measurements consists of 0.1 M MgCl2 in 10 mM Tris-HCl buffer, with a pH of 7.2. 3. Results and Discussion 3.1. Polymerization of Pyrrole The electrical conduction of PPy is the result of electron movement within delocalized orbitals and positive charge defects known as polarons [46]. In conjugated polymers other than polyacetylene, electrons added or removed from the delocalized π-bonded backbone ini- tially produce polarons (radical ions coupled to a spa- tially extended distortion of the bond lengths), which subsequently combine to form dianions or dications (spinless bipolarons), respectively [47]. The applications of co nducting polymers in biosensing have been detailed in a recent review [48]. During the electrop olymerizatio n of pyrrole, the forming polymer backbone was charged positively. The positive charge is compensated by anions from the electrolyte solution via an incorporation of these anions int o the po lymer film. 3.1.1. Cyclic Voltammetry Characteristics of Polymerization of Pyrrole The PPy films were deposited on the working Au elec- trode by scanning the potential between positive (0.8 V) and negative (–0.3 V) limits repeatedly for 20 cycles. The anodic oxidation of PPy, where the anions, Cl– from MgCl2 compensate the positive charge of the produced PPy+, enhanced the electrochemical polymerisation of the PPy film and the formation of PPy film was evident from the growth of oxidation current after each cycle. The growth of the PPy can be controlled by varying the Figure 2. Cyclic voltammograms before (a) and after polymerization process (b). applied positive potential, scan rate , number of scans and also the concentration of monomer/electrolyte ratio. Figure 2 shows the cyclic voltammogram for the PPy film prepared by electrochemical polymerization on Au electrode. After the bare Au electrode was modified with PPy film, the oxidation current increased significantly. The increase in oxidation current provides the evidence to demonstrate the existence of PPy, which is later con- firmed b y us ing EIS. 3.1.2. Impedance Characteristics of Polymerization of Pyrrole EIS measurements impedance is generally expressed as a complex number, which is the combination of the real component, mainly from the Ohmic resistance and the imaginary component, from the capacitive reactance. The impedance spectra of the conducting polymer were used to evaluate film conductivity and the charge transport at the PPy film/electrolyte interface. Figure 3 shows the Nyquist diagram for the bare Au electrode and modified Au electrode (Au/PPy) after po- lymerization using CV. From the Nyquist plot the im- pedance (resistance) appears to be substantially larger for the bare Au electrode (Figure 3(a)) since there is no ac- (a) (b) Figure 3 . Nyquist pl ots befo re ( 3 a) and aft er pol y merizati on process (3b) on a gold electrode. 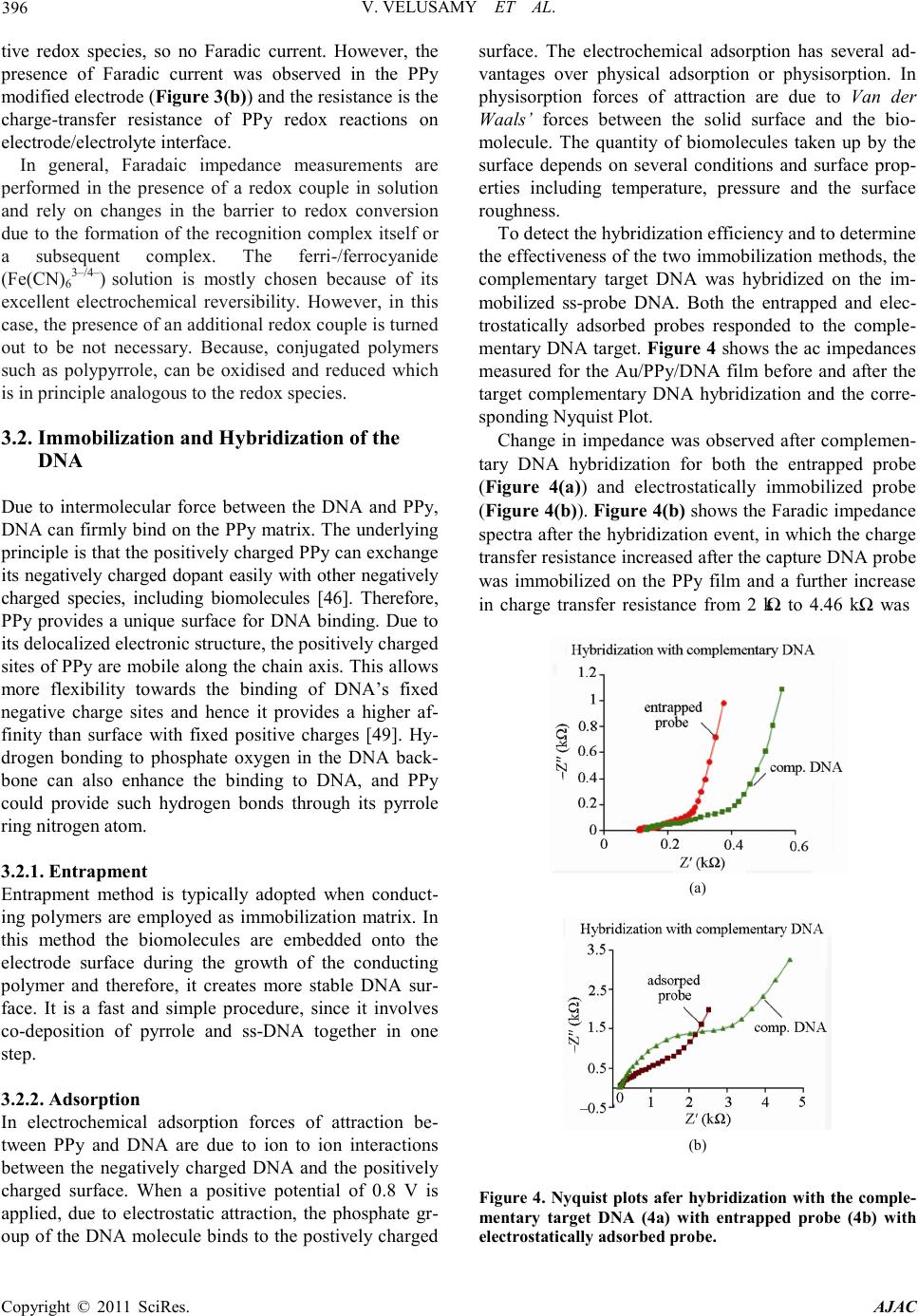 V. VELUSAMY ET AL. Copyright © 2011 SciRes. AJAC 396 tive redox species, so no Faradic current. However, the presence of Faradic current was observed in the PPy modified electrode (Figure 3(b)) and the resistance is the charge-transfer resistance of PPy redox reactions on electrode/electrolyte interface. In general, Faradaic impedance measurements are performed in the presence of a redox couple in solution and rely on changes in the barrier to redox conversion due to the formation of the recognition co mplex itself or a subsequent complex. The ferri-/ferrocyanide (Fe(CN)63–/4–) solution is mostly chosen because of its excellent electrochemical reversibility. However, in this case, the presence of an additional redox couple is turned out to be not necessary. Because, conjugated polymers such as polypyrrole, can be oxidised and reduced which is in principle a nalogous to the redox species. 3.2. Immobilization and Hybridization of the DNA Due to intermolecular force between the DNA and PPy, DN A can fir mly bind on the PPy matr ix. The underlying principle is t hat the p ositi vel y cha rged PP y can e xc hang e its negatively charged dopant easily with other negatively charged species, including biomolecules [46]. Therefore, PPy provides a unique surface for DNA binding. Due to its delocalize d electronic structure, the positively charged sites of PPy are mobile along the chai n axis. T his allo ws more flexibility towards the binding of DNA’s fixed negative charge sites and hence it provides a higher af- finity than surface with fixed positive charges [49]. Hy- drogen bonding to phosphate oxygen in the DNA back- bone can also enhance the binding to DNA, and PPy could provide such hydrogen bonds through its pyrrole ring nitro gen at om. 3.2.1. Entrapment Entrapment method is typically adopted when conduct- ing polymers are employed as immobilization matrix. In this method the biomolecules are embedded onto the electrode surface during the growth of the conducting polymer and therefore, it creates more stable DNA sur- face. It is a fast and simple procedure, since it involves co-deposition of pyrrole and ss-DNA together in one step. 3.2.2. Adsorption In electrochemical adsorption forces of attraction be- tween PPy and DNA are due to ion to ion interactions between the negatively charged DNA and the positively charged surface. When a positive potential of 0.8 V is applied, due to electrostatic attraction, the phosphate gr- oup of the DNA molecule binds to the postively charged surface. The electrochemical adsorption has several ad- vantages over physical adsorption or physisorption. In physisorption forces of attraction are due to Van der Waals’ forces between the solid surface and the bio- molecule. The quantity of biomolecules taken up by the surface depends on several conditions and surface prop- erties including temperature, pressure and the surface roughness. To detec t the hybridizatio n efficiency and to determine the ef fective ness of the t wo immobiliza tion methods, the complementary target DNA was hybridized on the im- mobilized ss-probe DNA. Both the entrapped and elec- trostatically adsorbed probes responded to the comple- mentary DNA target. Figure 4 shows the ac impedances measured for the Au/PPy/DNA film before and after the target complementary DNA hybridization and the corre- sponding Nyquist Plot. Change in impedance was observed after complemen- tary DNA hybridization for both the entrapped probe (Figure 4(a)) and electrostatically immobilized probe (Figure 4(b)). Figure 4(b) shows the Faradic impedance spectra after the hybridization event, in which the charge transfer resistance increased after the capture DNA probe was immobilized on the PPy film and a further increase in charge transfer resistance from 2 kΩ to 4.46 kΩ was (a) (b) Figure 4. Nyquist plots afer hybridization w ith the co mp le - mentary target DNA (4a) with entrapped probe (4b) with electrostat ically adsorbed pr obe. 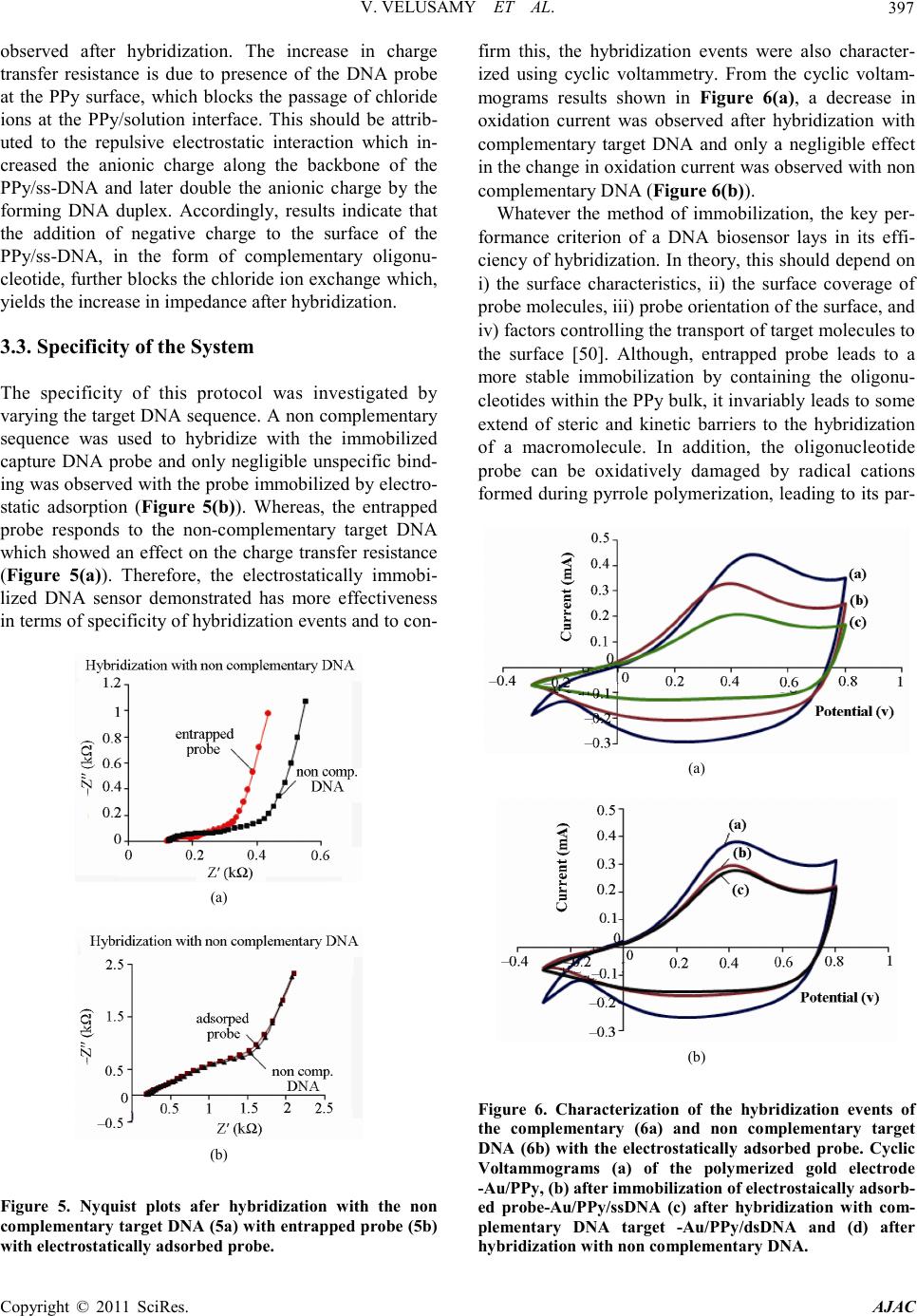 V. VELUSAMY ET AL. Copyright © 2011 SciRes. AJAC 397 observed after hybridization. The increase in charge transfer resistance is due to presence of the DNA probe at the PPy surface, which blocks the passage of chloride ions at the PPy/solution interface. This should be attrib- uted to the repulsive electrostatic interaction which in- creased the anionic charge along the backbone of the PPy/ss-DNA and later double the anionic charge by the forming DNA duplex. Accordingly, results indicate that the addition of negative charge to the surface of the PPy/ss-DNA, in the form of complementary oligonu- cleotide, further blocks the chl orid e ion exc hange which, yields the increase in impedance after hybridization. 3.3. Specificity of the System The specificity of this protocol was investigated by varying the target DNA sequence. A non co mplementary sequence was used to hybridize with the immobilized capture DNA probe and only negligible unspecific bind- ing was observed with the probe immobilized by electro- static adsorption (Figure 5(b)). Whereas, the entrapped probe responds to the non-complementary target DNA which showed an effect on the charge transfer resistance (Figure 5(a)). Therefore, the electrostatically immobi- lized DNA sensor demonstrated has more effectiveness in ter ms of specificity of hybridization events and to co n- (a) (b) Figure 5. Nyquist plots afer hybridization with the non complementary target DNA (5a) with entrapped probe (5b) with electr os tatically adsorbed probe. firm this, the hybridization events were also character- ized using cyclic voltammetry. From the cyclic voltam- mograms results shown in Figure 6(a), a decrease in oxidation current was observed after hybridization with complementary target DNA and only a negligible effect in the chan ge in oxid a tio n cur r ent wa s o bs er ved wit h no n complementary DNA (Figure 6(b)). Whatever the method of immobilization, the key per- formance criterion of a DNA biosensor lays in its effi- ciency of hybridization. In theory, this should depend on i) the surface characteristics, ii) the surface coverage of probe molecules, iii) probe orientation of the surface, and iv) factors con trollin g the tra nspo rt o f target molec ules to the surface [50]. Although, entrapped probe leads to a more stable immobilization by containing the oligonu- cleotides within the P Py bulk, it i nvariably leads to so me extend of steric and kinetic barriers to the hybridization of a macromolecule. In addition, the oligonucleotide probe can be oxidatively damaged by radical cations formed during p yrrole p olymerization, leading to its pa r- (a) (b) Figure 6. Characterization of the hybridization events of the complementary (6a) and non complementary target DNA (6b) with the electrostatically adsorbed probe. Cyclic Voltammograms (a) of the polymerized gold electrode -Au/PPy, (b) after immobilization of electrost ai cal ly adsorb- ed probe-Au/PPy/ssDNA (c) after hybridization with com- plementary DNA target -Au/PPy/dsDNA and (d) after hybridiz a tio n with no n co mplementary DNA.  V. VELUSAMY ET AL. Copyright © 2011 SciRes. AJAC 398 tial degrada tion [51]. In electrochemical adsorption, by application of a po- tential to the electro de, the delocalized positive charge of the oxidized PPy electrostatically attracts the negatively charged phosphate groups of the DNA. Also, contro lle d adsorption of DNA onto the PPy surface can be obtained since the positively charged PPy substrate exerts electro- static attraction o n the DNA mole cules. T he q uantit y and strength of adsorption depends on the nature of the anion dopant (electrolyte, in this case MgCl2), type and the ionic strength of the buffer used, solution pH, and on the DNA itself. While the DNA surface coverage can be controlled, the DNA conformation and orientation on the surface is difficult to modulate. Studies [52-54] suggest that electrostatically driven DNA adsorption results in orientation of the molecules parallel to, rather than per- pendicular to the surface, with base pairing sites exposed to the liquid medium. Howev er, the results sho w that t he orientation is appropriate to target DNA hybridization and does not limit specificity of the biosensor . 4. Conclusions Electrochemically assisted adsorption of DNA on the PPy modified electrode surface leads to more active and functional DNA layers. This method takes advantage over the entrapment method. Therefore, the DNA-PPy surface interaction is stronger and more stable when a potential is applied during adsorption. Moreover, the measurements performed in an inert electrolyte solution resulted in much higher impedance due to the presence of redox polymer and were reproducible. Therefore, the application of redox conductive polymer such as PPy as an immobilization matrix is an attractive approach in which the presence of external redox probe or active species can be totally eliminated. A further study of nu- cleic acid interaction on transducer interfaces and the investigation of conformation of DNA on the electrode surface can generate the insights on the design surfaces for more effective hybridization. 5. Acknowledgemen t s This project is funded by Science Foundation Ireland (SFI) Research Frontiers Programme, ID no: 07RPF- ENEF500. The presented research work was co funded by SFI-Short Term Travel Fellowship award. 6. References [1] M. V. Murugendrappa and M. V. N. Ambika Prasad, “Dielectric Spectroscopy of Polypyrrole-[Gamma] -Fe2O3 Composites,” Materials Research Bulletin, Vol. 41, No. 7, 2006, p p. 1364-1369. doi:10.1016/j.materresbull.2005.12.011 [2] B. D. Malhotra, A. Chaubey and S. P. Singh, “Prospects of Conducting Polymers in Biosensors,” Analytica Chimi ca Acta, Vol. 578, No. 1, 2006, pp. 59-74. doi:10.1016/j.aca.2006.04.055 [3] R. J. Geise, J. M.Adams, N. J. Barone and A. M. Ya- c ynyc h , “Electropolymerized Films to Prevent In terfer- ences and Electrode Fouling in Biosensors,” Biosensors & Bioelectronics, Vol. 6, No. 2, 1991, pp. 151-160. doi:10.1016/0956-5663(91)87039-E [4] N. J. Trujillo, M. C. Barr, S. G. Im and K. K. Gle a so n , “Oxidative Chemical Vapor Deposition (oCVD) of Pat- terned and Functional Grafted Conducting Polymer Nanostructures,” Journal of Materials Chemistry, Vol. 20, No. 19, pp. 3968-3972. doi:10.1039/b925736e [5] S. Liu, J. Q. Wang, D. Zhang, P. L. Zhang, J. F. Ou, B. Liu and S. R. Yan g, “Inve stigat ion on Cell Biocompatible Behaviors of Polyaniline Film Fabricated via Electroless Surface Polymerization,” Applied Surface Science, Vol. 256, No. 11, pp. 3427-3431. doi:10.1016/j.apsusc.2009.12.046 [6] F. Wei, W. Liao, Z. Xu, Y. Yang, D. T. Wong and C. M. Ho, “Bio/Abiotic Interface Constructed from Nanoscale DNA Dendrimer and Conducting Polymer for Ultrasensi- tive Biomolecular Diagnosis”, Small, Vol. 5, No . 15, 2009, pp . 1784-1790. doi:10.1002/smll.200900369 [7] H. S. Kim, H. L. Hobbs, L. Wang, M. J. Rutten and C. C. Wamser, “Biocompatible Composites of Polyaniline Nanofibers and Collagen,” Synthetic Metals, Vol. 159, No. 13, 2009, pp. 1313-1318. doi:10.1016/j.synthmet.2009.02.036 [8] N. K. E. Guimard, J. L.Sessler and C. E. Schmidt, “To- ward a Biocompatible and Biodegradable Copolymer In- corporating Electroactive Oligothiophene Units,” Mac- romolecules, Vol. 42 , No . 2, 2009, pp. 502-511. doi:10.1021/ma8019859 [9] A. Ramanaviciene and A. Ramanavicius, “Application of Polypyrrole for the Creation of Immunosensors,” Crit ical Reviews in Analytical Chemistry, Vol. 32, No. 3, 2002, pp. 245-252. doi:10.1080/10408340290765542 [10] A. Ramanavicius, A. Ramanaviciene and A. Malinaus kas, “Electrochemical Sensors Based on Conducting Poly- mer-Polypyrrole,” Electrochimica Acta, Vol. 51, No. 27, 2006, pp . 6025-6037. doi:10.1016/j.electacta.2005.11.052 [11] M. V. Murugendrappa and M. Prasad, “Chemical Syn- thesis, Characterization, and Dir ec t-Current Conductivity Studies of Polypyrrole/Gamma-Fe2O3 Composites,” Journal of Applied Polymer Science, Vol. 103, No. 5, 2007, pp . 2797-2801. doi:10.1002/app.23868 [12] S. Uzun and M. Can, “Oxidizing Effect on Chemical Polymerization of Pyrrole Monomer in Anhydrous Me- dia,” Asian Journal of Chemistry, Vol. 22, No . 2, 2010, pp. 1321-1330. [13] S. Cavallaro, A. Colligiani and G. Cum, “Oxidative Chemical Po l yme r ization of Pyrrole-Calorimetric and Kinetic Measurements,”Journal of Thermal Analysis and Calori metry, Vo l. 38 , No. 12, 1992, pp. 2649-2655. doi:10.1007/bf01979741  V. VELUSAMY ET AL. Copyright © 2011 SciRes. AJAC 399 [14] J. Duchet, R. Legras and S. Demoustier-Champagne, “Chemical Synthesis of Polypyrrole: Structure-Properties Relationship,” Synth etic Met al s, Vol. 98 , No. 2, 1998 , pp . 113-122. doi:10.1016/S0379-6779(98)00180-5 [15] P. Dutta and S. K Mandal, “Charge Transport in Chemi- cally Syn t h esized, DNA-Doped Po l ypyrr o le ,” Journal of Physics D-Applied Physics, Vol. 37, No. 20, 2004, pp. 2908-2913. doi:10.1088/0022-3727/37/20/019 [16] A. K. Sharma, J. H. Kim and Y. S. Lee, “An Efficient Synthesis of Polypyrrole/Carbon Fiber Composite Nano- thin Films,” International Journal of Electrochemical Scien ce, Vol. 4, No. 11, 2009, pp. 1560-1567. [17] S. Stankovic, R. Stankovic, M. Ristic, O. Pavlovic and M. Vojnovic, “Some Aspects of the Electrochemical Synt he- sis of Polypyrrole,” Reactive & Functional Polymers, Vol. 35, No . 3, 1997, pp. 145-151. doi:10.1016/S1381-5148(97)00090-4 [18] K. Ghanbari, S. Z. Bathaie and M.F. Mousavi, “Electro- Chemically Fabricated Polypyrrole Nanofiber-Modified Electrode as a New Electrochemical DNA Biosensor,” Biosensors & Bioelectronics, Vol. 23, No . 12, 2008, pp. 1825-1831. doi:10.1016/j.bios.2008.02.029 [19] O. Fichet, F. Tran-V an , D. Te yss ie and C. Chevrot, “In- terfacial Polymerization of a 3, 4-Ethylenedioxythiophene Derivative Using Lang- mui r -Blodgett Technique. Spectroscopic and Electro- chemical Character izations,” Thin Solid Films, Vol. 411, No. 2, 2002, pp. 280-288. doi:10.1016/S0040-6090(02)00271-7 [20] M. P. Srinivasan and F. J. Jing, “Composite Langmuir - Blodgett Films Containing Polypyrrole and Polyimide,” Thin Solid Films, Vol. 327-329, 1998, pp. 127-130. doi:10.1016/S0040-6090(98)00613-0 [21] S. M. Shang, X. M. Yang, X. M. Tao and S. S. Lam, “Vapor-Phase Polymerization of Pyrrole on Flexible Substr ate at Low Temperat ure an d its Application in Heat Generation,” Polymer International, Vol. 59, No. 2, 2010 , p p. 204-211. [22] H. Goktas, F. G. Ince, A. Iscan, I. Yildiz, M. Kurt and I. Kaya , “The Molecular Structure of Plasma Polymerized Thiophene and Pyrrole Thin Films Produced by Double Discharge Technique,” Synthetic Metals, Vol. 159, No . 19-20, 2009, pp. 2001-2008. doi:10.1016/j.synthmet.2009.07.003 [23] Z. Zhang, P. Liang, X. Zheng, D. Peng, F. Yan, R. Zh ao and C.-L. Feng, “DNA Immobilization/Hybridization on P l a sma -Polymerized Pyrrole,” Biomacro m olecules, Vol. 9, No. 6, 2008, p p. 1613-1617. doi:10.1021/bm800111a [24] Z. B. Bahsi, A. Buyukaksoy, S. M. Olmezcan , F. Simsek, M. H. Asl an and A. Y.Oral, “A Novel Lab el-Free Op tical Biosensor Using Synthetic Oligonucleotides from E. coli O157:H7: Elementary Sensitivity Tests,” Sensors, Vol . 9, No. 6, 2009, pp. 4890-4900. doi:10.3390/s90604890 [25] J. Liu, and Y. Lu, “Colorimetric and Fluorescent Biosen- sors Based on Directed Assembly of Nanomaterials with Functional DNA,” Integrated Analytical Systems, Springer, New York, 2009, pp. 155-178. doi:10.1007/978-0-387-73711-9_6 [26] A. K. Singh, D. Senapati, S. G. Wan g, J. Griffin, A. Neely, P. Candice, K. M. N a yl o r, B. Varisli, J. R. Kalluri and P. C. Ray, “Go ld Nanorod Based Selective Identifi- cation of Escherichia coli Bacteria Using Two-Photon Rayleigh S catterin g Spectr oscop y,” Acs Nano , Vol. 3 , No . 7, 2009, p p. 1906-1912. doi:10.1021/nn9005494 [27] P. S. Petrou, S. E. Kakabakos and K. Misiakos, “Silicon Optocouplers for Biosensing,” International Journal of Nanotechnology, Vol. 6, No. 1-2, 2009, pp. 4-17. [28] S. J. P. Canete, W. W. Yang and R. Y. Lai, “Fold- ing-Based Electrochemical DNA Sensor Fabricated by ‘Click’ Chemistry,” Chemical Communications, No. 32, 2009, pp . 4835-4837. [29] J. Jiang, D. Jiang, Y. Li, J. Wang and K. L. P. Sung, “P o l ypyr r ol e -Based Genolelectronics: Electrochemical DNA Biosensors,” Science & Technology Review, No. 11, 2009 , pp. 93-101. [30] V. Velusamy, K. Arshak, O. Korostynska, K. Oliwa and C. Adley, “Design of a Real Time Biorecognition System to Detect Foodborne Pathogens-DNA Biosen sor,” in SAS 2009. New Orleans, USA, 2009, pp. 38-41. [31] P. I. Reyes, Z. Zhang, H. H. Ch en , Z. Q. Du a n , J. Zhong, G. Saraf, Y. C. Lu, O. Taratula, E. Galo ppini and N. N. Boustany, “A ZnO Nanostructure-Based Quartz Crystal Microbalance Device for Biochemical Sensing,” IEEE Sensors Journal, Vol. 9, No. 10, 2009, pp . 1 30 2-1307. doi:10.1109/JSEN.2009.2030250 [32] A. Tsortos, G. Papadaki s and E. Gizeli, “Shear Acoustic Wave Biosensor for Detecting DNA Intrinsic Viscosity and Conformation: A study with QCM-D,” Bi osensors & Bioelectron ics, Vol. 24 , No. 4, 2008, pp. 836 -841. doi:10.1016/j.bios.2008.07.006 [33] J. Wang, G. Rivas, C. Parr ado, X. H. Cai and M. N. Flair, “Electrochemical Biosensor for Detecting DNA Se- quen ces fro m the Pathogenic Protozoan Cryptosporidium parvum,” Talanta, Vol . 4 4, No. 11, 1997, pp. 2003-2010. doi:10.1016/S0039-9140(96)02191-1 [34] D. Zhang and E. C. Alocilja, “Characterization of Nano - porous Sili co n-Based DNA Biosensor for the Detection of Salmonella Enteritidis,” IEEE Sensors Journal, Vol. 8 , No. 5-6, 2008, pp. 77 5-780. doi:10.1109/JSEN.2008.923037 [35] V. Vyskocil, J. Labuda and J. Barek, “Voltammetric De- tection of Damage to DNA Caused by Nitro Derivatives of Fluorene Using an Electrochemical DNA Biosensor,” Analytical and Bioanalytical Chemistry, Vol. 397, No. 1, pp. 233-241. doi:10.1007/s00216-010-3517-y [36] A. M. Nowi c k a, A. Kowalczyk, Z. Stojek and M. Hepel, “Nanogravimetric and Voltammetric DNA-Hybridization Biosensors for Studies of DNA Damage by Common Toxicants and Pollutants,” Biophysical Chemistry, Vol. 146, No. 1, pp. 42-53. doi:10.1016/j.bpc.2009.10.003 [37] S. L. Yang, B. Y. Xia, X. D. Zeng, S. L. Luo, W. Z. Wei and X. Y. Liu, “Fabrication of DNA functionalized car- bon nanotubes/Cu2+ complex by one-step electrodeposi- tion and its sensitive determination of nitrite,” Analytica Chimi ca Acta, Vol. 667, No. 1-2, pp. 57-62. doi:10.1016/j.aca.2010.03.063  V. VELUSAMY ET AL. Copyright © 2011 SciRes. AJAC 400 [38] H. J. Qiu, Y. L. Sun, X. R. Huan g and Y. B. Qu, “A Sen- sitive Nanoporous Gold-Based Electrochemical Apta- sensor for Thrombin Detection,” Colloids and Surfaces B-Bioin terfaces, Vo l. 79 , No . 1, pp. 304-308. doi:10.1016/j.colsurfb.2010.04.017 [39] A. M. Tencaliec, S. Laschi, V. Magearu and M. Mascini, “A Comparison Study between a Disposable El ectro- chemical DNA Biosensor and a Vibrio Fischeri-Based Luminescent Sensor for the Detection of Toxicants in Water Samp l es,” Talanta, Vol. 69, No. 2, 2006, pp. 365-369. doi:10.1016/j.talanta.2005.09.042 [40] G. Bagni, T. Baussant, G. Jonsson, J. Barsiene and M. Mascini, “Electrochemical Device for the Rapid Detec- tion of Genotoxic Compounds in Fish Bile Samples,” Analytical Letters, Vol. 38, No. 15, 2005 , pp. 26 39-2652. doi:10.1080/00032710500371105 [41] J. Y. Park and S. M.Park, “DNA Hybridization Sensors Based on Electrochemical Impedance Spectroscopy as a Detection Tool,” Sensors, Vol. 9, No. 12, 2009, pp. 9513-9532. doi:10.3390/s91209513 [42] V. Velusamy, K. Ars h ak , O. Korostynska, K. Oliwa and C. Adley, “An Overview of Foodborne Pathogen Detec- tion: In the Perspective of Biosensors,” Biotechnology Adva nces, Vol. 28, No. 2, 2010, pp. 232-254. doi:10.1016/j.biotechadv.2009.12.004 [43] A. Bogomolova, E. Komarova, K. Reber, T. Gerasimo v, O. Yavuz, S. Bhatt and M. Aldissi, “Challenges of Elec- trochemical Impedance Spectroscopy in Protein Biosens- ing,” Analytical Chemistry, Vol. 81, No. 10, 2009, pp. 3944-3949. doi:10.1021/ac9002358 [44] C. Gautier, C. Esnault, C. Cougnon, J.-F. Pilard, N. Casse, and B. Chénais, “Hybridization-induced interfacial changes detected by non-Faradaic impedimetric meas- urements compared to Faradaic approach,” Journal of Electroanalytical Chemistry, Vol. 610, No. 2,20 07, pp. 227-233. doi:10.1016/j.jelechem.2007.07.013 [45] C. Adley, K. Arsh a k, C. Molnar, K. Oliwa and V. Velu- samy, “Design of Specific DNA Primers to Detect the Bacillus Cereus Group Species,” Sensors Applications Symposium, New Orleans, USA, 2009, pp. 236-239. doi: 10.1109/SAS.2009.4801807 [46] F. Devreux, F. Genoud, M. Nechts chein and B. Villeret, “Electro n-spin-resonance investigation of polarons and bipolarons in conducting polymers-the case of polypyr- role,” Synthetic Metals, Vol. 18, No. 1-3, 1987, pp. 89-94. doi:10.1016/0379-6779(87)90859-9 [47] A. O. Patil, A. J. Heeger and F. Wudl, “Optical-Prop- erties of Conducting Polymers,” Chemical Reviews, Vol. 88, No. 1, 1988, pp. 183-200. doi:10.1021/cr00083a009 [48] K. Arshak, V. Velusamy, O. Korost ynska, K. Oliwa-Stasiak and C. Adl ey, “Conducting Polymers and Their Applications to Biosensors: Emphasizing on Food- borne Pathogen Detection,” IEEE Sensors Journal, Vol. 9, No. 12, 2009 , pp. 19 42-1951. doi:10.1109/JSEN.2009.2032052 [49] D. S. Minehan, K. A. M ar x and S. K. Tripathy, “DNA Binding to Electropolymerized Polypyrrole: The De- pendence on Film Characteristics,” Journal of macromo- lecular science. Pure and applied chemistry, Vol. 38 2001, pp . 1245-1258. doi:10.1081/MA-100108381 [50] K. L.Hanson, l. Filipponi and D. V. Nicolau, “Bio- molecules and cells on surfaces: Fundamental concepts,” Uwe R. Müller and Dan V. Nicolau, Editors, Microarray Technology: Fundamentals, Fabrication and Applica- tions, Springer Verlag, 2004, pp. 23-44. [51] L. A. Thompson, J. Kowalik, M. Josowicz and J. Janata, “Label-Free DNA Hybridization Probe Based on a Con- ducting Polymer,” Journal of the American Chemical So- ciety, Vol. 125, No. 2, 2002, pp. 324-325. doi:10.1021/ja027929z [52] V. Chan, D. J. Graves, P. Fortina and S. E. McKenzie, “Adsorption and Surface Diffusion of DNA Oligonucleo- tides at Liquid/Solid Interfaces,” Langmuir, Vol. 13, No. 2, 1997, pp. 320-329. doi:10.1021/la960670b [53] V. Chan, S. E. McKenzie, S. Sur r e y, P. Fortina and D. J. Graves, “Effect of Hydrophobicity and Electrostatics on Adsorption and Surface Diffusion of DNA Oligonucleo- tides at Liquid/Solid Interfaces,” Journal of Colloid and Interface Scien ce, Vol. 203, No. 1, 1998, pp . 197-207. doi:10.1006/jcis.1998.5495 [54] A. M. Oliveira Brett, and A. -M. Chiorcea, “Atomic Fo rce Microscopy of DNA Immobilized onto a Highly Oriented Pyrolytic Graphite Electrode Sur face,” Langmuir, Vol. 19, No. 9, 2003, pp. 3830-3839. doi:10.1021/la027047d
|