 American Journal of Molecular Biology, 2011, 1, 52-61 doi:10.4236/ajmb.2011.12007 Published Online July 2011 (http://www.SciRP.org/journal/ajmb/ AJMB ). Published Online July 2011 in SciRes. http://www.scirp.org/journal/AJMB Characterization of Asian and North American avian H5N1 Wei Hu Department of Computer Science, Houghton College, New York, USA. E-mail: wei.hu@houghton.edu Received 15 March 2011; revised 13 May 2011; accepted 30 May 2011. ABSTRACT Since the emerge of the highly pathogenic avian H5N1 virus in Asia in 1996, the possibility for this virus to cross species barriers to infect humans and its ability to cause large outb reaks in birds have been a public health concern. This virus has been spread- ing from Asia to Europe and Africa by migratory birds with North America as its next possible stop. In this study, an ensemble of computational techniques including Random Forests, Informational Spectrum Method, Entropy, and Mutual Information were em- ployed to unravel the distinct characteristics of Asian and North American avian H5N1 in comparison with human and swine H5N1. Critical differences were identified in the HA cleavage and binding sites, the HA receptor selection, the interaction patterns of HA and NA, and NP, PA, PB1, and PB2, and the impor- tant sites in the influenza proteins including HA, NA, M1, M2, NS1, NS2, NP, PA, PB1, PB1-F2, and PB2. Keywords: H5N1; Hemagglutinin, Influenza; Informational Spectrum Method; Mutation; Random Forests; Receptor Binding Specificity 1. INTRODUCTION Wild birds are a natural reservoir of all known influenza A subtypes, and many of these viruses cause only mild symptoms in birds. The highly pathogenic avian H5N1 virus was first detected in China in 1996 [1] and was also the cause of two subsequent outbreaks in migratory birds at Qinghai Lake in China in 2005 and 2006. The first group of H5N1 human infections were reported in Hong Kong in 1997 [2]. However, there is no evidence of efficient human-to-human transmission of the highly pathogenic avian H5N1 virus. This virus has spread to other Asian countries including Indonesia, Japan, Korea, Thailand, Vietnam, and Malaysia, and most recently to Europe and Africa. Therefore, there is a growing concern over the potential for migratory birds to introduce the highly pathogenic Asian H5N1 strain into North Amer- ica. A strategic plan was developed for the early detec- tion of highly pathogenic avian H5N1 in the United States [3]. In addition to the possible spread of highly pathogenic avian H5N1 by migratory birds, viral mutations and re- assortment of avian, human and swine viruses could generate a new strain capable of transmission among humans. In particular, swine can serve as a “mixing ves- sel” in the generation of a novel virus, because they are susceptible to infection with both avian and human vi- ruses. A recent report [4] showed that swine H1N1, H1N2, and H3N2 viruses are currently co-circulating in China, and the highly pathogenic avian H5N1 virus might be able to contribute genes to swine H3N2 virus, demonstrating the continued risk for further reassortment of swine virus and continued spread of pandemic 2009 H1N1 virus worldwide. Another fear is that if swine can carry both H5N1 and 2009 H1N1, the viruses can com- bine and mutate into a novel strain of high virulence that can transmit efficiently among humans. The influenza A virus genome encodes for 11 genes. Four proteins, HA, NS1, PB1-F2, and PB2, are recog- nized as major determinants for pathogenicity of influ- enza, with PB1-F2 as the most recently discovered pro- tein. Although the host barrier for avian viruses to spread in humans is multigenic, the receptor binding specificity of HA is a major obstacle for direct transmission of avian viruses to humans. In general, human influenza viruses tend to bind to SA α2, 6Gal receptors, whereas avian viruses favor SA α2, 3Gal receptors. Adaptation of avian virus to humans likely requires a shift in receptor binding specificity of the virus from avian-type to hu- man-type. In humans, the SA α2, 6Gal receptor is ex- pressed mainly in the upper airway, but the SA α2, 3Gal receptor is expressed in alveoli and the terminal bron- chiole [5]. Clinical data illustrated that an influenza virus that could bind to both SA α2, 3Gal and SA α2, 6Gal receptors is highly pathogenic. The receptor binding site of HA comprises three sec-  W. Hu / American Journal of Molecular Biology 1 (2011) 52-61 53 ondary structure elements: the 190 helix (residues 190 - 198), the 130 loop (residues 135 - 138) and the 220 loop (residues 221 - 228) that form the sides of the site with the base made up of the conserved residues Tyr 98, Trp 153, His 183 and Tyr 195 (H3 numbering) [6]. The re- ceptor binding affinity of HA could be altered by several key residues, and a wide variety of such mutations have been identified. A single mutation D225G changed the binding of one strain of 1918 H1N1 from pure human- type binding to human and avian types (dual binding) [7]. Amino acids at positions 226 and 228 (H3 numbering) could affect binding preference of several subtypes in- cluding H2, H3, H4 and H9, and on the other hand the changes at positions 190 and 225 could influence H1 subtype. Computing modeling also indicated that HA amino acid residues, Tyr 98, Val 135, Ser 136, Ser 137, Trp 153, Ile 155, His 183, Glu 190, Leu 194, and Gln 226, are important for the avian-type binding of H5N1 HA, with Gln 226 as a very critical residue in this regard. Further, amino acid residues, Leu 133, Val 135, Trp 153, Ile 155, Glu 190, and Lys 193, are important for the hu- man-type binding of H5N1 HA [8]. Interestingly, a bio- informatics approach, termed informational spectrum method (ISM), was applied to the HA1 domain of HA in the study of its receptor binding specificity [9-13]. Besides the role of HA plays in host range restriction, the cooperative contributions to human adaptation from other proteins of avian and swine influenza were also investigated. Support vector machines, entropy, and mu- tual information were utilized to uncover the unique mo- lecular features of these viruses [14-22]. Most recently, Random Forests [23] were applied successfully to tackle the same problem, where novel host markers in the pro- teins and genes of pandemic 2009 H1N1 were identified [24,25]. These findings highlighted that host adaptation of influenza viruses is a complex and polygenic trait. While there have been extensive studies on host markers in general influenza species including avian and swine viruses, the purpose of this study was to narrow the fo- cus by identifying the distinct characteristics of the highly pathogenic avian H5N1 from Asia and the low pathogenic avian H5N1 from North America, in com- parison with human and swine H5N1. 2. MATERTIALS AND METHDOS 2.1. Sequence Data The sequences of influenza were retrieved from the In- fluenza Virus Resource of the National Center for Bio- technology Information (NCBI). Only the full length and unique sequences were selected. All sequences used in this study were aligned with MAFFT [26]. 2.2. Informational Spectrum Method The informational spectrum method (ISM) is a bioin- Ta b l e 1 . Characteristic IS frequencies of HA proteins in 2009 H1N1, swine H1N1/H1N2, avian H1N1, and A/South Caro- lina/1/18 (H1N1). Subtype 2009 H1N1 Swine H1N2/H1N1 Avian H5N1 Human H1N1 A/South Caro- lina/1/18 (H1N1) Frequency F(0.295) F(0.055)F(0.076)F(0.236)F(0.258) formatics technique that can be used to analyze protein sequences [27]. The idea is to translate the protein se- quences into numerical sequences based on electron-ion interaction potential (EIIP) of each amino acid. Then the Discrete Fourier Transform (DFT) can be applied to these numerical sequences, and the resulting DFT coef- ficients are used to produce the energy density spectrum. The informational spectrum (IS) comprises the frequen- cies and the amplitudes of this energy density spectrum. According to the ISM theory, the peak frequencies of IS of a protein sequence reflect its biological or biochemi- cal functions. The ISM was successfully applied to quantify the effects of HA mutations on the receptor binding preference in [10] and reveal the change of re- ceptor binding selection caused by the mutations identi- fied in [28] and [12]. Table 1 shows several common IS frequencies identified in [12,13]. It was observed in [11] that some of the influenza strains displayed dual HA receptor binding preference. Consequently, in this study we used top two IS frequen- cies, one primary and one secondary, to describe the HA receptor selection. 2.3. Entropy and Mutual Information In information theory [29,30], entropy is a measure of the uncertainty associated with a random variable. Let x be a discrete random variable that has a set of possible values {a1, a2, a3,…, an} with probabilities{p1, p2, p3,…, pn} where P (x = ai) = pi. The entropy H of x is log ii i xp p The mutual information of two random variables is a quantity that measures the mutual dependence of the two variables or the average amount of information that x conveys about y, which can defined as , , xyHx Hy Hxy where H(x) is the entropy of x, and H(x, y) is the joint entropy of x and y. I(x, y) = 0 if and only if x and y are independent random variables. In the current study, each of the n columns in a multi- ple sequence alignment of a set of influenza protein se- quences of length N is considered as a discrete random variable xi (1 ≤ i ≤N) that takes on one of the 20 (n = 20) C opyright © 2011 SciRes. AJMB  W. Hu / American Journal of Molecular Biology 1 (2011) 52-61 54 amino acid types with some probability. H(xi) has its Table 2. Amino acids near and at the cleavage site in HA con- sensus sequences of different origins. Virus Cleavage site Number of basic amino acids Human H1N1 NIPS – – – – IQSRGLF 1 2009 H1N1 NVPS – – – – IQSRGLF 1 A/goose/Guandong/1/96 (H5N1) NTPQRERRRKKRGLF 7 North American avian H5N1 NVPQ – – – – RETRGLF 2 2010: Asian avian H5N1 NSPQREGRRRKRGLF 6 2009: Asian avian H5N1 NSPQRERRRRKRGLF 7 2008: Asian avian H5N1 NSPQRERRRKKRGLF 7 2007: Asian avian H5N1 NSPQRERRRKKRGLF 7 2006: Asian avian H5N1 NSPQRERRRKKRGLF 7 2005: Asian avian H5N1 NSPQRERRRKKRGLF 7 2004: Asian avian H5N1 NSPQRERRRKKRGLF 7 2010: Human H5N1 NSPQGERRRKKRGLF 6 2009: Human H5N1 NSPQGERRRKKRGLF 6 2008: Human H5N1 NSPLRERRRKKRGLF 7 2007: Human H5N1 NSPQRERRRKKRGLF 7 2006: Human H5N1 NSPQRESRRKKRGLF 6 2005: Human H5N1 NSPQRERRRKKRGLF 7 2004: Human H5N1 NSPQRERRRKKRGLF 7 minimum value 0 if all the amino acids at position i are the same, and achieves its maximum if all the 20 amino acid types appear with equal probability at position i, which can be verified by the Lagrange multiplier tech- nique. A position of high entropy means that the amino acids are often varied at this position. While H (xi) measures the genetic diversity at position i in our current study, I (x, y) measures the correlation between amino acid substitutions at positions i and j. 3. RESULTS 3.1. Cleavage Site in HA Avian H5N1 can be divided into two groups, highly and low pathogenic viruses based on their difference in viru- lence. The HA protein playing a key role in pathogenic- ity has two domains, HA1 and HA2, which are cleaved from their precursor HA0 by cellular proteases. Nor- mally, mammalian and low pathogenic avian viruses carry an HA cleavage site with a monobasic motif, whereas high pathogenic avian viruses possess a K/R polybasic HA cleavage site, which is hydrolyzed by a broad range of proteases in the host cells. Therefore, the polybasic HA cleavage site is a salient virulence feature. Removal of the polybasic HA cleavage site results in a drastic decrease in pathogenicity. However, introduction of a polybasic motif into the HA cleavage site of a low pathogenic avian strain might or might not transform it into a highly pathogenic strain, indicating the existence of additional virulence determinants in HA or other pro- teins [31]. It turned out the amino acids V346 and S346 (323 in H3 numbering) adjacent to the cleavage site played a central role in virulence as well [32] (Table 2), which demonstrated experimentally that the presence of V346 reduced the virulence whereas S346 produced the opposite results. Clearly, North America avian H5N1 had V346 and its Asian counterpart had S346 (Table 2), in addition to the difference in their cleavage site. 3.2. Receptor Binding Specificity 3.2.1. Mutations in HA Capable of Changing Binding Preference It is well established that avian viruses will have to ac- quire human-type receptor preference for sustained rep- lication and transmission in humans. The receptor bind- ing affinity is primarily determined by the amino acids at the receptor binding domain (RBD) along with other critical sites [9-11]. According to [33], the amino acids at the sites implicated in receptor specificity were listed in Ta b le 3 (H3 numbering), which showed that the amino acid differences between avian, human, and swine H5N1 only occurred at sites 193 and 216, and their key sites 190E and 225G retained the avian-type binding prefer- ence. This finding supported that viruses with avian spe- cific receptor binding properties could replicate and cause infection in humans and swine, but could not do so efficiently. To offer a comparison with human viruses, the related amino acids in the human H1N1 HA consen- sus sequence from 1918 to 2008 was added to the table. H5 HA mutation K193R increased the human-type bin- ding, however the virus with mutation R216K or S227N did not bind to human-type receptor [34]. The frequent occurrences of mutations K193R and R216K observed in Ta b le 3 implied the inclination of H5N1 to increase its human-type binding, which could be a concern. North American avian H5N1 had E216 and P221, but Asian avian, human, and swine H5N1 all had K(R) 221 and S221. Considering the corresponding amino acids at the same sites in human H1N1 HA, the two amino acids E216 and P221 in North American avian H5N1 appeared to favor human-type binding. In addition to the critical mutations discussed above, avian H5N1 HA mutations L133V, A137V [35], N186K, Q196R [36], E190D, G225D [37], G228S [38] were shown to enhance its human-type binding, and mutations Q226L and G228S to reduce its avian-type binding af- finity [35], and mutation S227N to reduce its avian-type binding and increase its human-type binding affinity [39]. Even though pandemic 2009 H1N1 was known for its efficient transmission among humans, its HA was of classical swine lineage [40]. Its HA carried D190, E216, P221, D225, and E227 (Table 3), the first two were a feature for avian-type binding whereas the second two C opyright © 2011 SciRes. AJMB 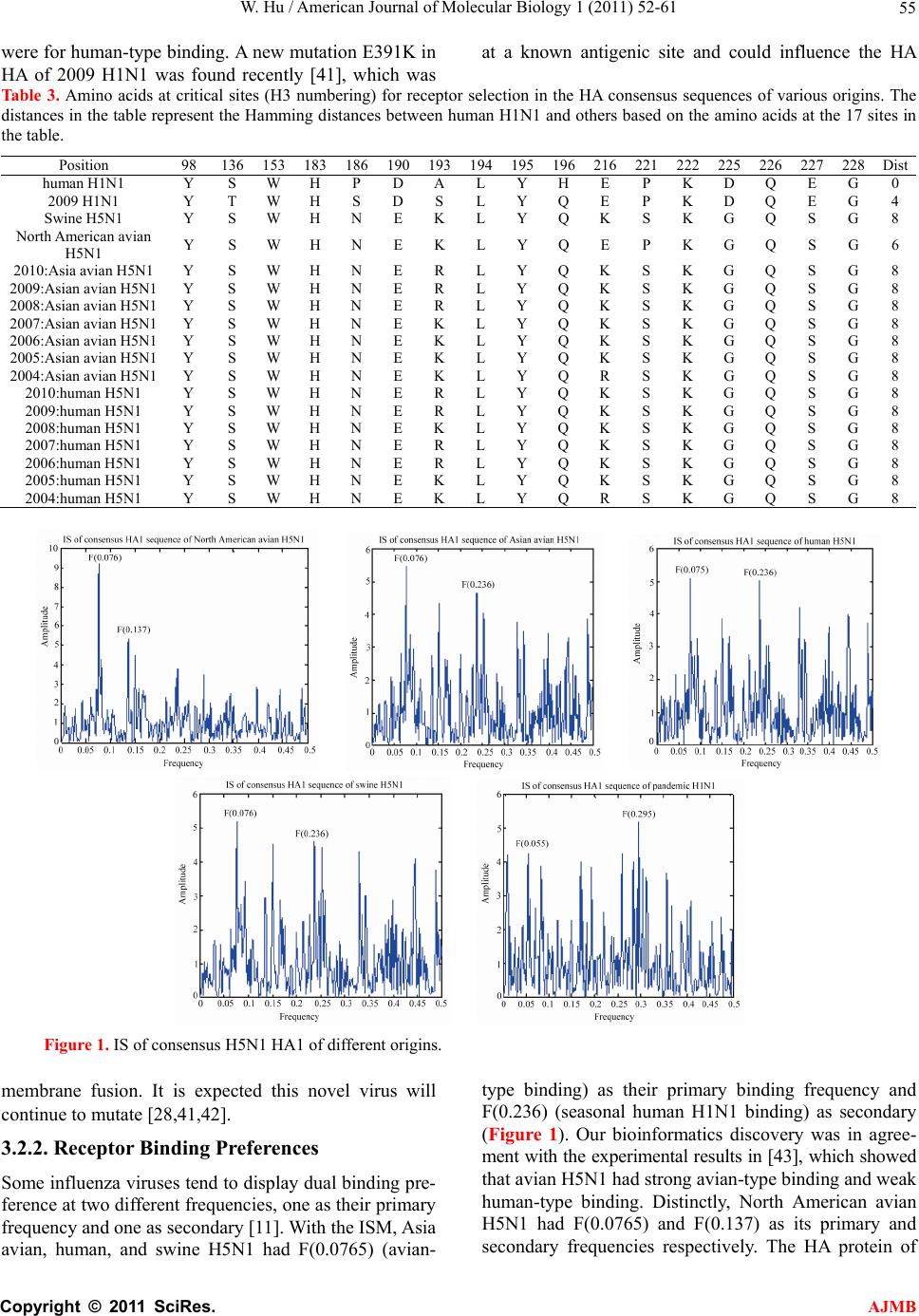 W. Hu / American Journal of Molecular Biology 1 (2011) 52-61 Copyright © 2011 SciRes. 55 were for human-type binding. A new mutation E391K in HA of 2009 H1N1 was found recently [41], which was at a known antigenic site and could influence the HA Ta ble 3. Amino acids at critical sites (H3 numbering) for receptor selection in the HA consensus sequences of various origins. The distances in the table represent the Hamming distances between human H1N1 and others based on the amino acids at the 17 sites in the table. Position 98 136 153 183 186 190 193 194 195 196216 221 222 225 226 227 228Dist human H1N1 Y S W H P D A L Y H E P K D Q E G 0 2009 H1N1 Y T W H S D S L Y Q E P K D Q E G 4 Swine H5N1 Y S W H N E K L Y Q K S K G Q S G 8 North American avian H5N1 Y S W H N E K L Y Q E P K G Q S G 6 2010:Asia avian H5N1 Y S W H N E R L Y Q K S K G Q S G 8 2009:Asian avian H5N1 Y S W H N E R L Y Q K S K G Q S G 8 2008:Asian avian H5N1 Y S W H N E R L Y Q K S K G Q S G 8 2007:Asian avian H5N1 Y S W H N E K L Y Q K S K G Q S G 8 2006:Asian avian H5N1 Y S W H N E K L Y Q K S K G Q S G 8 2005:Asian avian H5N1 Y S W H N E K L Y Q K S K G Q S G 8 2004:Asian avian H5N1 Y S W H N E K L Y Q R S K G Q S G 8 2010:human H5N1 Y S W H N E R L Y Q K S K G Q S G 8 2009:human H5N1 Y S W H N E R L Y Q K S K G Q S G 8 2008:human H5N1 Y S W H N E K L Y Q K S K G Q S G 8 2007:human H5N1 Y S W H N E R L Y Q K S K G Q S G 8 2006:human H5N1 Y S W H N E R L Y Q K S K G Q S G 8 2005:human H5N1 Y S W H N E K L Y Q K S K G Q S G 8 2004:human H5N1 Y S W H N E K L Y Q R S K G Q S G 8 Figure 1. IS of consensus H5N1 HA1 of different origins. membrane fusion. It is expected this novel virus will continue to mutate [28,41,42]. 3.2.2. Receptor Binding Preferences Some influenza viruses tend to display dual binding pre- ference at two different frequencies, one as their primary frequency and one as secondary [11]. With the ISM, Asia avian, human, and swine H5N1 had F(0.0765) (avian- type binding) as their primary binding frequency and F(0.236) (seasonal human H1N1 binding) as secondary (Figure 1). Our bioinformatics discovery was in agree- ment with the experimental results in [43], which showed that avian H5N1 had strong avian-type binding and weak human-type binding. Distinctly, North American avian H5N1 had F(0.0765) and F(0.137) as its primary and secondary frequencies respectively. The HA protein of AJMB  W. Hu / American Journal of Molecular Biology 1 (2011) 52-61 56 North America avian H5N1 didn’t carry the highly pa- thogennic avian signature amino acids RERR at the its cleavage site. Adding these four amino acids to the clea- vage site of North America avian viruses didn’t change Table 4. Hamming distances between the concatenated protein sequences of H5N1 of different origins. Due to limited number of sequences for Swine (“S”) and North American H5N1 (“N”), we formed one consensus from different years. Asian H5N1 (“A”) had seven consensuses by year (2004-2010), while hu- man H5N1 (“H”) had five consensuses by year (2004-2008). Year\Dist (N, A) (S, A) (N, H) (S, H) (A, H) 2010 240 112 2009 197 110 2008 186 44 200 65 40 2007 186 28 206 56 50 2006 163 61 214 35 88 2005 162 48 196 32 51 2004 171 68 195 47 28 their primary and secondary frequencies, implying that these amino acids were not relevant for HA binding. As a comparison, the primary F(0.295) (human-type) and the secondary F(0.055) (swine-type) frequencies of 2009 H1N1 were included in Figure 1. This kind of subtle difference in receptor binding pre- ference as reflected in the secondary frequency was also observed in the two clusters of 2009 H1N1 found in [44], where both of them shared the same primary binding frequency while differed in their secondary frequency. One had swine secondary frequency and the other had 1918 Spanish flu frequency [45]. As a whole group, the 2009 H1N1 HA sequences showed its swine-type recep- tor selection in the early months of its run in 2009, but this selection gradually disappeared in the late months [28]. As an extension of the work in [28], a recent report using affinity propagation identified six clusters of 2009 H1N1 HA sequences collected from May 2010 to Feb- ruary 2011 and found the HA receptor preference of each cluster [46]. 3.3. Hamming Distances between H5N1 Sequences of Different Origins One easy way to compare two sequences is to calculate their Hamming distance. To present a holistic view for the similarities of influenza sequences under the current study, we concatenated the consensus sequences of dif- ferent proteins from each species including HA, NA, M1, M2, NS1, NS2, NP, PA, PB1, PB1-F2, and PB2. Because there were many Asian avian H5N1 sequences available, the consensus was formed by year. Table 4 presents Hamming distances of consensus sequences of different origins. When no sequences were available in a particular year, we left the table entry empty in that year. The distances between Asian avian, human, and swine H5N1 were close to each other. However, there was a clear increase in the distances of swine and Asian avian H5N1 in 2009 and 2010. Unfortunately, there were no data for distances of swine and human as well as swine and Asian avian H5N1 in 2009 and 2010. Finally, the distances from North American avian H5N1 to other species were the largest in Table 4. In summary, human and swine H5N1 sequences were closer to Asian avian H5N1 than to North American avian H5N1. 3.4. Characteristic Sites between Asian and North American Avian H5N1 Here we first summarized some amino acid differences in HA, NA, NS1, and PB2 between Asian and North American avian H5N1 because of their crucial functions in the pathogenicity of influenza. As discussed in section 3.1, Asian avian H5N1 HA possessed a series of basic amino acids (RRKKR) at the cleavage site, a determi- nant of high pathogenicity for avian virus and a marker its North American counterpart lacked. In addition, all five consensuses of human H5N1 NS1 (2004-2008) car- ried a deletion of 5 amino acids at positions 80 - 84 and all swine H5N1 strains but one had this deletion, while Asian avian H5H1 contained this deletion in 2007, 2008, and 2010. In contrast, North American avian H5H1 had full length NS1. This deletion in NS1 seems to increase pathogenicity in chickens and mice [47]. Large-scale sequence analysis of avian viruses identified a PDZ li- gand domain of the X-S/T-X-V type at the C-terminus of NS1. Typically, highly pathogenic H5N1 NS1 has ESEV or EPEV while most low pathogenic human viruses contain a different motif RSKV or RSEV, which cannot bind PDZ-containing proteins. Most of the Asian and North American avian, human, and swine H5N1 strains in our dataset had an avian-type motif ESEV All North American avian and swine H5N1 strains had E at PB2 627, but both human and Asian avian H5N1 had a mixture of E and K at 627. In general, 627K is a marker for human influenza and 627E is for avian viruses. PB2-627K confers to avian H5N1 the advantage of efficient growth in the upper and lower respiratory tracts of mammals [48]. Many Asian avian, human, and swine H5N1 strains carried a deletion of 20 amino acids in the NA stalk (re- sidues 49 - 68), which is also a indicator for high viru- lence. But only two of the North American avian strains contained a deletion of 21 amino acids in the NA stalk (residues 54 - 74). The active site of NA is lined by sev- eral conserved residues (117 - 119, 133 - 138, 146 - 152, 156, 179, 180, 196 - 200, 223 - 228, 243 - 247, 277, 278, 293, 295, 344 - 347, 368, 401, 402, and 426 - 441) that participate in recognition of its substrate [40]. In [49] the role of second active site in NA was assessed. It found C opyright © 2011 SciRes. AJMB  W. Hu / American Journal of Molecular Biology 1 (2011) 52-61 Copyright © 2011 SciRes. 57 that in avian NA the interaction between this second site and the primary site is essential for NA function, and the NA of 2009 H1N1 has retained some of the important features of the second site. Following [49], we listed the amino acids at the positions at are lined with the sec- th Figure 2. AT/CG density curves of PB1-F2 of different origins with sliding window size = sequence length/20. Figure 3. Top 10 important positions in each protein of H5N1 that could distinguish North American and Asian avian H5N1. AJMB  American Journal of Molecular Biology, 2011, 1, 52-61 doi:10.4236/ajmb.2011.12007 Published Online July 2011 (http://www.SciRP.org/journal/ajmb/ AJMB ). Published Online July 2011 in SciRes. http://www.scirp.org/journal/AJMB ond site (N2 numbering) (Ta b l e 5 ). It appeared that the main amino acid difference occurred at position 369 for H5N1 and A/California/04/2009(H1N1) (Cal_04_09). The GC content of PB1-F2 of 2009 H1N1 is more similar to swine influenza than to humans [50], although its PB1 gene segment was derived from human H3N2 Table 5. Amino acid comparison at second active site residues in NA of H5N1 and Cal_04_09. The conserved residues are highlighted (N2 numbering). 366 - 373 399 - 403 430 - 433 Asia KSTNSRSG AITDWS RPKE North America KSTSSRSG AITDWS RPKE Human KSTNSRSG AITDWS RPKE Swine KSTNSRSG AITDWS RPKE Cal_04_09 KSISSRNG GINEWS RPKE Ta b l e 6 . Consensus amino acids at top 10 important positions in each protein of H5N1 distinguishing Asian and North Ame- rican avian viruses. There were several positions that had two frequent amino acids. A “-” stands for a deletion. Position/HA 36 45 96 105108 163 169 275294504 North American E K S MT T V DVD Asian T N, D N LI G, S Q NIS Position/NA 23 42 45 4853 67 75 82234241 North American V S Y TV I I PII Asian I, M N Q P-, I - L SVV Position/M1 15 27 36 101166 207 224 230232240 North American V R N RV S S KDY Asian I K N KA N N RNY Position/M2 11 14 18 2127 28 66 829095 North American T G R DV I E SHE Asian T E R DV V E,A SHE Position/NS1 80 81 82 8384 91 118 127171217 North American T I A SV T R NDK Asian -, A, T -, I -,A -, S-, S, V T K TSN Position/NS2 14 22 23 3455 60 71 89113115 North American M G S QL S Q IIT Asian V A S QF I Q IIA Position/NP 34 77 105 146373 377 430 450482497 North American G K M AT S T NSD Asian S R V AA N T NND Position/PA 58 101 129 204261 269 272 348400631 North American G E I RL K D LPG Asian G, S D, E I, T R, K L, M R D IS, PG, S Position/PB1 14 113 149 191215 384 386 511642744 North American A V V VR S R SNM Asian V I I VR L K SNM Position/PB1-F2 28 33 35 4850 55 57 586670 North American L H L QD T S LSE Asian Q P S PG I Y WNG Position/PB2 64 108 147 295339 340 368 390478649 North American M T I VK R R DIV Asian I T, A T, I VT, K K Q, R N, DVV [40]. This discovery prompted us to look at the GC con- tent of PB1-F2 of the viruses under the current study. It turned out the three PB1-F2s of Asian avian, human, and swine H5N1 were similar to each other in their GC con- tent, but none of them was close to the GC content of general avian, human, or swine viruses determined in [50] (Figure 2). However, the GC content of PB1-F2 of North American avian H5N1 was similar to the general avian pattern found in [50]. The high pathogenicity of 1918 H1N1 reminded us to analyze the GC content of its PB1-F2 (A/Brevig Mission/1/1918), which turned out to be different from all the other viruses in Figure 2. Finally, Random Forests [23] were applied to identify Figure 4. Correlated pair counts within and between H5N1 HA and NA of different origins. top 10 positions in each protein that could differentiate Asian and North American avian H5N1 with high con- fidence (Figure 3). The two surface proteins HA and NA had their top 10 positions with similar importance, while the internal proteins had their importance more varied and some of these positions displayed very high impor- tance values. To further elucidate the difference observed in Figure 3, we presented the consensus amino acids at the top 10 positions in Table 6, which could complement the information for the amino acids at the HA binding site in section 3.2. The difference in amino acids at these positions and those in Tab le 3 contributed to the differ- ent HA receptor selections revealed in Figure 1. 3.5. Correlations within and between HA and NA, and NP, PA, PB1, and PB2 Respectively A right balance between the HA receptor binding and the release of progeny virions by NA requires the close co- operation of these two surface proteins. To reveal the correlation patterns for the two proteins of H5N1, their mutual information was calculated. We only recorded the positions in these proteins that had a positive MI value. The correlated position pairs of a positive MI value were counted according to their locations in the proteins, and then averaged by the number of sequences in each species. It was evident that the correlation be- tween HA and NA was higher than within across differ- ent species. The overall interaction patterns for HA and  W. Hu / American Journal of Molecular Biology 1 (2011) 52-61 Copyright © 2011 SciRes. 58 NA of Asian avian and human H5N1 were alike, whe- reas the North American avian and swine H5N1 dis- played distinct patterns (Figure 4). The interactions among NP and the three proteins PA, PB1, and PB2 of the viral polymerase are essential for virus replication and host adaptation. The mutual infor- mation was also computed for these four proteins of H5N1. As in the HA and NA situation, in general inter- protein correlations of the four proteins were stronger than intra-protein across different species (Figure 5). Although the sequences identities of Asian avian, human, and swine H5N1 were close to each other (Table 4), only the NP, PA, PB1, and PB2 of Asian avian and swine ere similar in their interaction patterns (Figure 5). It is w Figure 5. Correlated pair counts within and between H5N1 NP, PA, PB1, and PB2 of different origins. interesting to note that the most similar two sequences does not always exhibit the most similar correlation pat- terns [51]. 4. CONCLUSIONS It is of prime importance to learn the molecular dis- tinctions between the highly and low pathogenic avian H5N1 viruses as we develop the knowledge for avian influenza. This study took Asian and North American avian H5N1 as examples of highly and low pathogenic avian viruses respectively. We sought to investigate several crucial aspects of these viruses including HA receptor preference and cleavage site, NA second ac- tive site, interaction patterns of HA and NA, and NP, PA, PB1, and PB2, and important sites in the proteins of these viruses. It is believed that a switch from SA α2, 3Gal to SA α2, 3Gal receptor specificity is a critical step in the adaptation of avian viruses to a human host. The SA α2, 3Gal specificity of avian influenza viruses makes it difficult for these viruses to be easily transmitted from human to human after avian to human infection. The bioinformatics technique ISM provided an efficient way of revealing the HA receptor selections of avian AJMB  W. Hu / American Journal of Molecular Biology 1 (2011) 52-61 60 H5N1. The IS analysis on the consensus HA1 sequen- ces of Asian avian, human, and swine H5N1 demon- strated two dominant frequencies: F(0.076) as primary frequency and F(0.236) as secondary, while North American avian H5N1 had F(0.076) and F(0.137). Se- quence examination also showed that amino acid dif- ference at two critical sites (H3 numbering) for recep- tor selection were 193K and 216E for North American avian H5N1 and 193R and 216K for Asian avian H5N1 in recent years. The frequent occurrences of mutations K193R and R216K observed in Asian avian H5N1 highlighted the selection pressure on this virus to in- crease its human-type binding. The different amino acids at sites 193 and 216 plus the top 10 important HA sites identified by Random Forests accounted for the difference in HA receptor specificity of Asian and North American avian H5N1 revealed in this study. 5. ACKNOWLEDGEMENTS We thank Houghton College for its financial support. REFERENCES [1] Xu, X., Subbarao, K., Cox, N.J. and Guo, Y. (1999) Genetic characterization of the pathogenic influenza A/ Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagg-lutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology, 261, 15-19. doi:10.1006/viro.1999.9820 [2] Claas, E.C.J., Osterhaus, A.D.M.E., van Beek, R., de Jong, J.C., Rimmelzwaan, G.F., et al. (1998) Human influenza a H5N1 virus related to a highly pathogenic avian influenza virus. The Lancet, 351, 472-477. doi:10.1016/S0140-6736(97)11212-0 [3] www.usda.gov/documents/wildbirdstrategicplanpdf.pdf [4] Bi, Y., Fu, G., Chen, J., Peng, J., Sun, Y., Wang, J., et al. (2010) Novel swine influenza virus reassortants in pigs, China. Emerging Infectious Diseases. http://www.cdc. gov/EID/content/16/7/1162.htm [5] Shinya, K., Ebina, M., Yamada, S., Ono, M., Kasai, N. and Kawaoka, Y. (2006) Avian flu: Influenza virus re- ceptors in the human airway. Nature, 440, 435-436. doi:10.1038/440435a [6] Skehel, J.J. and Wiley, D.C. (2000) Receptor binding and membrane fusion in virus entry: The influenza hema- gglutinin. Annual Review of Biochemistry, 69, 531-569. doi:10.1146/annurev.biochem.69.1.531 [7] Glaser, L., Stevens, J., Zamarin, D., Wilson, I.A., García- Sastre, A., Tumpey, T.M., Basler, C.F., Taubenberger, J.K. and Palese, P. (2005) A single amino acid substi- tution in 1918 influenza virus hemagglutinin changes receptor binding specificity. Journal of Virology, 79, 11533-11536. doi:10.1128/JVI.79.17.11533-11536.2005 [8] Li, M. and Wang, B. (2006) Computational studies of H5N1 hemagglutinin binding with SA-alpha-2,3-Gal and SA-alpha-2,6-Gal. Biochemical and Biophysical Rese- arch Communications, 347, 662-668. doi:10.1016/j.bbrc.2006.06.179 [9] Hu, W. (2010) Identification of highly conserved do- mains in gemagglutinin associated with the receptor binding specificity of influenza viruses: 2009 H1N1, avian H5N1, and swine H1N2. Journal of Biomedical Science and Engineering, 3, 114-123. doi:10.4236/jbise.2010.32017 [10] Hu, W. (2010) Quantifying the effects of mutations on receptor binding specificity of influenza viruses. Jour- nal of Biomedical Science and Engineering, 3, 227-240. [11] Hu, W. (2010) Highly conserved domains in hemagg- lutinin of influenza viruses characterizing dual receptor binding. Natural Science, 2, 1005-1014. doi:10.4236/ns.2009.29123 [12] Veljko, V., Henry, L.N., Sanja, G., Nevena, V., Vladimir, P. and Claude, P.M. (2009) Identification of hema- gglutinin structural domain and polymorphisms which may modulate swine H1N1 interactions with human receptor. BMC Structural Biology, 9, 62. doi:10.1186/1472-6807-9-62 [13] Veljkovic, V., Veljkovic, N., Muller, C.P., Müller, S., Glisic, S., Perovic, V. and Köhler, H. (2009) Charac- terization of conserved properties of hemagglutinin of H5N1 and human influenza viruses: Possible conse- quences for therapy and infection control. BMC Struc- tural Biology, 7, 9-21. [14] Hu, W. (2009) Analysis of correlated mutations, stalk motifs, and phylogenetic relationship of the 2009 influ- enza a virus neuraminidase sequences. Journal of Bio- medical Science and Engineering, 2, 550-558. [15] Hu, W. (2010) The interaction between the 2009 H1N1 influenza a hemagglutinin and neuraminidase: Muta- tions, co-mutations, and the NA stalk motifs. Journal of Biomedical Science and Engineering, 3, 1-12. doi:10.4236/jbise.2010.31001 [16] Chen, G.-W., Chang, S.-C., Mok, C.-K., Lo, Y.-L., Kung, Y.-N., et al. (2006) Genomic signatures of human versus avian influenza a viruses. Emerging Infectious Diseases, 12, 1353-1360. [17] Chen, G.-W. and Shih, S.-R. (2009) Genomic signatures of influenza a pandemic (H1N1) 2009, virus. Emerging Infectious Diseases, 15, 1897-1903. [18] Pan, C., Cheung, B., Tan, S., Li, C., Li, L., et al. (2010) Genomic signature and mutation trend analysis of pandemic (H1N1) 2009, influenza A virus. PLoS ONE, 5, Article ID e9549. doi:10.1371/journal.pone.0009549 [19] Miotto, O., Heiny, A., Tan, T.W., August, J.T. and Brusic, V. (2008) Identification of human-to-human trans- missibility factors in PB2 proteins of influenza A by large-scale mutual information analysis. BMC Bioinfor- matics, 9, S18. doi:10.1186/1471-2105-9-S1-S18 [20] Miotto, O., Heiny, A.T., Albrecht, R., García-Sastre, A., Tan, T.W., August, J.T. and Brusic, V. (2010) Complete- proteome mapping of human influenza A adaptive mutations: Implications for human transmissibility of zoonotic strains. PLoS ONE, 5, Article ID e9025. doi:10.1371/journal.pone.0009025 [21] Finkelstein, D.B., Mukatira, S., Mehta, P.K., Obenauer, J.C., Su, X., Webster, R.G. and Naeve, C.W. (2007) Persistent host markers in pandemic and H5N1 influenza viruses. Journal of Virology, 81, 10292-10299. doi:10.1128/JVI.00921-07 C opyright © 2011 SciRes. AJMB  W. Hu / American Journal of Molecular Biology 1 (2011) 52-61 61 [22] Allen, J.E., Gardner, S.N., Vitalis, E.A. and Slezak, T.R. (2009) Conserved amino acid markers from past influenza pandemic strains. BMC Microbiology, 9, 77. doi:10.1186/1471-2180-9-77 [23] Breiman, L. (2001) Random forests. Machine Learning, 45, 5-32. doi:10.1023/A:1010933404324 [24] Hu, W. (2010) Novel host markers in the 2009 pande- mic H1N1 influenza A virus. Journal of Biomedical Science and Engineering, 3, 584-601. [25] Hu, W. (2010) Nucleotide host markers in the influenza A viruses. Journal of Biomedical Science and Engineer- ing, 3, 684-699. [26] Katoh, K., Kuma, K., Toh, H. and Miyata, T. (2005) MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Research, 33, 511- 518. doi:10.1093/nar/gki198 [27] Cosic, I. (1997) The Resonant Recognition Model of Macromolecular Bioreactivity, Theory and Application. Birkhauser Verlag, Berlin. [28] Hu, W. (2011) Receptor binding specificity and origin of 2009 H1N1 pandemic influenza virus. Natural Science, 3, 234-248. [29] Cover, T.A. and Thomas, J.A. (1991) Elements of Information Theory. John Wiley and Sons, New York. doi:10.1002/0471200611 [30] MacKay, D. (2003) Information Theory, Inference, and Learning Algorithms. Cambridge University Press, Cam- bridge. [31] Bogs, J., Veits, J., Gohrbandt, S., Hundt, J., Stech, O., et al. (2010) Highly pathogenic H5N1 influenza viruses carry virulence determinants beyond the polybasic he- magglutinin cleavage site. PLoS ONE, 5, Article ID e11826. doi:10.1371/journal.pone.0011826 [32] Gohrbandt, S., Veits, J., Hundt, J., Bogs, J., Breithaupt, A., Teifke, J.P., Weber, S., Mettenleiter, T.C. and Stech, J. (2011) Amino acids adjacent to the haemagglutinin cleavage site are relevant for virulence of avian influenza viruses of subtype H5. Journal of General Virology, 92, 51-59. doi:10.1099/vir.0.023887-0 [33] Stevens, J., Blixt, O., Tumpey, T.M., Taubenberger, J.K., Paulson, J.C. and Wilson, I.A. (2006) Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science, 312, 404-410. doi:10.1126/science.1124513 [34] Wang, W., Lu, B., Zhou, H., Suguitan, A.L. Jr., Cheng, X., Subbarao, K., Kemble, G. and Jin, H. (2010) Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. Journal of Virology, 84, 6570-6577. doi:10.1128/JVI.00221-10 [35] Auewarakul, P., Suptawiwat, O., Kongchanagul, A., et al. (2007) An avian influenza H5N1 virus that binds to a human-type receptor. Journal of Virology, 81, 9950-9955. doi:10.1128/JVI.00468-07 [36] Yamada, S., Suzuki, Y., Suzuki, T., Le, M.Q., Nidom, C.A., et al. (2006) Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human- type receptors. Nature , 444, 378-382. doi:10.1038/nature05264 [37] Neumann, G., Chen, H., Gao, G.F., Shu, Y.-L. and Kawaoka, Y. (2010) H5N1 influenza viruses: Outbreaks and biological properties. Cell Research, 20, 51-61. doi:10.1038/cr.2009.124 [38] Ayora-Talavera, G., Shelton, H., Scull, M.A., Ren, J., Jones, I.M., et al. (2009) Mutations in H5N1 influenza virus hemagglutinin that confer binding to human tra- cheal airway epithelium. PLoS ONE, 4, Article ID e7836. doi:10.1371/journal.pone.0007836 [39] Gambaryan, A., Tuzikov, A., Pazynina, G., Bovin, N., Balish, A. and Klimov, A. (2006) Evolution of the rece- ptor binding phenotype of influenza a (H5) viruses. Virology, 344, 432-438. doi:10.1016/j.virol.2005.08.035 [40] Scalera, N.M. and Mossad, S.B. (2009) The first pande- mic of the 21st century: A review of the 2009 pan-demic variant influenza a (H1N1) virus. Postgraduate Medicine, 121, 43-47. doi:10.3810/pgm.2009.09.2051 [41] Maurer-Stroh, S., Lee, R.T., Eisenhaber, F., Cui, L., Phuah, S.P. and Lin, R.T. (2010) A new common mu- tation in the hemagglutinin of the 2009 (H1N1) influ- enza A virus. PLoS Currents Influenza, 1, 162. doi:10.1371/currents.RRN1162 [42] Barr, I.G., Cui, L., Komadina, N., Lee, R.T., Lin, R.T., Deng, Y., Caldwell, N., Shaw, R. and Maurer-Stroh, S. (2010) A new pandemic influenza A(H1N1) genetic vari- ant predominated in the winter 2010 influenza season in Australia, New Zealand and Singapore. Euro Surveill, 15, 19692. [43] Stevens, J., Blixt, O., Chen, L.M., Donis, R.O., Paulson, J.C. and Wilson, I.A. (2008) Recent avian H5N1 viruses exhibit increased propensity for acquiring human recep- tor specificity. Journal of Molecular Biology, 381, 1382- 1394. doi:10.1016/j.jmb.2008.04.016 [44] Fereidouni, S.R., Beer, M., Vahlenkamp, T. and Starick, E. (2009) Differentiation of two distinct clusters among currently circulating influenza A(H1N1) viruses. Euro Surveill, 14, 19409. [45] Hu, W. (2010) Subtle differences in receptor binding specificity and gene sequences of the 2009 pandemic H1N1 influenza virus. Advances in Bioscience and Biotechnology, 1, 305-314. doi:10.4236/abb.2010.14040 [46] Hu, W. (2011) New mutational trends in the HA protein of 2009 H1N1 pandemic influenza virus from May 2010 to February 2011. Natural Science, 3, 379-387. [47] Long, J.X., Peng, D.X., Liu, Y.L., Wu, Y.T. and Liu, X.F. (2008) Virulence of H5N1 avian influenza virus enhan- ced by a 15-nucleotide deletion in the viral nonstructural gene. Virus Genes, 36, 471-478. doi:10.1007/s11262-007-0187-8 [48] Hatta, M., Hatta, Y., Kim, J.H., Watanabe, S., Shinya, K., et al. (2007) Growth of H5N1 influenza a viruses in the upper respiratory tracts of mice. PLoS Pathog, 3, Article ID e133. doi:10.1371/journal.ppat.0030133 [49] Sung, J.C., van Wynsberghe A.W., Amaro, R.E., Li, W.W. and McCammon, J.A. (2010) Role of secondary sialic acid binding sites in influenza N1 neuraminidase. Journal of the American Chemical Society, 132, 2883- 2885. doi:10.1021/ja9073672 [50] Hu, W. (2010) Host markers and correlated mutations in the overlapping genes of influenza viruses: M1, M2; NS1, NS2; and PB1, PB1-F2. Natural Science, 2, 1225-1246. doi:10.4236/ns.2010.211150 C opyright © 2011 SciRes. AJMB  W. Hu / American Journal of Molecular Biology 1 (2011) 52-61 Copyright © 2011 SciRes. 62 AJMB [51] Hu, W. (2010) Correlated mutations in the four influen- za proteins essential for viral RNA synthesis, host adaptation, and virulence: NP, PA, PB1, and PB2. Natural Science, 2, 1138-1147. doi:10.4236/ns.2010.210141
|