 Advances in Microbiology, 2015, 5, 780-786 Published O n li ne November 2015 in SciRes. http://www.scirp.org/journal/aim http://dx.doi.org/10.4236/aim.2015.512082 How to cite this paper: Halwani, M.A., Tashkandy, N.A.J., Aly, M.M., Al Masoudi, S.B. and Dhafar, O.O. (2015) Incidence of Antibiotic Resistance Bacteria in Jeddah’s Ministry of Health Hospitals, Saudi Arabia. Advances in Microbiology, 5, 780-786. http://dx.doi.org/10.4236/aim.2015.512082 Incidence of Antibiotic Resistance Bacteria in Jeddah’s Ministry of Health Hospitals, Saudi Arabia Muhammad A. Halwani1, Nidal A. J. Tashkandy2, Magda M. Aly3*, Saad B. Al Masoudi3, Osama O. Dhafar2 1Department of Microbiology, Faculty of Medicine, Al Baha University, Al Baha, Saudi Arabia 2Health Affairs, Jeddah, Saudi Arabia 3Biology Department, Faculty of Science, King Abdu laziz University, Jeddah, Saudi Arabia Received 8 October 2015; accepted 13 Novemb er 2015; published 16 November 2015 Copyright © 2015 by authors and Scientific Research Publishing Inc. This work is licensed under the Creative Commons Attribution International License (CC BY). http://creativecommons.org/licenses/by/4.0/ Abstract This study aimed to determine the emergence and spread of resi s tant bacteria in Jeddah Ministry of Health hospitals. Sixteen month fo llow-up (January 2010 to April 2011) study was carried out and clinical isolates of hos pit ali zed patients were collected, identified and their antimicrobial re- sistance was determined using two automated systems, Phoen i x and Vitek 2. Results revealed that 6195 isolates were identifie d of whi ch 94% (5846/6195) were Gram negatives. In Escherichia coli, the resistance was 40% (681/1703) to ciprofloxacin, 30% (511/1703) to cefepime, 29% (494/1703) to c eft azidi me , 8 . 5% (145/1703) to tazocin and amikacin, 40% (681/1703) to gentami ci n and ce- furoxime. In Klebsiella pneumonia , the resistance was 48% (550/1147) to ceftazidime, 49% (565/1147) to cefuroxime, 45.5% (522/1147) to cefepime, 38% (436/1147) to g ent ami cin, 30% (344/1147) to ciproflox aci n, 19% (218/1147) to tazocin, 7.5% (86/1147) to amikacin and 2.4% (27/1147) to imipenem/meropenem. In Acinetobacter bumannii, 79% (850/1076) were resistant to ciprofloxacin, 68. 5% (737/1076) to tazocin, 6 7% (721/1076) to cefepime, 66% (710/1076) to gen t amicin and imipenem /mer ope nem, 65% (699/1076) to ce ftaz idi me , 68% (735/1076) to amikacin and no resistance to colistin was reported. In Pseudomonas aeruginosa, almost 34% (555/1632) were resistant to ceftazidi me, 31% (506/1632) to c ip roflox acin, 29% (473/1632) to cefepime, 26.5% (434/1638) to gentamicin, 19% (310/1632) to i mi penem /mer ope nem, 17% (277/1632) to amikacin , and 15.5% (253/1632) were resistant to tazocin. In Gram positive iso- lates, MRSA counted only for 4.6% (302/6552) and no vancomycin intermediate Staphylococcus aureus (VISA) were detected. In conclusion, the resistance detected in this study is considered high an d antibiotic Stewardship Programs is inevitably required. *  M. A. Halwani et al. Keywords Resistance, Clinical Isolates, Jeddah, Klebsiella, Antibiotic 1. Introduction In Kingdom of Saudi Arabia, Jeddah is the second largest city with commercial importance and with a popula- tion of 3.2 million people. It is the gateway to the holy cities of Makkah and Medina [1]. The Ministry of Health oversees Jeddah’s health system and provides free medical care to Saudi citizens and legal residents. The Minis- try of Health in Saudi Arabia is a major source of medical services in the country and has 12 different hospitals distributed among the city with approximately a total of 2400 beds. The infection prevention and control pro- gram in those hospitals is covered by one specialized administration that runs a scientifically based standardized program. All microbio log y results a nd antibiotic susceptibilities are sen t to the administration on a monthly basis for follow-up and anal ysi s. Dat a are checke d for val idly and ac curac y wit h the m ic robiol ogist s in the hospi tal s. Antimicrobial drugs decreased death and illness associated with infectious diseases in however there is an emergence and spr ead of drug-resistance isolates among bacteria [2]. An tibacterial resistant isolates caused out- breaks, and more involved in health care-associated infections: including bacteremia, pneumonia, meningitis, urinary tract infection, and wound infection and greatly limited th e therapeutic options for infected patients [3]. Moreover, the developed resistance for a major class of antimicrobial drugs appeared as short as 1 year to >10 years [4]. What’s mor e, there is a major increase in the emergence and spread of multidrug-resistant bacteria to newer compounds, such as cephalosporins and fluoroquinolones [5]. The more problematic drug-resistant pathogens include multidrug-resistant Acinetobacter baumannii, Kleb- siella pneumoniae, Escherichia coli, and Pseudomonas aeruginosa among the gram-negative bacteria and me- thicillin-resistant Staphylococcus aureus, penicillin-resistant Streptococcus pneumoniae, and vancomycin-resi- stant enterococci among the gram-positive bacteria [6]. Some E. co li isolates caused urinary tract infection, meningitis, peritonitis intestinal and extr a-intestinal infec- tions and their resistance for the longest used antimicrobial agents was consistently the highest [7]. During a 12-year period (1971-1982), the susceptibility of E. coli, isolated from hospitals, showed no major change in re- sistance to all tested antimicrobial drugs [7 ]. In contrast, E. coli collected during 1997-2007 from urine speci- mens showed increasing resistanc e to ciprofloxacin, trimethoprim/sulfamethoxazole, and amoxicillin/clav ulanic acid [8]. In a 30-year (1979-2009) follow-up study in Sweden, E. coli showed an increasing resistance towards ampicillin, sulfona mide, tr imeth oprim, and gentamicin [9] and this yet was a great concern. In intensive care units, Klebsiella spp. is among the most common isolated pathogens and the emergence of K. pneumoniae resistance to antibiotics is well documented [10]. The study of Sanchez et al. [11] showed that K. pneumoniae antimicrobial drug resistance increased for every antimicrobial class st udied except tetracyclines and cross-resistance among imipenem-resistant K. pneum oniae was high for cipr ofloxa cin but lowe r fo r am ikaci n an d tetracycline. Pseudomonas aeruginosa, associated with nosocomial infections, rapidly developed resistance to multiple classes of antibiotics and the import of resistance mechanisms on mobile genetic elements was always a concern [12]. Multi-drug -resistant P. aeruginosa was intermediate or resistant to at least three drug classes: β-lactams, carbapenems, aminoglycosides, and fluoroquinolones and the reported rates varied from 0.6% - 32% based on geographic location and the type of surveillance study [13]. Increasing emergence and spread of resistant Acine- tobacter strains to the first and second generati on cephalospo rins starte d since 1975 but during 1980s to 19 90s, the worldwide spread of imipenem resistant strains was recorded that could combat many severe Acinetobacter in- fections. Furthermore, carbapenem resistance in the strains of A. baumannii decreased the therapeutic options and made the life of treating physicians and patients really miserab le [14]. The aim of the study was to detect the in- cidence of resistance in clin ic al bacterial isolates that was identified in 12 hos pitals at Jeddah. 2. Material & Methods Bacterial Identification and Sensitivity The study was conducted for 16 months (January 2010 to April 2011) in 12 Ministry of Hea lth hospitals in Jed- 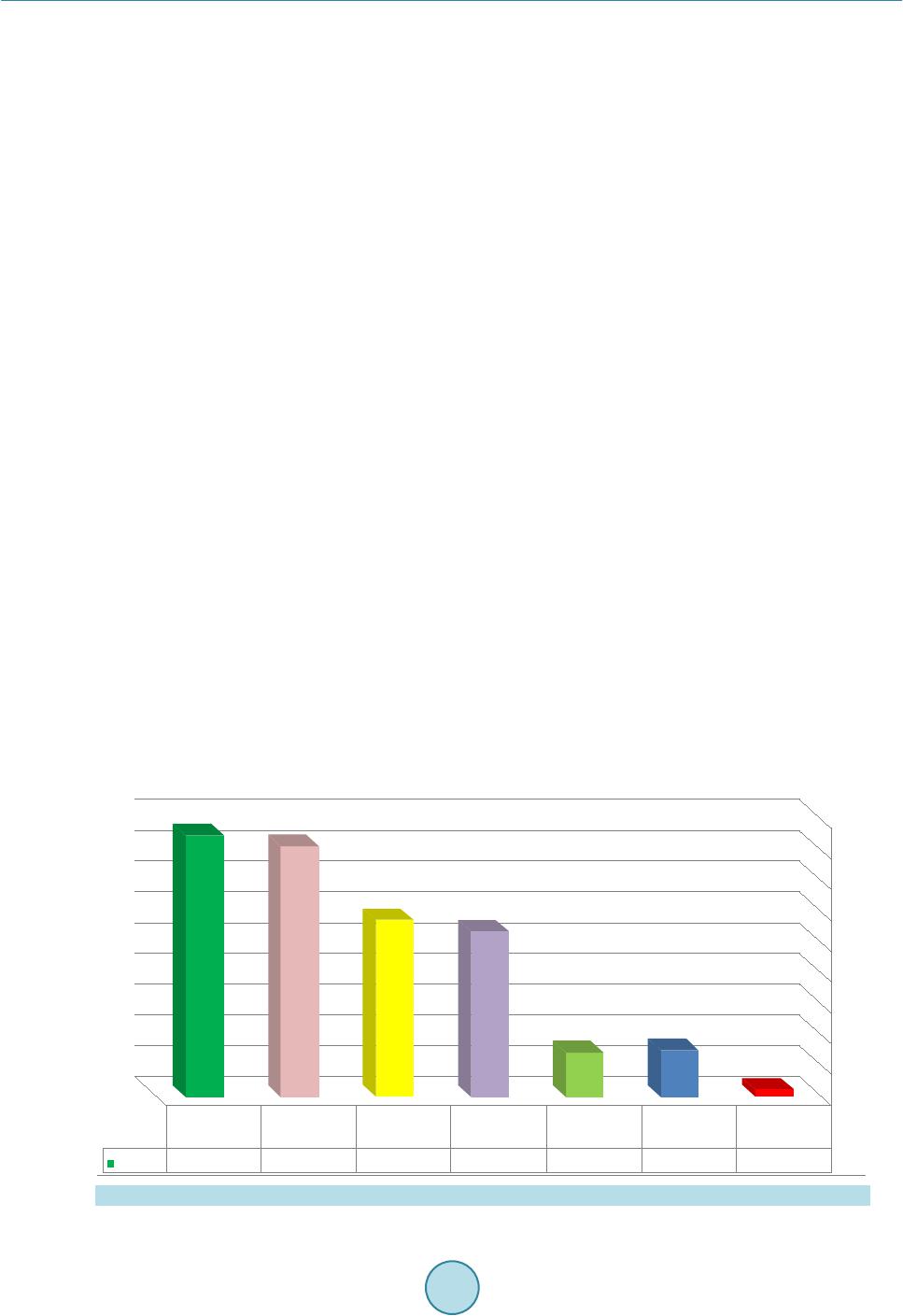 M. A. Halwani et al. dah, Saudi Arabia. During that period, 6195 non-repetitive clinical isolates from various specimens of hospita- lized patients wer e collected and tested against the routinely used antibiotics. Two main automated systems are available and used in Jeddah hospitals laboratories; Phoenix (Becton Dickinson Diagnostic Systems, Sparks, MD, USA) and Vitek 2 (bioMérieux, Marcy l’Étoile, France). Susceptibilities of the Gram negative isolates were tested to the following antibacterial agents: Ceftazidime, Ciprofloxacin, Cefepime, Amikacin, Tazocin, Gentamicin, Cefuroxime and Imipene m. Colistin was tested only when an isolate showed a complete pattern of resistance to all tested antibiotics. All bacterial isolates were tested using standard bacteriological procedures and according to the procedures recommended by the manufacturers. No attempt was made to compare between the results of the two methods in this study. 3. Results Sixteen month follow-up (January 2010 to April 2011) of the clinical isolates, obtained from hospitalized pa- tients, in the 12 Ministry of Health hospitals was carried out. The counts and resistance of some of the Gram positive and negativ e bacterial isolates were id entified using in-house diagnostic microbiology methods. Figure 1 revealed that 6195 isolates were identified of which 94% (5846/6195) were Gram negatives and 6% (637/6195) were Gram positive. Furthermore, E. coli was the most isolated organism and counted for 27% (1703/6195) of all identified isolates followed by P. aerignosa 26% (1632/6195), K. pneumoniae 18% (1147/6195), Acineto- bacter bumannii 17% (1076/6195) and Enterobacter cloacae 4.6% (288/6195). MRSA counted for almost 5% (349/6195) and Streptococcus pneumoniae for 0.7% (47/6195). The rate of resistance in Gram negative isolates is shown in Figures 2-5. In Escherichia coli; 40% (681/1703) were resistance to ciprofloxacin, gentamicin or cefuroxime, 30% (511/1703) were resistant to cefepime, 29% (494/1703) to ceftazidime, 8.5% (145/1703) to tazocin and amikacin, 40% (681/1703 ) to (Figure 2). In Klebsiella pneumonia isolates, 48 % (550/1147) were resistant to ceftazidime, 49% (565/1147) to cefuroxime, 45.5% (522/1147) to cefepime, 38% (436/1147) to gentamicin, 30% (344/1147) to ciprofloxacin, 19% (218/1147) to tazocin, 7.5% (86/1147) to amikacin and 2.4% (27/1147) to imipenem/me- ropenem (Figure 3). In A. bumannii, 79% (850/1076) were resistant to ciprofloxacin, 68.5% (737/107 6) to tazo- cin, 67% (721/1076) to cefepime, 66% (710/1076) to gentamicin and imipenem/meropenem, 65% (699/1076) to ceftazidime, 68% (735/1076) to amikacin and no resistance to colistin was reported (Figure 4). In Pseudomonas aeruginosa, almost 34% (555/1632) were resistant to ceftazidime, 31% (506/1632) to ciprofloxacin, 29% (473/1632) to cefepime, 26.5% (434/1638) to gentamicin, 19% (310/1632) to imipenem/meropenem, 17% (277/1632) to amikacin, and 15.5% (253/1632) were resistant to tazocin (Figure 5). In Gram positive isolates; out of 357 Staphylococcus aureus isolates collected, 302 (89%) w ere MRSA wh ich counted only for 4.6% (302/6552) of the total collected isolates. No vancomycin intermediate Staphylococcus Figure 1. The different pathogenic bacterial isolates and their rates. 0 200 400 600 800 1000 1200 1400 1600 1800  M. A. Halwani et al. Figure 2. The overall resistance rate of E. coli isolates (N = 1703) to different used antibiotics. Figure 3. The overall resistance rate of K. pnermoniae isolates (N = 1147) to different used antibiotics. Figure 4. The overall resistance rate in A. bumannii isolates (N = 1076) to different used antibiotics. 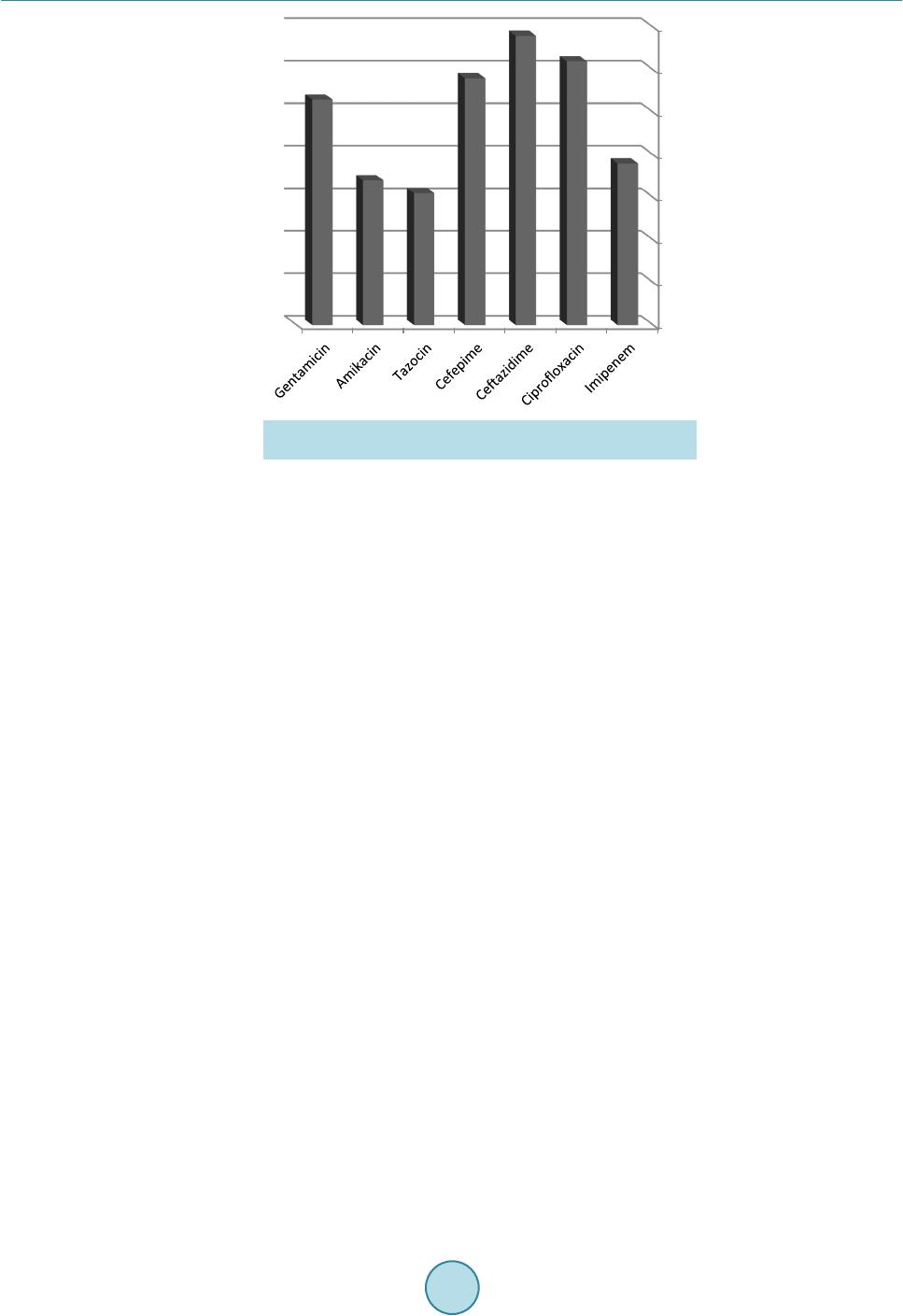 M. A. Halwani et al. Figure 5. The overall resistance rate in P. aeruginosa isolates (N = 1632) to different used antibiotics. aureus (VISA) were detected. Furthermore, only 10% (5/47) Streptococcus pneumonia isolates were resistant to penicillin (data not shown). 4. Discussion Bacterial pathogens have a great ability to adapt and overcome the ch allenges of antib iotics in their environment that threaten to move us into the “post-antibiotic era” of infectious diseases. The therapy for resistant bacterial infections becomes more problematic than ever and the infection prevention is now ver y essential [15 ] [16]. Our study has some limitations where identification of the multi-drug resistance isolates is performed with two dif- ferent automated systems and this may have given slight variations of the antibiotics’ break points. Hence, stan- dardization of lab identification methodologies in all hospital labs might be desirable. Our study rev ealed E. coli is the dominating organism (27%) in comparison to other Gram negatives bacteria. In addition, P. aeruginosa was found to be the second isolated organism (26%) which might highlight the heavy involvement of hospital environment. Lower percentages 18% and 17% were recorded in K. pneumoniae and A. baumonnii, respectively. In a study that was conducted by Wagenlehner et al. [ 17 ], E. coli counted for 50% of the yielded isolates, Proteus spp. 15%, Klebsiella spp. 15% and P. aeruginosa 5%. The resistance rates reported in this study was disturbing to ciprofloxacin, gentamicin or cefuroxime (40%) in E. coli, to cefuroxime (49%) and ceftazidime (48%) in K. pneumonia. Similarly, Livermore et al. [18] reported the changes in susceptibility pattern of E. coli and during 2011 in Central Greece, a significant increase in E. coli ciprofloxacin resistance (21%) has occurred [19]. Th e emergence of extended r esistant Acinetobacter spp. was rapidly spreading to even newer antimicrobials and they acquired resistance faster than other Gram-negative organisms due to their ease of survival in the hospital environment, their immense potential to cause nosocomial outbreaks and their biofilm forming abil it y whic h pl a ye d a c ruc i al rol e in the ir in-vitro and in-vivo survival [20]. This study showed that ≥66% of A. bumannii isolates were resistant to almost all tested antibiotics and this clearly ind icated how problematic to have patients infected with this organism and the minimal chances available to find proper treatment. Increasing resistance of P. aeruginosa to the various antipseudomonal agents has been reported worldwide which cause a serious problem in infection management. Although up to 34% of P. aeruginosa in our study iso- lates were resistant to ceftazidime and 31% to Ciprofloxacin, other alternative antibiotics are available. This high resistance clearly indicates the overuse of the latter antibiotics and insists the demand of a strictly respected comprehensive antibio tic stewardship program in our hospitals. I n United States, a dramatic antimicrobial r esis- tance increase to ciprofloxacin (from 15% to 32%) and ceftazidime (from 15% to 19%) over the ten-year period was recorded [21]. In Kuala Lumpur, Malaysia, approximately 10% of clinical isolates of P. aeruginosa were resistant to imipenem, while 11% showed resistance to ciprofloxacin, piperacillin and ceftazidime [22] [23].  M. A. Halwani et al. Despite the increasing r esistance to ceftazidime, it was still in use against P. aeruginosa infections as the resis- tance did not exceed 35% in our study. Staphylococcus aureus cause health care-associated infections for both hospitalized patients with healthy or decreased host defenses. In the 1980s, methicillin-resistant S. aureus (MRSA) emerged as a major clinical and epidemiologic problem in hospitals and spreaded out of the hospitals into communities. In the present study, MRSA counted only for 4.6% of the total collected isolates and 89% of S. aureus isolates which probab ly indi- cated the need for better understanding to infection control measures in the hospitals. Prevalence of MRSA in hospitals increased from 2.1% in 1975 to 35% in 1991 [24] and it was 46% in Western Pacific region, 5.7% in Canada, varied from less than 2% in the Netherlands to 54.4% in Portugal, from 23.6% in Australia to more than 70% in Japan and Hong Kong [25] [26] and 30% - 60 % in Thailand [27]. For serious MRSA infec tions, vanco- mycin is the treatment of choice and MRSA strains with reduced susceptibility to vancomycin have been re- ported [28 ] [ 29 ] but in this study no vancomycin resistant isolates were found. Only 10% of S. pneumonia iso- lates were resistant to penicillin which might indicate that the resistance to this bacteria in particular was not very high. However, this rate might increase if more awareness were not really considered at this stage. Hof- mann et al. [ 30 ] reported that 25% of the isolates were resistant to penicillin. This may indicates that more re- sistant S. pneumonia isolates are on the way. Acknowledgements Thanks to all the staff members and the technicians of the Microbiology Lab., KAUH, Jeddah, Saudi Arabia. References [1] http://en.wikipedia.org/wiki/Jeddah [2] Aarestrup, F.M., Wegener, H.C. and Collignon, P. (2008) Resistance in Bacteria of the Food Chain: Epidemiology and Control Strategies. Expert Review of An ti-Infective Therapy, 6, 733-750. http://dx.doi.org/10.1586/14787210.6.5.733 [3] Maragakis, L.L. and Perl, T.M. (2008) Acinetobacter baumannii: Epidemiology, Antimicrobial Resistance, and Treat- ment Options. Clinical Infectious Diseases, 46, 1254-1263. http://dx.doi.org/10.1086/529198 [4] Walsh, C.T. (2003) Antibiotics: Actions, Origins, Resistance. American Society for Microbiology, Washington DC. http://dx.doi.org/10.1128/9781555817886 [5] Levy, S.B. and Marshall, B. (2004) Antibacterial Resistance Worldwide: Causes, Challenges and Responses. Nature Medicine, S122-S129. http://dx.doi.org/10.1038/nm1145 [6] Atkinson, B.A. and Lorian, V. (1984) Antimicrobial Agent Susceptibility Patterns of Bacteria in Hospitals from 1971 to 1982. Journal of Clinical Microbiology, 20, 791-796. [7] von Baum, H. and Marre, R. (2005) Antimicrobial R esistance of Escherichia coli and Therapeutic Implications. Inter- national Journal of Medical Microbiology, 295, 503-511. http://dx.doi.org/10.1016/j.ijmm.2005.07.002 [8] Kronvall, G.A. (2010) Antimicrobial Resistance 1979-2009 at Karolinska Hospital, Sweden: Normalized Resistance Interpretation during a 30-Year Follow-Up on Staphylococcus aureus and Escherichia coli Resistance Development. APMIS, 118, 621-639. http://dx.doi.org/10.1111/j.1600-0463.2010.02660.x [9] Blaettler, L., Mertz, D., Frei, R., Elzi, L., Widmer, A.F., Bat t e gay, M., e t al. (2009) Secular Trend and Risk Factors for Antimicrobial Resistance in Escherichia coli Isolates in Switzerland 1997-2007. Infection, 37, 534-539. http://dx.doi.org/10.1007/s15010-009-8457-0 [10] Naas, T., Nordmann, P., Vedel, G. and Poyar t , C. (2005) Plasmid-Mediated Carbapenem-Hydrolyzing Beta-Lactamase KPC in a Klebsiella pneumoniae Isolate from France. Antimicrobial Agents and Chemotherapy, 49, 4423-4424. http://dx.doi.org/10.1128/AAC.49.10.4423-4424.2005 [11] Schwaber, M.J. and Carmeli , Y. (2008) Carbapenem-Resistant Enterobacteriaceae: A Potential Threat. JAMA, 300, 2911-2913. http://dx.doi.org/10.1001/jama.2008.896 [12] Won, S.Y., Munoz-Price, L.S., Lolans, K., Ho ta, B., Weinstein, R.A. and Hayden, M.K. ( 2011) Emergence and Rapid Regional Spread of Klebsiella pneumonia Carbapenemase-Producing Enterobacteriaceae. Clinical Infectious Diseases, 53, 532-540. http://dx.doi.org/10.1093/cid/cir482 [13] Sanchez, G.V., Master, R.N., Clark, R.B., Fyy az, M., Duvvuri, P., Ekta, G. and Bordon, J. (2013) Klebsiella pneumo- niae Antimicrobial Drug Resistance, United States, 1998-2010. Emerging Infectious Diseases, 19, 133-136. http://dx.doi.org/10.3201/eid1901.120310 [14] Obritsch, M.D., Fish, D.N., MacLaren, R. and Jung, R. (2005) Nosocomial Infections Due to Multidrug-Resistant  M. A. Halwani et al. Pseudomonas aeruginosa: Epidemiology and Treatment Options. Pharmacotherapy, 25, 1353-1364 [15] Go, E.S., Urban, C., Burns, J., Kreiswirth, B., Eisner, W., Mariano, N., et al. (1994) Clinical and Molecular Epidemi- ology of Acinetobacter Infections Sensitive Only to Polymyxin B and Sulbactam. Lancet, 344, 1329-1332. http://dx.doi.org/10.1016/S0140-6736(94)90694-7 [16] Butler, M.S., Blaskovich, M.A. and Cooper, M.A. (2013) Antibiotics in the Clinical Pipeline in 2013. The Journal of Antibiotics, 66, 571-591. http://dx.doi.org/10.1038/ja.2013.86 [17] Wagenlehner, F.M., Weidner, W. and Naber, K.G. (2007) Pharmacokine tic Characteristics of Antimicrobials and Op- timal Treatment of Urosepsis. Clinical Pharmacokinetics, 46, 291-305. http://dx.doi.org/10.2165/00003088-200746040-00003 [18] Livermore, D.M., Nichols, T., Lamagni, T.L., Potz, N., Reynolds, R. and Duckworth, G. (2003) Ciprofloxacin-Resis- tant Escherichia coli from Bacteraemias in England; Increasingly Prevalent and Mostly from Men. Journal of Antimi- crobial Chemotherapy, 52, 1040-1042. http://dx.doi.org/10.1093/jac/dkg479 [19] Mavroidi, A., Miriagou, V., Liakopoulos, A., Tzelep i, Ε., Stefos, A., Dalekos, G.N. and Petinak, E. (2012) Ciproflox- acin-Resistant Escherichia coli in Central Greece: Mechanisms of Resistance and Molecular Identification. BMC In- fectious Disea ses, 12, 371. http://www.biomedcentral.com/1471-2334/12/371 [20] Manchanda, V., Sanchaita, S. and Singh, N.P. (2010) Mult idrug Resistant Acinetobacter. Journal of Global Infectious Diseases, 2, 291-304. http://dx.doi.org/10.4103/0974-777X.68538 [21] Obritsch, M.D., Fish, D.N., MacLaren, R. and Jung, R. (2004) National Surveillance of Antimicrobial Resistance in Pseudomonas aeruginosa Isolates Obtained from Intensive Care Unit Patients from 1993 to 2002. Antimicrobial Agents and Chemotherapy, 48, 4606-4610. http://dx.doi.org/10.1128/AAC.48.12.4606-4610.2004 [22] Raja, N.S. and Singh, N.N. (2007) Antimicrobial Susceptibility Pattern of Clinical Isolates of Pseudomonas aeruginosa in a Tertiary Care Hospital. Journal of Microbiology, Immunology and Infection, 40, 45-49. [23] Manno, G., Cruciani, M., Romano, L., Scapolan, S., Mentasti, M., Lorini, R., et al. (2005) Antimicrobial Use and Pseudomonas aeruginosa Susceptibility Profile in a Cystic Fibrosis Centre. International Journal of Antimicrobial Agents, 25, 193-197. http://dx.doi.org/10.1016/j.ijantimicag.2004.11.009 [24] Panlilio, A.L., Culver, D.H., Gaynes, R.P., Banerjee, S., Henderson, T.S., Tolson, J.S., et al. (1992) Methicillin Resis- tant Staphylococcus aureus in US Hospita ls, 1975-1991. Infection Control and Hospital Epidemiology, 13, 582-586. http://dx.doi.org/10.2307/30148460 [25] Mekviwattanawong, S., Srifuengfung, S., Chokepaibulkit, K., Lohsiriwat, D. and Thamlikitkul, V. (2006) Epidemio l- ogy of Staphylococcus aureus Infections and the Prevalence of Infection Caused by Community Acquired Methicil- lin-Resistant St aphylococcus aureus in Hospitalized Patients at Siriraj Hospital. Journal of the Medical Association of Thailand, 89, S106-S117. [26] Diekema, D.J., Pfaller, M.A., Schmitz, F.J., Smayevsky, J., Bell, J., Jones, R.N., et al. (2001) Survey of Infections Due to Staphylococcus Species: Frequency of Occurrence and Antimicrobial Susceptibility of Isolates Collected in the United States, Canada, Latin America, Europe, and the Western Pacific Region for the SENTRY Antimicrobial Sur- veillance Program, 1997-1999. Clinical Infectious Diseases, 32, S114-S132. [27] Thamlikitkul, V., Jintanothaitavorn, D., Sathitmethakul, R., Vaithaya phichet, S., Trakulsomboon, S. and Danchaivijitr, S. (2001) Bacterial Infections in Hospitalized Patients in Thailand in 1997 and 2000. Journal of the Medical Associa- tion of Thailand, 84, 666-673. [28] Hiramatsu, K., Hanaki, H., Ino, T., Yabuta, K., Oguri, T. and Tenover, F.C. (1997) Methicillin-Resistant Staphylococ- cus aureus Clinical Strain with Reduced Vancomycin Susceptibility. Journal of Antimicrobial Chemotherapy, 40, 135- 136. http://dx.doi.org/10.1093/jac/40.1.135 [29] Ploy, M.C., Grelaud, C., Martin, C., de Lumley, L. and Denis, F. (1998) First Clinical Isolate of Vancomycin-Inter- mediate Staphylococcus aureus in a French Hospital. Lancet, 351, 1212. http://dx.doi.org/10.1016/S0140-6736(05)79166-2 [30] Hofmann, J., Cetron, M.S., Farley, M.M., Baughman, W.S., Facklam, R.R., Elliott, J., Deaver, K.A. and Breiman, R.F. (1995) Prevalence of Drug-Resistant Streptococcus pneumoniae in Atlanta. The New England Journal of Medicine, 333, 481-486. http://dx.doi.org/10.1056/NEJM199508243330803
|