 American Journal of Molecular Biology, 2011, 1, 114-121 AJMB doi:10.4236/ajmb.2011.12012 Published Online July 2011 (http://www.SciRP.org/journal/ajmb/). Published Online July 2011 in SciRes. http://www.scirp.org/journal/AJMB An investigation of 10 Y-STR loci and the detection of specific haplotype frequencies in Turkish population Aydın Rüstemoglu1, Güvem Gümüs Akay2, Halil Gürhan Karabulut3, Ahmet Kadıkıran4, Işık Bökesoy3 1Gaziosmanpaşa University, Faculty of Medicine, Department of Medical Biology, Tokat, Turkey; 2Ankara University, Brain Studies Application and Research Center, Ankara, Turkey; 3Ankara University, Faculty of Medicine, Department of Medical Genetics, Ankara, Turkey; 4Ankara University, Faculty of Science, Department of Molecular Biology, Ankara, Turkey. E-mail: arustamov@gop.edu.tr Received 26 May 2011; revised 16 June 2011; accepted 30 June 2011. ABSTRACT This study is to survey 10 Y-STR loci in 241 males from Turkey. In this study, the 241 healthy and unrelated males living in different parts of Turkey for at least three generations were included. Genomic DNAs were iso- lated from peripheral blood samples by standard phenol-chloroform extraction method. 10 Y-STR loci including DYS19, DYS385a/b, DYS388, DYS389I/II, DYS390, DYS391, DYS392, DYS393, and YCAIIa/b were analyzed by using PCR and denaturing PAGE. Allele frequencies, gene diversities and haplotype frequencies were analyzed. Gene diversity per locus varied from 0.5788 (DYS388) to 0.8903 (DYS385a/b). The numbers of haplotypes in minHt recommended by YCC and Ht10 have been 208 and 186, respec- tively. When our minHt haplotypes frequencies com- pared with the other seven populations, we have found statistically significant differences between our results and other populations (p < 0.01) except that Czech population (p > 0.05). We suggest that an al- ternative haplotype designated as aHt maybe alterna- tive to minHt in respect of its Y-STR content with the highest gene diversity value. The aHt haplotype has found a higher discriminatory potential than minHt haplotype with a better Pd combined value (0.9999936 vs 0.9999836) and has higher average gene diversity per locus (0.7834 vs 0.7518) in Turkish population. aHt haplotype can be proposed as an alternative to minHt in paternity testing and forensic medicine ap- plications involving Turkish male population. This study has also provided additional information to the framework of variation involving 10 Y-STR loci as well as a further contribution to the Y-STR database for Turkish male population. Keywords: Y Chromosome; Y-STR; Polymorphism; Haplotype; Turkey 1. INTRODUCTION Human Y chromosome has been known to display com- paratively low levels of polymorphisms in contrast with autosomal chromosomes. Nonetheless, there are many types of Y-chromosome specific polymorphisms identi- fied in the non-recombining region of Y (NRY) includ- ing RFLPs, Y Alu and microsatellite polymorphisms [1, 2]. Microsatellites or short tandem repeats (STRs) are dispersed throughout the genome and are known to be highly polymorphic; hence, each microsatellite locus generally has many alleles due to variable number of repeat units [3,4]. Allelic genotyping of STRs does not require the use of complex molecular techniques, since amplifications and visualization of PCR products make it easy. Y-chromo- some specific STRs (Y-STRs) are chosen as more infor- mative in paternity testing, forensic applications and the study of population histories due to the haploid state of Y chromosome which ensures both the transmittance by the paternal lineages and the lack of recombination in NRY, excluding pseudoautosomal regions (PARs) [5- 11]. Allelic and haplotypic distributions of Y-STRs have shown significant differences in different geographical regions, ethnical groups and communities [12-18]. There- fore, allelic and haplotypic frequencies of Y-STRs should be determined in a male population prior to any interpretations of forensic analysis and paternity testing [6,8-11,19]. In this study, allelic and haplotypic frequent-  A. Rüstemoglu et al. / American Journal of Molecular Biology 1 (2011) 114-121 Copyright © 2011 SciRes. AJMB 11 5 cies involving 10 Y-STR loci: 8 Y-STR loci as recom- mended by Y Chromosome Consortium (YCC) plus DYS388 and YCAIIa/b-have been determined with such a necessity in a representative group of Turkish popula- tion in order to make comparisons with other popula- tions. 2. MATERIALS AND METHODS 10 Y-STR loci were analyzed which included eight loci recommended by YCC for minimal haplotype (minHt)- DYS19, DYS385a/b, DYS389I/II, DYS390, DYS391, DYS392, and DYS393 [20,21] and the additional two: DYS388 and YCAIIa/b. Healthy and unrelated 241 males living in different parts of Turkey for at least three generations were in- cluded in this study. The written informed consents were obtained from the study subjects and the study protocol was approved by the ethics committee of Ankara Uni- versity Medical Faculty. Genomic DNAs were isolated from peripheral blood samples by standard phenol-chloroform extraction me- thod [22]. Four multiplex PCR analyses were carried out with locus specific primers in a total volume of 25 µl reaction mixture, containing; 50 - 150 ng genomic DNA, 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 0.1 - 0.8 µM of each primer, 200 µM of each dNTP (Sigma), 5 µg BSA and 1 unit Taq DNA polymerase (Invitrogen) [23]. Y-STRs amplified in combination in multiplex PCR are as follows: DYS389I, DYS389 II and DYS390 in multiplex I; DYS392 and DYS393 in multiplex II; DYS19 and DYS388 in multiplex III; DYS385a/b and DYS391 in multiplex IV; while YCAIIa/b loci were am- plified separately. Cycling conditions were as follows: 32 cycles of 94˚C - 1 min, 54˚C - 1 min, 72˚C - 1 min for multiplex I, II, IV and YCAIIa/b; 35 cycles of 94˚C - 1 min, 55˚C - 1 min, 72˚C - 60 min for multiplex III. An initial denaturation at 94˚C - 2.5 min and a final exten- sion at 72˚C - 10 min were performed before and after each cycling reactions. Female DNA was used as a neg- ative control in every run. Amplified products were separated by 6% denaturing polyacrylamide gel electrophoresis (PAGE) for 3 h at 1500 V. Visualization of PCR products was carried out by a modification of the silver staining method of Santos, et al. [23]. Gels were fixed for 15 min at room tempera- ture in 10% (v/v) ethanol which were treated with 1% (v/v) nitric acid for 3 min with agitation thereafter. Gels were then rinsed in deionized water for 1 min and treated with 0.2% (w/v) silver nitrate and 0.1% (v/v) formalde- hyde solution for 25 min with agitation. Gels were rinsed for a few seconds in deionized water, and developed in an aqueous solution of 3% Na2CO3 (w/v), 100 µl/L 2% (w/v) Na2S2O3 and 0.1% formaldehyde until the bands were well visualized. Staining was ended with a fixative solution. Allelic genotyping were carried out by using the de- fined DNA size markers (Fermentase, pUC Mix Marker, 8; ΦX174 RF DNA/BsuRI [HaeIII] Marker, 9) and self- made ladders as standards. Allele frequencies, gene diversities (H), haplotype frequencies and genetic differentiation between popula- tions were computed using Arlequin 3.1.1 software. Al- lele frequencies were calculated by gene counting, while H was computed for each locus according to the by for- mula (1), where n is the number of samples, k is the number of haplotypes, and pi is the frequency of i-th haplotype [24]. 2 1 1 1 k i n p n (1) The Combined Power of Discrimination (Pd combined) for haplotypes was calculated using the by formula (2), where Pdi is Power of Discrimination of i-th locus [25]. 1 1π1 n d combineddi i PP (2) 3. RESULTS AND DISCUSSION 10 Y-STRs have been analyzed for diversity in 241 healthy and unrelated male individuals from Turkey. Observed allele or genotype frequencies of the 10 Y-STR loci have been given in Table 1. Variations in the number of individuals for certain loci have been brought about by some technical problems not anticipated. Gene diversity values for each 10 Y-STR loci have been given in Table 1. The lowest gene diversity (0.5788) has been found in DYS388 locus, wherein the most fre- quent allele has been allele 13 with a frequency of 62.08%. This result has been in accord with the data reported by YCC [21]. The highest gene diversity (0.8903) has been found in DYS385 locus, wherein the most frequent allele has been allele 14 with a frequency of 17.62% (Table 1). The observed number of haplotypes and their fre- quencies involving minHt and Ht10 haplotypes in this current survey have been tabulated in Table 2. The number of haplotypes detected for minHt is 208 and 186 for Ht10. Each haplotype belonging to Ht10 have been found to be unique while the same holds for minHt with the exception of H15 which has been detected in two individuals. Gene diversity, average gene diversity per locus and Combined Power of Discrimination (Pd combined) values for Ht10 and minHt haplotypes have been given in Table 3. minHt haplotypes detected in this study group have been compared with seven other populations: Croatian 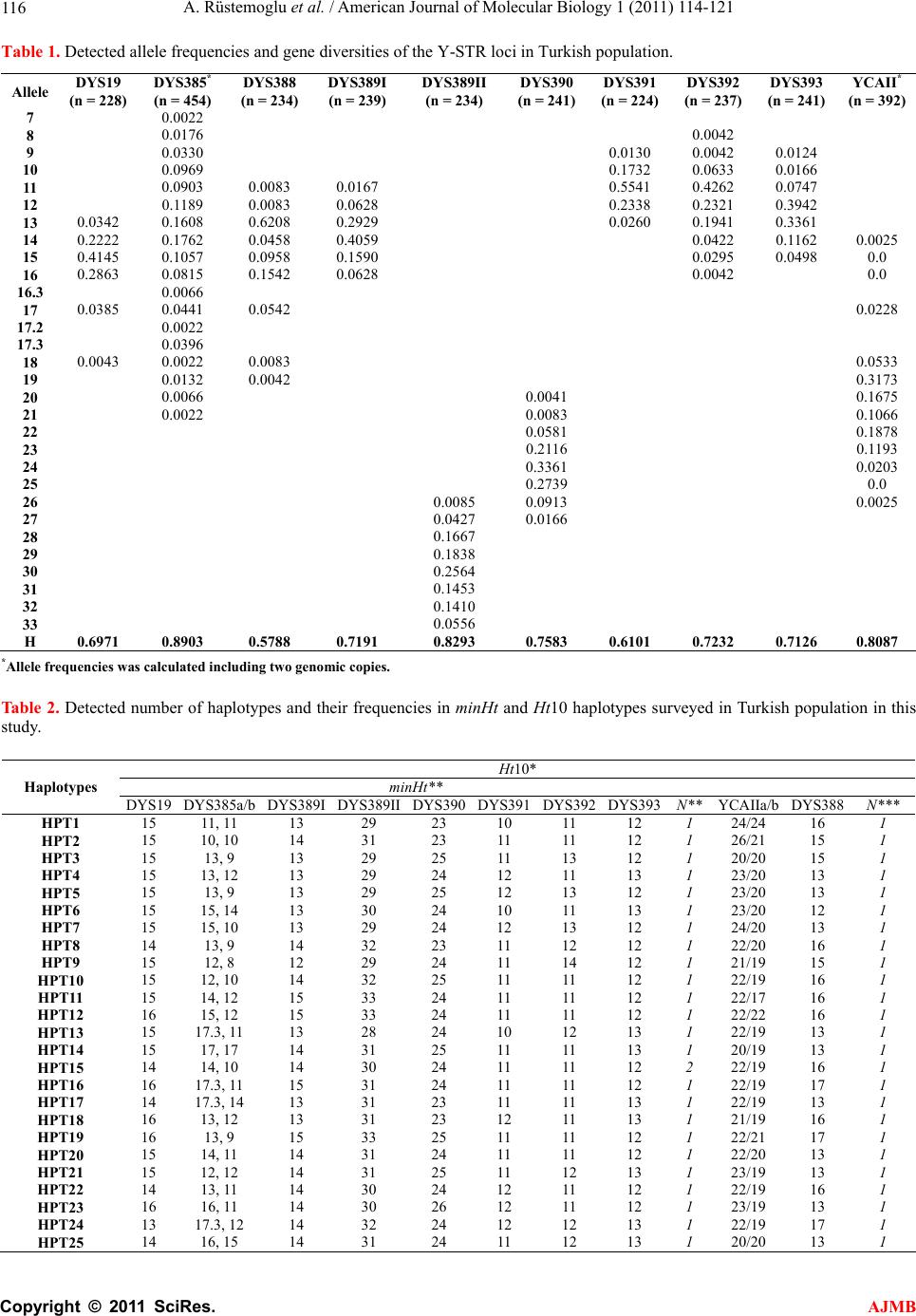 A. Rüstemoglu et al. / American Journal of Molecular Biology 1 (2011) 114-121 Copyright © 2011 SciRes. AJMB 116 Table 1. Detected allele frequencies and gene diversities of the Y-STR loci in Turkish population. Allele DYS19 (n = 228) DYS385* (n = 454) DYS388 (n = 234) DYS389I (n = 239) DYS389II (n = 234) DYS390 (n = 241) DYS391 (n = 224) DYS392 (n = 237) DYS393 (n = 241) YCAII* (n = 392) 7 0.0022 8 0.0176 0.0042 9 0.0330 0.0130 0.0042 0.0124 10 0.0969 0.1732 0.0633 0.0166 11 0.0903 0.0083 0.0167 0.5541 0.4262 0.0747 12 0.1189 0.0083 0.0628 0.2338 0.2321 0.3942 13 0.0342 0.1608 0.6208 0.2929 0.0260 0.1941 0.3361 14 0.2222 0.1762 0.0458 0.4059 0.0422 0.1162 0.0025 15 0.4145 0.1057 0.0958 0.1590 0.0295 0.0498 0.0 16 0.2863 0.0815 0.1542 0.0628 0.0042 0.0 16.3 0.0066 17 0.0385 0.0441 0.0542 0.0228 17.2 0.0022 17.3 0.0396 18 0.0043 0.0022 0.0083 0.0533 19 0.0132 0.0042 0.3173 20 0.0066 0.0041 0.1675 21 0.0022 0.0083 0.1066 22 0.0581 0.1878 23 0.2116 0.1193 24 0.3361 0.0203 25 0.2739 0.0 26 0.0085 0.0913 0.0025 27 0.0427 0.0166 28 0.1667 29 0.1838 30 0.2564 31 0.1453 32 0.1410 33 0.0556 H 0.6971 0.8903 0.5788 0.7191 0.8293 0.7583 0.6101 0.7232 0.7126 0.8087 *Allele frequencies was calculated including two genomic copies. Table 2. Detected number of haplotypes and their frequencies in minHt and Ht10 haplotypes surveyed in Turkish population in this study. Ht10* minHt** Haplotypes DYS19 DYS385a/b DYS389I DYS389II DYS390 DYS391 DYS392 DYS393N** YCAIIa/b DYS388N*** HPT1 15 11, 11 13 29 23 10 11 12 1 24/24 16 1 HPT2 15 10, 10 14 31 23 11 11 12 1 26/21 15 1 HPT3 15 13, 9 13 29 25 11 13 12 1 20/20 15 1 HPT4 15 13, 12 13 29 24 12 11 13 1 23/20 13 1 HPT5 15 13, 9 13 29 25 12 13 12 1 23/20 13 1 HPT6 15 15, 14 13 30 24 10 11 13 1 23/20 12 1 HPT7 15 15, 10 13 29 24 12 13 12 1 24/20 13 1 HPT8 14 13, 9 14 32 23 11 12 12 1 22/20 16 1 HPT9 15 12, 8 12 29 24 11 14 12 1 21/19 15 1 HPT10 15 12, 10 14 32 25 11 11 12 1 22/19 16 1 HPT11 15 14, 12 15 33 24 11 11 12 1 22/17 16 1 HPT12 16 15, 12 15 33 24 11 11 12 1 22/22 16 1 HPT13 15 17.3, 11 13 28 24 10 12 13 1 22/19 13 1 HPT14 15 17, 17 14 31 25 11 11 13 1 20/19 13 1 HPT15 14 14, 10 14 30 24 11 11 12 2 22/19 16 1 HPT16 16 17.3, 11 15 31 24 11 11 12 1 22/19 17 1 HPT17 14 17.3, 14 13 31 23 11 11 13 1 22/19 13 1 HPT18 16 13, 12 13 31 23 12 11 13 1 21/19 16 1 HPT19 16 13, 9 15 33 25 11 11 12 1 22/21 17 1 HPT20 15 14, 11 14 31 24 11 11 12 1 22/20 13 1 HPT21 15 12, 12 14 31 25 11 12 13 1 23/19 13 1 HPT22 14 13, 11 14 30 24 12 11 12 1 22/19 16 1 HPT23 16 16, 11 14 30 26 12 11 12 1 23/19 13 1 HPT24 13 17.3, 12 14 32 24 12 12 13 1 22/19 17 1 HPT25 14 16, 15 14 31 24 11 12 13 1 20/20 13 1  A. Rüstemoglu et al. / American Journal of Molecular Biology 1 (2011) 114-121 Copyright © 2011 SciRes. AJMB 11 7 HPT26 14 17.3, 13 13 30 25 11 13 13 1 20/19 13 1 HPT27 15 17.3, 15 14 31 23 11 11 12 1 22/19 16 1 HPT28 15 14, 10 14 30 25 12 13 12 1 23/19 13 1 HPT29 14 14, 10 13 30 25 11 11 13 1 23/19 16 1 HPT30 15 16, 14 14 30 24 11 13 13 1 23/23 13 1 HPT31 15 15, 13 14 30 26 11 11 12 1 22/19 13 1 HPT32 16 14, 10 14 32 25 10 11 13 1 23/19 13 1 HPT33 15 21, 12 14 33 24 12 11 12 1 22/19 15 1 HPT34 15 14, 10 15 30 25 12 11 13 1 22/19 13 1 HPT35 15 17.3, 13 14 30 24 11 12 11 1 22/18 16 1 HPT36 14 16, 12 13 30 26 11 11 12 1 23/19 13 1 HPT37 15 11, 9 14 30 26 11 8 13 1 22/19 15 1 HPT38 16 14, 14 12 29 23 11 10 14 1 22/19 13 1 HPT39 15 17.2, 15 13 30 25 11 10 14 1 22/18 16 1 HPT40 16 14, 14 13 30 23 11 10 13 1 23/19 13 1 HPT41 15 14, 10 14 30 24 12 11 13 1 22/19 15 1 HPT42 15 20, 9 13 30 24 12 10 12 1 22/19 13 1 HPT43 15 19, 17 13 29 25 11 10 13 1 22/18 16 1 HPT44 15 15, 10 14 31 25 12 11 12 1 23/19 13 1 HPT45 15 16, 12 14 31 24 12 10 12 1 22/19 15 1 HPT46 15 9, 9 14 31 25 12 11 13 1 22/19 13 1 HPT47 15 16, 12 14 33 23 10 11 12 1 22/18 16 1 HPT48 16 11, 10 14 32 24 11 10 12 1 23/19 13 1 HPT49 15 13, 11 13 30 24 11 12 13 1 22/19 15 1 HPT50 13 14, 12 13 32 22 11 11 15 1 22/19 13 1 HPT51 14 15, 12 15 33 24 11 12 12 1 22/18 16 1 HPT52 14 17, 11 12 29 24 10 11 12 1 23/19 13 1 HPT53 14 15, 14 12 30 23 10 11 13 1 22/19 15 1 HPT54 14 17, 13 14 32 24 10 11 12 1 22/19 13 1 HPT55 14 17, 14 14 32 25 10 13 13 1 22/18 16 1 HPT56 14 17, 12 14 32 23 10 12 12 1 23/19 13 1 HPT57 16 12, 9 13 31 26 11 11 13 1 22/19 15 1 HPT58 14 10, 10 14 32 24 11 11 12 1 22/19 13 1 HPT59 14 14, 11 13 31 26 10 13 12 1 22/18 16 1 HPT60 15 17, 13 14 32 24 10 12 11 1 23/19 13 1 HPT61 16 14, 14 14 31 24 10 12 13 1 22/19 15 1 HPT62 14 13, 11 15 32 25 11 13 12 1 22/19 13 1 HPT63 15 15, 12 13 31 22 12 11 15 1 22/18 16 1 HPT64 15 19, 12 14 32 26 12 13 13 1 23/19 13 1 HPT65 16 13, 11 14 29 25 11 10 14 1 22/19 15 1 HPT66 14 14, 12 14 31 25 11 11 12 1 22/19 13 1 HPT67 15 14, 12 14 33 25 11 12 14 1 22/18 16 1 HPT68 14 13, 10 14 31 26 12 13 12 1 23/19 13 1 HPT69 13 15, 15 14 32 25 12 11 13 1 21/19 13 1 HPT70 14 16, 13 14 32 24 11 12 12 1 22/17 16 1 HPT71 14 19, 13 14 33 24 11 12 11 1 22/19 19 1 HPT72 16 14, 10 15 33 25 12 12 13 1 23/19 13 1 HPT73 15 16, 13 13 30 25 10 10 12 1 20/19 16 1 HPT74 14 13, 12 15 29 26 9 11 12 1 22/20 16 1 HPT75 14 14, 12 15 28 24 10 12 12 1 22/19 16 1 HPT76 16 14, 13 16 29 24 11 12 14 1 20/19 13 1 HPT77 15 14, 10 13 27 22 10 13 15 1 20/19 13 1 HPT78 15 15, 12 14 28 24 10 13 11 1 22/19 16 1 HPT79 14 13, 10 15 29 23 12 13 11 1 21/19 17 1 HPT80 13 14, 12 14 30 25 10 12 12 1 22/20 13 1 HPT81 15 14, 7 15 28 23 10 13 10 1 21/20 13 1 HPT82 15 11, 10 14 27 23 10 11 14 1 20/19 13 1 HPT83 13 15, 13 15 28 25 10 11 11 1 20/19 13 1 HPT84 14 12, 12 15 29 25 12 12 9 1 24/19 13 1 HPT85 14 15, 14 16 29 24 11 11 13 1 21/19 13 1 HPT86 14 14, 11 14 28 23 10 10 13 1 21/19 13 1 HPT87 15 16, 16 14 27 25 11 11 11 1 22/21 13 1 HPT88 13 17.3, 17 14 28 25 11 11 11 1 22/18 13 1 HPT89 16 14, 14 13 26 24 11 11 13 1 20/20 13 1 HPT90 16 13, 11 14 28 26 11 11 11 1 21/19 13 1 HPT91 15 14, 14 13 27 23 11 11 15 1 19/18 13 1 HPT92 13 17.3, 12 14 28 25 11 11 9 1 22/19 16 1 HPT93 15 15, 12 14 28 23 11 11 15 1 19/17 13 1 HPT94 14 15, 12 14 27 25 12 12 11 1 23/19 13 1  A. Rüstemoglu et al. / American Journal of Molecular Biology 1 (2011) 114-121 Copyright © 2011 SciRes. AJMB 118 HPT95 14 12, 11 15 28 25 11 12 11 1 20/19 13 1 HPT96 15 14, 12 14 28 25 12 12 9 1 23/19 13 1 HPT97 16 14, 13 14 28 24 11 12 13 1 20/19 15 1 HPT98 17 15, 13 15 30 25 11 11 13 1 20/20 13 1 HPT99 14 17, 16 16 32 26 11 10 13 1 21/18 13 1 HPT100 15 14, 13 15 30 27 10 10 12 1 23/20 16 1 HPT101 16 16, 15 16 30 25 11 13 13 1 23/23 13 1 HPT102 16 16, 13 14 29 23 11 11 15 1 20/19 13 1 HPT103 16 15, 14 15 28 24 11 12 10 1 21/19 13 1 HPT104 15 14, 10 14 28 27 12 13 11 1 23/19 13 1 HPT105 15 17,13 15 29 25 11 12 10 1 22/19 16 1 HPT106 14 17, 15 14 29 23 10 11 11 1 20/18 13 1 HPT107 15 13, 12 15 29 26 11 13 10 1 23/19 13 1 HPT108 16 15, 13 14 28 22 11 13 15 1 20/19 13 1 HPT109 17 17.3, 17 16 31 23 11 12 15 1 21/19 13 1 HPT110 17 14, 13 14 28 23 12 11 14 1 20/19 13 1 HPT111 15 17.3, 14 16 30 25 11 12 12 1 22/19 14 1 HPT112 15 16, 15 14 28 24 11 12 13 1 21/19 15 1 HPT113 16 17.3, 13 15 29 24 11 13 12 1 22/19 16 1 HPT114 16 17, 11 15 29 25 11 11 12 1 22/19 16 1 HPT115 15 16, 15 16 30 26 10 12 14 1 22/19 13 1 HPT116 18 19, 13 16 29 25 11 11 13 1 21/19 13 1 HPT117 15 15, 14 15 28 25 12 12 12 1 22/19 13 1 HPT118 17 14, 10 16 31 27 11 11 13 1 23/19 13 1 HPT119 16 14, 10 15 29 25 13 12 13 1 22/19 13 1 HPT120 17 13, 13 14 29 25 10 11 13 1 23/22 14 1 HPT121 15 15, 13 15 29 24 11 11 12 1 22/19 16 1 HPT122 16 16, 13 14 28 23 10 11 14 1 20/20 13 1 HPT123 15 14, 10 16 30 24 12 12 11 1 22/19 13 1 HPT124 15 13, 12 14 28 24 11 13 13 1 19/14 13 1 HPT125 14 16, 10 13 29 23 11 11 13 1 22/19 13 1 HPT126 16 16.3, 13 14 29 23 11 11 13 1 22/19 16 1 HPT127 15 17.3, 13 14 28 24 11 11 12 1 22/19 16 1 HPT128 15 15, 13 14 29 24 12 11 12 1 22/22 18 1 HPT129 14 16.3, 14 14 29 23 13 13 13 1 19/19 13 1 HPT130 15 17.3, 17.3 14 29 24 12 11 12 1 22/21 15 1 HPT131 16 16.3, 11 14 28 24 12 11 12 1 22/19 15 1 HPT132 16 13, 10 13 29 24 11 11 14 1 20/20 13 1 HPT133 15 16, 10 13 28 23 11 9 13 1 20/20 15 1 HPT134 15 17.3, 15 15 32 24 11 12 13 1 20/20 13 1 HPT135 16 15, 10 14 29 26 10 12 14 1 23/19 14 1 HPT136 16 15, 13 14 28 24 11 13 15 1 20/19 14 1 HPT137 16 16, 15 14 28 23 11 11 12 1 23/20 13 1 HPT138 16 13, 12 14 27 24 10 11 12 1 19/18 17 1 HPT139 16 17, 13 13 28 25 11 11 12 1 20/19 13 1 HPT140 15 14, 11 16 29 26 12 12 12 1 23/22 13 1 HPT141 16 14, 13 15 30 23 11 11 14 1 20/19 13 1 HPT142 15 17, 16 16 30 24 11 11 13 1 22/21 13 1 HPT143 16 16, 13 16 31 24 10 11 13 1 22/19 13 1 HPT144 16 13,12 16 31 24 11 10 13 1 20/19 13 1 HPT145 16 17, 16 16 32 22 11 11 12 1 21/20 13 1 HPT146 16 14, 13 15 30 25 12 11 13 1 22/21 14 1 HPT147 16 14, 13 13 29 23 11 11 14 1 20/19 13 1 HPT148 16 11, 11 15 32 24 12 12 13 1 22/18 13 1 HPT149 15 14, 11 14 30 25 11 12 12 1 22/18 13 1 HPT150 15 17.3, 13 14 30 24 11 11 12 1 22/21 17 1 HPT151 15 14, 13 13 30 24 12 12 13 1 18/17 16 1 HPT152 16 14, 12 13 32 25 11 11 12 1 19/18 15 1 HPT153 15 14, 11 15 31 25 12 12 12 1 18/18 15 1 HPT154 15 15, 10 11 29 25 11 11 13 1 20/18 15 1 HPT155 16 16, 15 15 32 23 11 11 14 1 20/19 13 1 HPT156 16 16, 13 14 32 25 11 11 12 1 21/18 15 1 HPT157 16 14, 12 13 30 24 11 12 12 1 21/18 18 1 HPT158 14 15, 14 13 30 25 11 11 13 1 21/19 13 1 HPT159 16 13, 11 14 29 24 11 11 12 1 20/19 15 1 HPT160 15 16, 12 13 30 23 11 11 12 1 21/20 14 1 HPT161 14 15, 13 13 30 24 11 11 14 1 22/19 14 1 HPT162 14 14, 14 13 30 25 10 11 13 1 22/19 13 1 HPT163 14 15, 13 13 28 23 11 15 13 1 19/19 13 1  A. Rüstemoglu et al. / American Journal of Molecular Biology 1 (2011) 114-121 Copyright © 2011 SciRes. AJMB 11 9 HPT164 16 14, 12 13 30 25 10 11 13 1 20/20 13 1 HPT165 17 13, 12 14 30 26 11 11 13 1 23/19 13 1 HPT166 14 16, 13 14 30 25 11 11 13 1 22/19 13 1 HPT167 16 15, 8 13 28 23 11 14 12 1 21/19 13 1 HPT168 17 13, 13 13 29 24 9 12 11 1 22/20 13 1 HPT169 15 13, 11 14 30 24 13 11 12 1 21/19 13 1 HPT170 15 16, 12 13 32 24 13 11 12 1 23/19 15 1 HPT171 17 11, 10 13 28 27 12 13 13 1 23/19 13 1 HPT172 16 13, 13 11 28 25 12 13 13 1 22/20 13 1 HPT173 15 15, 11 14 33 26 12 13 12 1 22/19 13 1 HPT174 16 14, 8 13 28 23 12 15 12 1 21/20 13 1 HPT175 14 15, 14 11 28 25 11 13 12 1 22/19 13 1 HPT176 13 14, 12 13 31 23 11 13 12 1 22/19 16 1 HPT177 14 13, 10 13 28 26 11 14 12 1 23/19 13 1 HPT178 15 20, 15 12 28 23 9 15 11 1 20/19 13 1 HPT179 16 15, 13 14 28 25 12 13 13 1 23/22 14 1 HPT180 14 20, 14 14 31 23 12 16 14 1 22/19 13 1 HPT181 16 19, 14 12 29 23 11 15 11 1 21/17 13 1 HPT182 15 12, 9 13 27 26 11 15 13 1 18/18 13 1 HPT183 16 14, 13 12 27 23 11 14 14 1 19/19 13 1 HPT184 15 15, 10 14 30 24 11 13 14 1 23/21 13 1 HPT185 16 10, 10 13 31 24 11 13 14 1 24/23 12 1 HPT186 16 14, 10 13 27 23 12 13 14 1 24/21 13 1 HPT187 15 12, 10 12 26 26 11 11 13 1 HPT188 16 12, 9 15 33 26 12 11 14 1 HPT189 14 12, 9 14 29 25 11 13 12 1 HPT190 15 15, 12 15 30 24 11 15 14 1 HPT191 16 13, 11 14 30 24 11 11 12 1 HPT192 14 12, 11 13 29 23 12 14 13 1 HPT193 16 12, 10 12 30 23 13 13 12 1 HPT194 15 13, 11 14 32 24 10 13 12 1 HPT195 14 14, 10 13 33 25 11 15 12 1 HPT196 15 17, 13 13 30 23 13 13 12 1 HPT197 14 15, 11 13 32 24 11 13 13 1 HPT198 15 16, 9 14 31 24 11 14 12 1 HPT199 15 14, 8 13 30 21 12 12 13 1 HPT200 16 16, 8 13 30 25 10 12 13 1 HPT201 16 16, 13 12 31 23 11 12 14 1 HPT202 15 14, 10 12 29 22 12 14 12 1 HPT203 16 14, 8 13 32 23 11 12 14 1 HPT204 16 14, 11 14 32 20 11 12 15 1 HPT205 17 11, 8 13 30 25 12 12 13 1 HPT206 14 19, 16 14 31 24 11 12 13 1 HPT207 15 14, 11 13 30 25 11 12 12 1 HPT208 14 17, 10 12 30 24 11 12 12 1 *Haplotype containing 10 Y-STR loci; **Haplotype containing 8 Y-STR loci; ***Number of individuals bearing each haplotype. Table 3. Comparative presentation of haplotype numbers, gene diversities, average gene diversities and Pd combined values be- longing to three haplotypes surveyed in this study. Haplotypes n Gene diversityAverage gene diversity per locusPDc* Ht10 186 1.0000 ± 0.00060.7476 0.9999989 MinHt 208 1.0000 ± 0.00050.7518 0.9999869 aHt 186 1.0000 ± 0.00060.7834 0.9999936 *Combined Power of Discrimination. (n = 166) [26], Czech (n = 50) [27], German (n = 166), Indian (n = 108), Mozambican (n = 112) [12], Japanese (n = 161) [28], Turkish (Antalya) (n = 210) [29], and Turkish (n = 280) [30]. Haplotypic comparisons have highlighted that no significant difference has been ob- served with Czech population (p > 0.05) while compari- sons with all other populations have produced statisti- cally significant differences from Turkish population in this study (p < 0.01), as shown in Table 4. Our results suggest that an alternative haplotype (aH t), which differs slightly from minHt in respect of its Y-STR loci contents, maybe alternative for minHt in Turkish population. aHt has included the selected 8 Y-STR loci: DYS19, DYS385a/b, DYS389I, DYS389II, DYS390, DYS392, DYS393, and YCAIIa/b. The only difference between the minHt and the proposed aHt is the inclusion of YCAIIa/b locus in place of DYS391 locus in aHt due to its higher gene diversity value of 0.8087 as compared with 0.6101 of DYS391 locus (Table 1). We have found 186 unique haplotype in aHt (data not shown). The aHt has reflected a better Pd combined value when compared with minHt (0.9999936 vs 0.9999869) and, has higher average gene diversity per locus (0.7834 vs 0.7518) (Table 3). The data has exhibited that aHt has a higher discriminatory potential than that of minHt.  A. Rüstemoglu et al. / American Journal of Molecular Biology 1 (2011) 114-121 Copyright © 2011 SciRes. AJMB 120 Table 4. Exact test P values of populational genetic differentiation measures based on minHt haplotype frequencies between all pairs involving samples of eight populations and Turkish population in this study. Population Turkish (This study)Croatian CzechGermanIndianMozambican Japanese Turkish Turkish (Antalya) Turkish (This study) - Czech27 0.32474 ±0.0283 - Mozambican12 0.0000 0.00103 ±0.0010 - Croatian26 0.0000 0.0000 0.0000- Indian12 0.0000 0.0000 0.00000.0000 - Japanese28 0.00205 ±0.0017 0.01176 ±0.0026 0.00000.0000 0.0000- German12 0.0000 0.0000 0.00000.0000 0.00000.0000 - Turk ish30 0.0000 0.0000 0.00000.0000 0.00000.0000 0.0000 - Turkish (Antalya)29 0.0000 0.0000 0.00000.0000 0.00000.0000 0.0000 0.0000 - In conclusion this study results have yielded sufficient evidence that aHt can reliably be proposed as an alterna- tive to minHt in paternity testing and forensic medicine applications for Turkish population. Our data have also provided additional information to the framework of variation involving 10 Y-STR loci as well as a further contribution to the Y-STR database for Turkish popula- tion. 4. ACKNOWLEDGEMENTS This study was supported by Ankara University Scientific Research Projects with the project # 2005-08-09-012 HPD and approved by the Ankara University, Research Ethics Committee of Medical Faculty with the approval # 57-1393. This study was performed in Ankara University, Medical Faculty, Medical Biology Department. REFERENCES [1] de Knijff, P. (2000) Messages through bottlenecks: On the combined use of slow and fast evolving polymorphic markers on the human Y chromosome. The American Journal of Human Genetics, 67, 1055-1061. [2] Spurdle, A.B., Woodfield, D.G., Hammer, M.F. and Jenkins, T. (1994) The genetic affinity of Polynesians: Evidence from Y chromosome polymorphisms. Annals of Human Genetics, 58, 251-263. doi:10.1111/j.1469-1809.1994.tb01889.x [3] Dupuy, B.M., Stenersen, M., Egeland, T. and Olaısen, B. (2004) Y-chromosomal microsatellite mutation rates: Differences in mutation rate between and within loci. Hum Mut, 23, 117-124. doi:10.1002/humu.10294 [4] Gill, P., Brenner, C., Brinkmann, B., Budowle, B., Carracedo, A., Jobling, M.A., de Knijff, P., Kayser, M., Krawczak, M., Mayr, W.R., Morling, N., Olaisen, B., Pascali, V., Prinz, M., Roewer, L., Schneider, P.M., Sajantila, A. and Tyler-Smith, C. (2001) DNA Commi- ssion of the International Society of Forensic Genetics: Recommendations on forensic analysis using Y-chromo- some STRs. Forensic Science International, 124, 5-10. doi:10.1016/S0379-0738(01)00498-4 [5] Betz, A., Bäbler, G., Dietl, G., Syeil, X., Weyermann, G. and Pflug, W. (2001) DYS STR analysis with epithelial cells in a rape case. Forensic Science International, 118, 126-130. doi:10.1016/S0379-0738(00)00482-5 [6] Corach, D., Figueira, R.L., Marino, M., Penacino, G. and Sala, A. (2001) Routine Y-STR typing in forensic case- work. Forensic Science International, 118, 131-135. doi:10.1016/S0379-0738(00)00483-7 [7] Dekairelle, A.F. and Hoste, B. (2001) Application of a Y-STR-pentaplex PCR (DYS19, DYS389I and II, DYS390 and DYS393) to sexual assault cases. Forensic Science International, 118, 122-125. doi:10.1016/S0379-0738(00)00481-3 [8] Gill, P., Werrett, D.J., Budowle, B. and Guerrieri, R. (2004) An assessment of whether SNPs will replace STRs in national DNA databases—joint considerations of the DNA working group of the European Network of Forensic Science Institutes (ENFSI) and the Scientific Working Group on DNA Analysis Methods (SWGDAM). Science & Justice, 44, 51-3. doi:10.1016/S1355-0306(04)71685-8 [9] Honda, K., Tun, Z., Young, D. and Terao, T. (2001) Examination of Y-STR mutations in sex chromosomal abnormality in forensic case. Forensic Science Interna- tional, 118, 136-140. doi:10.1016/S0379-0738(00)00484-9 [10] Jobling, M.A., Pandya, A. and Tyler-Smith, C. (1997) The Y chromosome in forensic analysis and paternity testing. International Journal of Legal Medicine, 110, 118-124. doi:10.1007/s004140050050 [11] Kayser, M. and Sajantila, A. (2001) Mutations at Y-STR loci: Implications for paternity testing and forensic analy- sis. Forensic Science International, 118, 116-121. doi:10.1016/S0379-0738(00)00480-1 [12] Alaves, C., Gusmao, L., Barbosa, J. and Amorim, A. (2003) Evaluating the informative power of Y-STRs: A comparative study using European and new African haplotype data. Forensic Science International, 134, 126- 133. doi:10.1016/S0379-0738(03)00127-0 [13] Gusmao, L., Sanchez-Diz, P., Alves, C., Beleza, S., Lopes, A., Carracedo, A. and Amorim, A. (2003) Group- ing of Y-STR haplotypes discloses European geografic clines. Forensic Science International, 134, 172-179. doi:10.1016/S0379-0738(03)00160-9 [14] Koyama, H., Iwasa, M., Maeno, Y., Tsuchimochi, T.,  A. Rüstemoglu et al. / American Journal of Molecular Biology 1 (2011) 114-121 Copyright © 2011 SciRes. AJMB 121 Isobe, I., Yoshimi, S.N., Jun, M.O., Matsumoto, T., Horii, T. and Nagao, M. (2001) Y-chromosomal STR haplotype in the Japanese population. Forensic Science Interna- tional, 124, 221-223. doi:10.1016/S0379-0738(01)00572-2 [15] Nakagome, Y., Young, S.R., Akane, A., Numabe, H., Jin, D.K., Yamori, Y., Seki, S., Tamura, T., Nagafuchi, S., Shiono, H. and Nakahori, Y. (1992) A Y-associated alele may be characteristic of cetain ethnic groups in Asia. Annals of Human Genetics, 56, 311-314. doi:10.1111/j.1469-1809.1992.tb01158.x [16] Rodig, H., Grum, M. and Grimmecke, H.-D. (2007) Population study and evaluation of 20 Y-chromosome STR loci in Germans. International Journal of Legal Medicine, 121, 24-27. doi:10.1007/s00414-005-0075-5 [17] Rustamov, A., Gümüş, G., Karabulut, H.G., Elhan, A.H., Kadıkıran, A. and Bökesoy, I. (2004) Y-STR polymor- phism in Central Anatolian region of Turkey. Forensic Science International, 139, 227-230. doi:10.1016/j.forsciint.2003.11.009 [18] Yan, J., Tang, H., Liu, Y., Jing, Y., Jiao, Z., Zhang, Q., Gao, J., Shang, L., Guo, H. and Yu, J. (2007) Genetic polymorphisms of 17 Y-STRs haplotypes in Chinese Han population residing in Shandong province of China. Legal Medicine, 9, 196-202. doi:10.1016/j.legalmed.2006.11.016 [19] Park, M.J., Lee, H.Y, Chung, U., Kang, S.-C. and Shin, K.-J. (2007) Y-STR analysis of degraded DNA using reduced-size amplicons. International Journal of Legal Medicine, 121, 152-157. doi:10.1007/s00414-006-0133-7 [20] Willuweit, S. and Roewer, L. (2007) Y chromosome haplotype reference database (YHRD): Update. Forensic Science International, 1, 83-87. doi:10.1016/j.fsigen.2007.01.017 [21] YHRD-Y Chromosome Haplotype Reference Database. (2011). http://www.yhrd.org [22] Sambrook, J. and Russell, D. (2001) Molecular Cloning: A Laboratory Manual. 3rd Edition, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, 543-554. [23] Santos, F.R., Pena, D.J. and Epplen, J.T. (1993) Genetic and population study of a Y-linked tetranucleotide repeat DNA polymorphism with a simple non-isotopic techni- que. Human Genetics, 90, 655-656. doi:10.1007/BF00202486 [24] Nei, M. (1996) Phylogenetic analysis in molecular evolu- tionary genetics. Annual Review of Genetics, 30, 371-403 doi:10.1146/annurev.genet.30.1.371 [25] Kimberly, A.H. (2001) Statistical analyses of STR data profiles in DNA. http://promega.de/profiles/103/Profile-sinDNA_103_14. pdf. [26] Ljubkovic, J., Stripisic, A., Sutlovic, D., Definis-Goja- novic, M., Bucan, K. and Andelinovic, S. (2008) Y-chro- mosomal short tandem repeat haplotypes in southern croatian male population defined by 17 loci. Croatian Medical Journal, 49, 201-206. doi:10.3325/cmj.2008.2.201 [27] Ehler, E., Marvan, R. and Vanek, D. (2010) Evaluation of 14 Y chromosomal short tandem repeat haplotype with focus on DYS449, DYS456, and DYS458: Czech population sample. Croatian Medical Journal, 51, 54-60. doi:10.3325/cmj.2010.51.54 [28] Hara, M., Kido, A., Takada, A., Adachi, N. and Saito, K. (2007) Genetic data for 16 Y-chromosomal STR loci in Japanese. Legal Medicine, 9, 161-170. doi:10.1016/j.legalmed.2006.11.002 [29] Demirçin, S. and Timur, S. (2010) Haplotype analysis of 8 Y-STR loci in the Antalya population. Romanian Journal of Legal Medicine, 18, 51-58. [30] Henke, J., Henke, L., Chatthopadhyay, P., Kayser, M., Dulmer, M., Cleef, S., Poche, H. and Felske, Z.H. (2001) Application of Y-chromosomal STR haplotypes to forensic genetics. Croatian Medical Journal, 42, 292- 297.
|