Journal of Cancer Therapy
Vol.2 No.2(2011), Article ID:5596,10 pages DOI:10.4236/jct.2011.22014
Applications of Fluorescence in Situ Hybridization (FISH) for Detecting Genetic Changes in Hematological Malignancies
![]()
1Vocational School of Health Services, Adiyaman University, Adiyaman, Turkey;
2Department of Medical Biology and Genetics, Çukurova University, Balcali, Adana, Turkey;
3Department of Hematology, Çukurova University, Balcali, Adana, Turkey.
Email: osdemir@cu.edu.tr
Received March 31st, 2011; revised May 8th, 2011; accepted May 16th, 2011.
Keywords: Flourescence in Situ Hybridization, BCR/ABL, Gene Rearrangements
ABSTRACT
Fluorescence in situ hybridization (FISH) has become an important tool both for defining initial chromosomal abnormalities within a disease process, and for monitoring response to therapy as well as minimal residual disease. We report the results of interphase FISH (iFISH) analysis of 92 patients. We have used five different FISH probes to detect common cytogenetic rearrangements associated with hematological malignancies. A total of 83 patients were screened for BCR/ABL gene rearrangements. Displayed iFISH patterns of BCR/ABL gene rearrangements in 37.3% of patients (31/83) ranged between 10% to 98%. In addition, while 3 patients and one patient with AML showed t(15; 17) (12.5%) and inv(16; 16) (8.3%) respectively, t(8; 21) was not found. Furthermore, secondary chromosomal aberrations (6.5% of all cases) were clearly non random in the present study. The diagnosis of BCR/ABL gene rearrangements are likely become an important tool for the monitoring of therapies in patients with CML. Atypical patterns also may have clinical prognostic implications. Further studies in larger groups of patients are needed in order to elucidate the role of AML1/ETO, PML/RARA, CBFB and p53, and to identify the specific chromosomal regions and interacting genes involved in this process.
1. Introduction
The recent World Health Organization (WHO) classification of tumors of hematopoietic and lymphoid tissues emphasizes the importance of chromosomal abnormalities for accurate diagnosis, appropriate treatment, and monitoring response to therapy [1]. Human malignancies may be developed by a variety of mechanisms, including inactivation of tumor suppressor genes, activation of oncogenes, and general genomic instability. Tumor-specific chromosomal translocations, and other genetic abnormalities, have been described for a large number of hematopoietic and lymphoid malignancies [2,3]. For many years, conventional karyotyping has been used as the sole diagnostic tool for hematologic malignancies. Flouresence In Situ Hybridization (FISH) provides an important adjunct to conventional cytogenetics and molecular studies in the evaluation of chromosome abnormalities associated with hematologic malignancies. In addition, FISH analysis offers one of the most sensitive, specific, and reliable strategies for identifying acquired genetic abnormalities such as characteristic gene fusions, aneuploidy, loss of a chromosomal region associated with hematologic disorders, and serving as a technique that can help in both the diagnosis of a genetic disease or suggesting prognostic outcomes. It is especially important for cells of patients with leukemia, where the quality of metaphases is often not so good, and is frequently used to monitor the response to therapy in various hematological malignancies [4]. Thus, FISH is widely used today in clinical practice to help in diagnosis and selects appropriate treatments for patients with hematological malignancies [5]. In research, FISH studies are used to investigate the origin and progression of hematological malignancies, and to establish which hematopoetic compartments are involved in neoplastic processes [6]. Interphase FISH (iFSH) is increasingly used for the identification of BCR/ABL gene rearrangements in CML, AML, ALL and CLL. The aim of this study was to determine the frequency of FISH patterns of BCR/ABL, AML1/ETO, PML/RARA, and p53 genes as a diagnostic tool to understand the pathophysiology, diagnosis, treatment, prognosis, and monitoring of disease activity in CML, AML, ALL, CLL and MDS cases.
2. Materials and Metods
2.1. Patients
A total of 92 patients with a known hematological disorder including 50 (54.34%) CML, 25 (27.2%) AML, 7 (7.6%) ALL, 4 (4.35%) CLL, and 6 (6.52%) MDS patients diagnosed at Departments of Hematology were referred to Department of Medical Biology and Genetics, Faculty of Medicine, Çukurova University for a routine iFISH analysis. There were 59 males and 33 females. Their ages ranged from 20 to 81 years, with a mean (±SD) age of 50, 45 ± 15, 19 years (Tables 1 and 2).
2.2. Slide Preparation and Flourescence in Situ Hybridization
A 2-ml venous blood taken from all patients in order to determine the t(9; 22), t(8; 21), t(15; 17), and/or inv(16) anomalies, and/or deletion of p53 gene. Standard techniques were used for harvesting and slide preparation without incubation. After incubating slides at room temperature overnight, fluorescence in situ hybridization was performed. For these purposes, LSI BCR/ABL ES Dual Color Translocation probe (Vysis), LSI PML/RARA Dual Color Dual Fusion Translocation Probe (Vysis), LSI AML1/ETO Dual Color Dual Fusion Translocation Probe (Vysis), LSI CBFB Dual Color Break Apart Rearrangement Probe (Vysis) and LSI p53, 17p13.1, Spectrum Orange Probe (Vysis) were used. Firstly slides were pretreated with 2XSSC for 5 min at room temperature and then immersed them in the solution contained HCl (1N), water and pepsin A (2:200:2 v/v/v) for 30 min at 37˚C. After the time, slides were washed with water immediately. Then, they were washed with PBS, PBS/MgCl2.6H2O, and PBS/MgCl2.6H2O with paraformaldehyde for 2 min, 2 min, and 10 min, respectively, and then passed through a dehydration series of 70, 85 and 100% ethanol for 3 min each. Then slides were left to dry. Simultaneously, 10 µl of each probe mixtures were applicated on slides immediately, and a coverslip was sealed onto the slides with rubber cement. The slides put in ThermoBrite Denaturation/ Hybridization System and denaturated 5 min at 95˚C and hybridized overnight at 37˚C. For posthybridization process, slides were washed with 0.4XSSC/0.3% Tween 20 for 2 min at 73˚C and 2XSSC/0.1% Tween 20 for 1 min at room temperature, respectively. After the slides were waited for drying at a dark room. In the next step, DAPI tube was vortexed and slides were counterstained with 10 µl of it, and then waited 30 min at –20˚C. At the end of time, slides were analysed at flourescent microscopy using red, green and DAPI filters. Interphase cells were analyzed using a BX51 Olympus fluorescence microscope equipped with Cytovision Probe Software (Applied Imaging, Santa Clara, CA). For each case and probe, a minimum of 100 interphase cells was evaluated for the signal patterns.
3. Results
In the present study, 92 hematological patients including CML [50 (54.34%)], AML [25 (27.17%)], ALL [7 (7.6%)], CLL [4 (4.35%)], and MDS [6 (6.52%)] were analyzed for some parameters [t(9; 22), t(8; 21), t(15; 17), inv (16; 16) and p53] (Table 1).
A total of 83 patients were screened for BCR/ABL gene rearrangements. The great majority of the patients analyzed including most CML (25/48, 52.1%), AML (4/22, 18.2%), ALL (1/7, 14.3%) and MDS (1/6, 16.7%) cases displayed iFISH patterns of BCR/ABL gene rearrangements (31/83, 37.3%) ranged between 10% to 98% (Tables 2 and 3). Fifty-two percent of CML patients demonstrated Ph translocation while 48% were negative for the Ph chromosome. Approximately 96.2% of Ph-positive patients displayed the typical FISH signal pattern. The iFISH patterns of BCR/ABL gene rearrangements showed four patterns. Pattern A, typical iFISH pattern, consisted of one fusion-der (22)-, one greennonrearranged 22- and two red-der (9) plus the nonrearranged chromosome 9- signals (1F2R1G) (29/83, 34.9%). Atypical patterns (1F1R1G and1F1R2G) among the Ph-positive patients included atypical BCR/ ABL fused gene rearrangements on chromosome 9 or 9q deletion of the rearranged chromosome 9; coexistence of der(9q) and der(22q) deletions (2/25, 7.7%) (Table 4) (Figure 1).
There are 27 patients- 23 CML, 2 AML, 1 ALL and one MDSshowed typical iFISH pattern (Pattern A) (Table 3, Figure 1). On the other hand, as illustrated in Figure 1 and shown in Table 4, pattern B (one fusion, one red and one green signals) was found in 3 patients (3.6%) included 1 CML and 2 AML. Pattern C (one fusion, one red and two green signals) was shown by
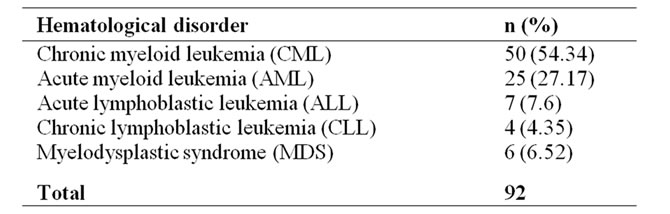
Table 1. The distribution of hematological cancers in the present study.



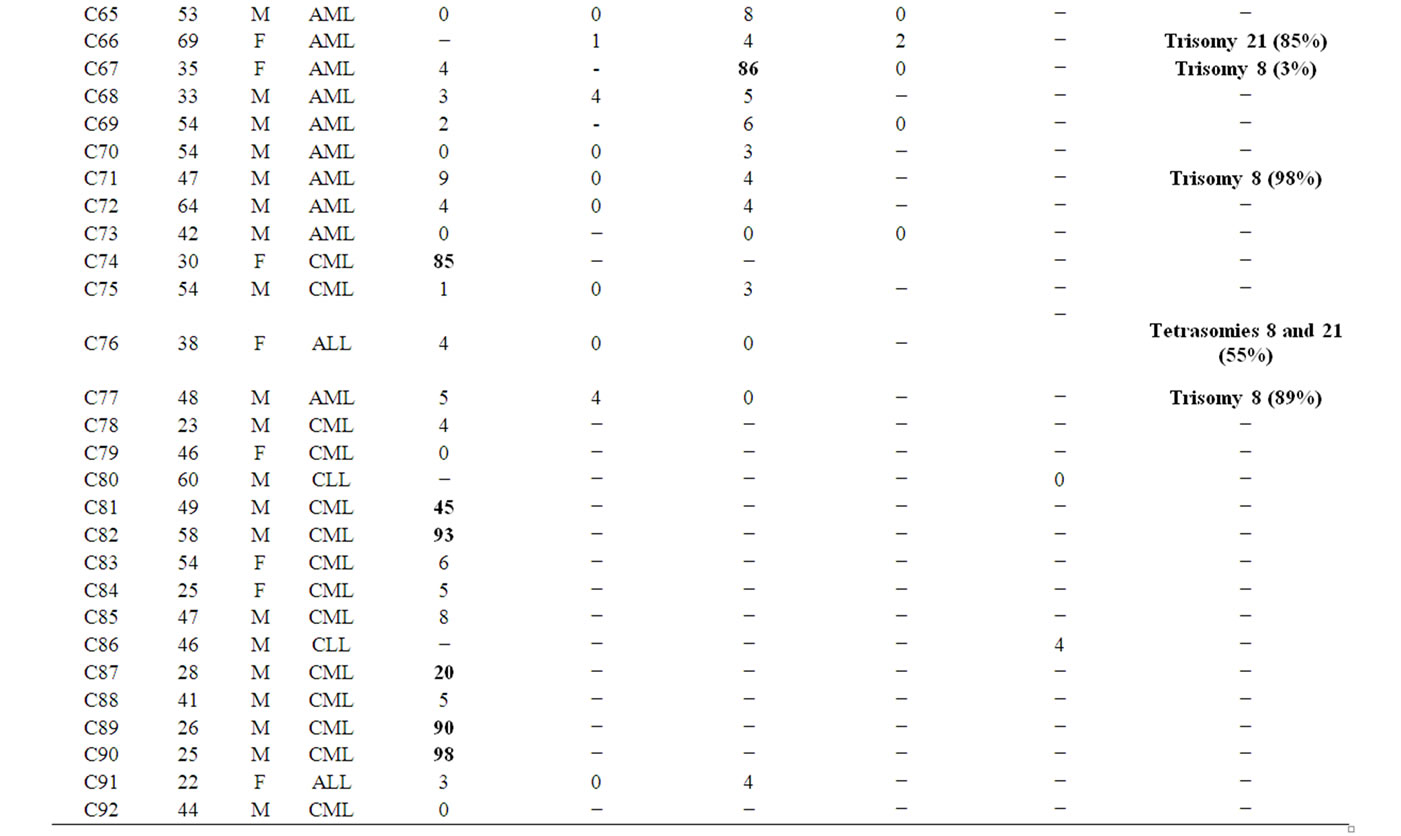
Table 2. The demographic information and iFISH results of the study population.

Table 3. The distribution of the patients according to the results of BCR/ABL, AML/ETO, PML/RARA, CBFB and P53.

Table 4. Distribution of typical and atypical iFISH patterns with the ES probe in BCR/ABL+ leukemias studied at diagnosis.
only one CML patients (2%) (Table 4, Figure 1). Although t(8;21) was not detected in our patients, the t(15;17) was seen in three AML patients [3/41 (7.31%) analyzed patients for PML/RARA], C28, C47, and C67, with the rates of 11%, 79%, and 86%, respectively (Tables 2 and 3, Figures 2(a) and (b)). Invertion (16; 16) was studied in 19 patients. Only one AML patient (C11) showed this invertion in 12/100 interphase cells analyzed (Tables 2 and 3, Figures 2(c) and (d)). The p53 gene deletion were screened in 7 patients included 2 CML, one AML, and 4 CLL. Only one CML patient (C54) showed positive result with 10% percentage. The other patients didn’t show any deletion in p53 gene (Tables 2 and 3).
On the other hand, some numerical chromosomal deficiencies or gains were observed in 8/92 of our patients. In C35, a 74-years old AML patient, monosomy 9 was seen in 70/100 analyzed cells. In C38 (MDS), C67 (AML), C71 (AML), and C77 (AML), trisomy 8 cells were obtained in 16%, 3%, 98%, and 89%, respectively. Also, trisomy 21 in C66 (AML) (85/100 cells), tetrasomies 8 and 21 in C76 (ALL) (55%) and, trisomy 8 together with monosomy 21 (37%) and monosomy 17 (25%) in C49 (AML) were seen in the study (Table 2, Figure 3).
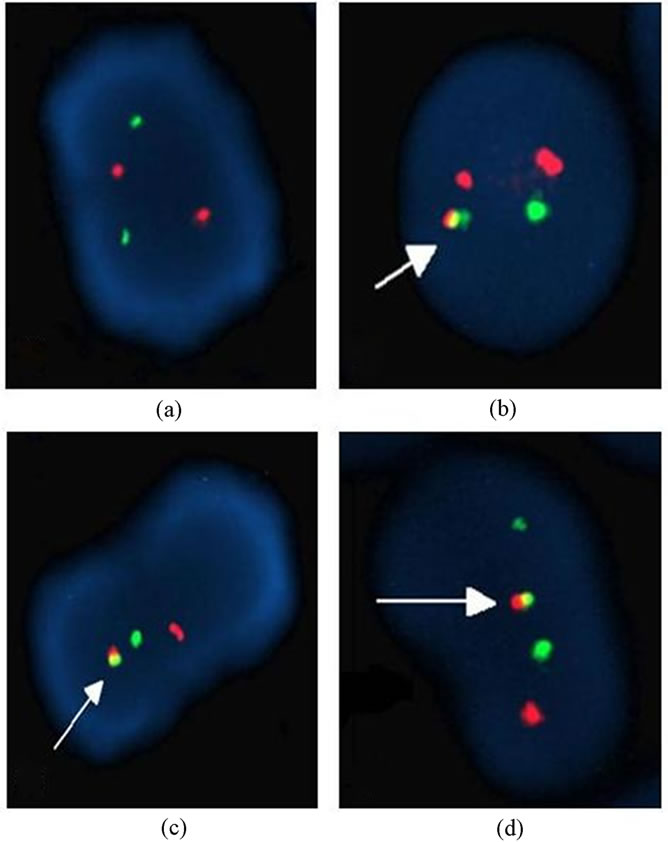
Figure 1. Different interphase FISH (iFISH) patterns found with the LSI BCR/ABL ES Dual Color Translocation probe (a) Normal nuclei, (b) 1F 1G 2R pattern (pattern A), (c) 1F 1G 1R pattern (pattern B), (d) 1F 2G 1R pattern (pattern C).
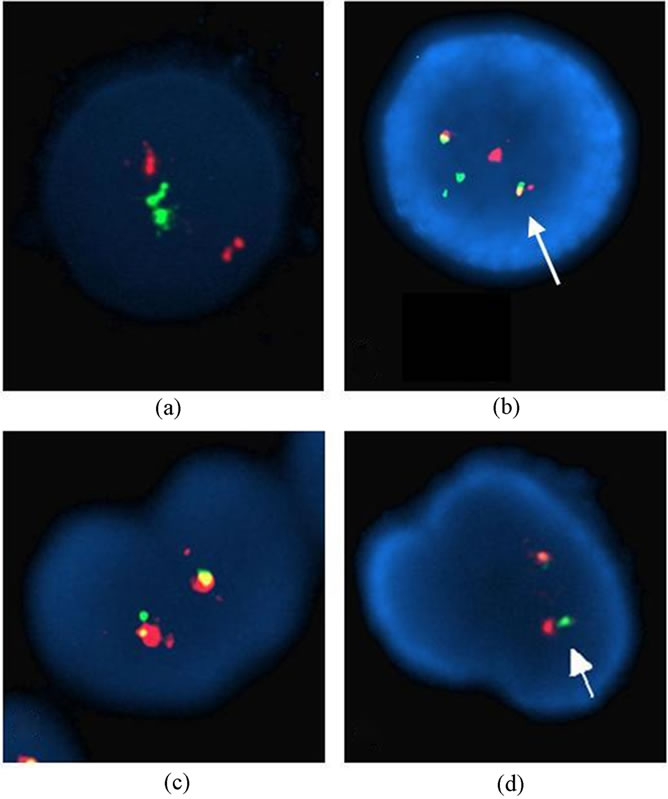
Figure 2. Interphase nuclei showing normal pattern and t(15;17) found with LSI PML/RARA Dual Color Dual Fusion Translocation Probe (a-b), normal pattern and inv(16) found with LSI CBFB Dual Color Break Apart Rearrangement Probe (c-d).
4. Discussion
In this study, we report the results of BCR/ABL, AML1/ETO, PML/RARA, CBFB and p53 gene rearrangements in hematological disorders including CML, AML, ALL, CLL and MDS as a genetic and molecular diagnostic model and try to understand their role in the pathophysiology, prognosis, and monitoring of these disease processes.
CML is the first hematological cancer type known to be associated with a specific clonal expansion of t(9; 22). All patients with CML have an abnormal clone with fusion of BCR and ABL1 loci; at least 90% of patients have a t(9; 22), and the rest have a complex or cryptic variant of this translocation. This finding was first reported in CML cases by Hagemeier et al. [7] and afterwards by other authors [8-11]. Our results show that among cases studied at diagnosis, the presence of positive BCR/ABL fusion results were more frequently observed in CML (52.1%) compared to AML (18.2%), ALL (14.3%) and MDS (16.7%) cases (Table 3). Among Chinese patients with CML, the 62% of 158 cases were Ph positive [12]. In another report, 87% of CML patients were positive for the Ph chromosome [13].
We detected a BCR/ABL1 fusion in 14.3% of adults
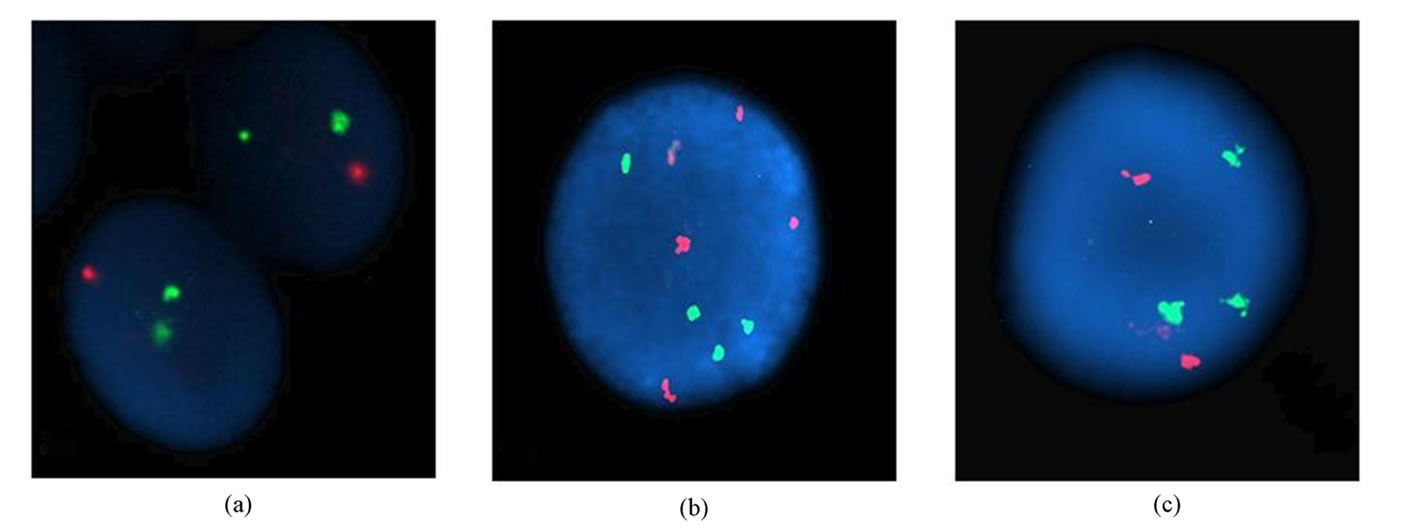

Figure 3. Abnormal FISH patterns (a) monosomy 9 with LSI BCR/ABL ES Dual Color Translocation probe, (b) tetrasomies 8 and 21, (c) trisomy 21, (d) trisomy 8, (e) trisomy 8 and monosomy 21 found with LSI AML1/ETO Dual Color Dual Fusion Translocation Probe, (f) del (17) found with LSI PML/RARA Dual Color Dual Fusion Translocation Probe.
with ALL and 18.2% of adults with AML. A similar BCR/ABL1 fusion occurs in 17% of adults with ALL and 1% of patients with AML [14]. Some investigators have suggested that these Ph-positive forms of acute leukemia may be similar to lymphoid or myeloid blast crisis of CML [15]. Such findings emphasize the importance of performing baseline FISH studies and that these patterns are verified using metaphases since the signal patterns may change during the course of the disease. Among the diagnostic studies, the use of BCR/ABL FISH ES probe was of more benefit in patients with CML among suspected hematologic malignancies and may confer a worse prognosis. Further studies in larger groups of patients are needed in order to elucidate the role of AML1/ETO, PML/RARA, CBFB and p53, and to identify the specific chromosomal regions and interacting genes involved in this process.
The establishment of signal patterns with FISH is important as atypical patterns may have clinical diagnostic and prognostic implications. Additional karyotypic changes may occur in BCR/ABL+ CML, ALL and AML cases [16-18]. These atypical iFISH patterns were most frequently seen as a result of additional numerical changes (most often gain or loss of chromosome 9 or 22). In the present study, the frequency (37.3%) of both typical and atypical BCR/ABL gene rearrangements was analyzed in 83 patients with CML, AML, ALL and MDS. The most frequently detected patterns with the ES probe corresponded to typical BCR/ABL gene rearrangement involving the 1F2R1G (one fusion, two red and one green signals). Among the 48 CML cases, 25 cases were Ph positive, of which 23 cases (92%) were typical FISH pattern (1F2R1G), the other 2 cases (8%) showed 2 different types of atypical FISH patterns (1F1R1G and 1F1R2G) (Table 4, Figure 1). Both patterns 1F1R1G and 1F1R2G were not found in ALL and MDS cases, but observed in CML and AML cases. In a study including Chinese patients with CML, Philadelphia positive cases (70.4%) demonstrated typical FISH pattern; 29.6% had atypical FISH pattern with 12 different types [12]. The frequency of atypical FISH patterns is comparable to the 15% reported by Cohen et al. [19] and Reid et al. [20]. In our study, 8% of patients with CML had atypical FISH patterns that the atypical FISH patterns were due to variant translocation; atypical BCR/ABL fused gene rearrangements on chromosome 9 or 9q deletion of the rearranged chromosome 9; coexistence of der(9q) and der(22q) deletions. Hagemeijer et al. [7] and Nacheva et al. [21] postulated a two-step rearrangement for the localisation of BCR/ABL gene on chromosome 9. A BCR/ABL translocation occurs initially, followed by a translocation between the derivatives 9 and 22. This results in the masking of the exchange, with the BCR/ABL transposition to chromosome 9. Alternatively, it may involve an insertion of the BCR gene from chromosome 22 into the ABL gene on chromosome 9 [22]. Indeed, Sinclair et al. [16] found that deletions of the proximal sequences on the derivative chromosome 9q were associated with a poor prognosis on standard drug therapy. In addition, it has been shown that the deletion is not acquired during disease progression as the deletion tends to be consistent throughout the course of the disease [19]. However, no distinctive clinical features were found in the patients with typical or atypical transcripts. At the same time, the typical 1F2R1G rearrangement of BCR/ABL in AML, ALL and MDS was observed in 4 cases (Table 4). The atypical patterns (1F1R1G) were also found in two AML cases (50%). ALL and MDS cases did not show an atypical iFISH pattern (Table 4). We did not observe extensive translocations, deletions and invertion of chromosomes 9 and 22 that can arise during follow up. However, in an other study, the most common atypical iFISH pattern among Ph-positive patients was seen to be 1F1R1G rearrangement [23]. Lawce et al. [24], for this subgroup of patients, detection of residual disease is difficult since the pattern is indistinguishable from juxtaposition artifacts as a result of random overlapping between chromosomes 9 and 22. Primo et al. [23] and Lim et al. [13] demonstrated the presence of such a pattern in about 12.5%, 9% and 10.4% respectively. For example, a 1F1R1G pattern at diagnosis may change to a 2F1R2G pattern as a result of a gain of a Ph chromosome, and this may inadvertently be misinterpreted as atypical iFISH abnormal pattern while in fact it has the clinical implication of secondary clonal changes. Molecular studies have shown that the poor prognosis is related to loss of BCR/ABL expression, increased BCR/ABL transcripts, or genetic instability [25]. It was shown recently that there were no difference in the survival rates between patients with and without deletions when imatinib treatment was given [26]. Nevertheless, the time to disease progression was significantly shorter for patients on imatinib treatment regardless of chronic or advanced phases. In our study, none of Ph-positive patients showed a gain of a Ph chromosome. This state is not usually associated with disease progression or secondary genetic changes, and is one of the major pathways of clonal evolution seen during blast crisis. Primo et al. [23] showed that the presence of additional Ph chromosomes was one of the most common underlying genetic abnormalities when using FISH to identify BCR/ABL rearrangements in CML and ALL. These gains were more frequently observed in ALL than in CML.
The AML1 gene has recently attracted a lot of interest in terms of its role in leukemogenesis. The translocation between chromosomes 8 and 21 is the most frequent abnormality seen in approximately 46% of patients with AML and an aneuploid karyotype. Amplification of AML1 has been reported in AML, where structural rearrangements resulted in partial gains of chromosome 21. Acquired trisomy 21, a frequent finding in childhood ALL, itself produces one extra copy of the gene. Structural rearrangements involving duplication of the long arm of chromosome 21, have also been described [27-29]. Interestingly, we have seen that a total 35 patients with AML (18 cases), CML (7 cases), ALL (5 cases) and MDS (5 cases) were not associated with different numbers of iFISH patterns with the usage of AML1/ETO [t(8;21)] Dual Color Dual Fusion Translocation probe. However, the PML/RARA [t(15; 17)] and CBFB[inv(16)] probes gave positive results only for 12.5% (3/24) and 8.3% (1/12) of patients with AML, respectively, and for p53 [del(17p13.1)] in one of two patients with CML (Table 3). It has been estimated that about 50% of all tumours have mutations in p53, and the p53 pathway may be nonfunctional for other reasons in many others. p53 is also known for its role in monitoring genomic stability, but the mechanisms underlying this function are not fully understood.
FISH can be a useful tool for monitoring remission status when clonal chromosome abnormalities have been identified at diagnosis. By conventional cytogenetic studies many of patients with myeloproliferative disorders, MDS or ALL are found to have an abnormal clone [eg, trisomy 8, monosomy 7, del(20q), etc.] that may help explain their clinical signs and symptoms. The addition of FISH to existing karyotyping procedures has led to a more definite assessment of the cytogenetic profiles of MDS and AML [30]. The sensitivity of FISH in the detection of numerical abnormalities in myeloid neoplasias has been the subject of several investigations. The most common numerical anomalies in myeloproliferative disorders are −Y, +8, +9, and −7. Trisomy 8 is a common cytogenetic abnormality found in the bone marrow of patients with myeloproliferative disorders, MDS or acute nonlymphocytic leukemia [31,32]. Just as, the secondary chromosomal aberrations (8.5% of all cases) are clearly nonrandom in the present study; with the most common chromosomal abnormalities being +8 (62.5% of cases with additional changes), +21(25%), del(17)(12.5%), and monosomy 9(12.5%) by interphase FISH. We suggest that all these aberrations, occurring in 75% of AML cases, 12.5% MDS and 12.5% of ALL with secondary changes, should be denoted major route abnormalities. Among the approximately 1600 CML cases with standard Ph and secondary changes published to date [33], +8, +Ph, i(17q) and +19 have been described in 35, 31, 21, and 14% of the cases, respectively. The incidence of trisomy 8 in 140 Chinese patients with CLL were found in only two patients (1.4%) [34]. Our study demonstrates that trisomy 8 was not found in CLL, and its role in prognosis of CLL remains unknown. Using conventional cytogenetic methods in which 20 to 30 metaphase cells are typically examined, it is possible to find 2 or more metaphases with trisomy 8 in many of these patients. According to the ISCN definition, 6 these specimens would have an abnormal clone. In our study, +8, −9 and +21 are clonal by ISCN criteria and may not reflect either artifact or the potential emergence of an abnormal clone. All other additional chromosomal abnormalities occur in less than 10% of the CML cases, the most frequent being –Y, +21, +17, –7, and –17 [35]. Anastasi et al. [36] demonstrated that of three patients with MDS associated with trisomy 8. Structural rearrangements involving duplication of 21q, dup(21q) have also been described [27-29]. Combining these myeloid disorder groups and our findings, the most common additional chromosomal changes are +8, +Ph, i(17q), del(17), +19, –Y, +21, +17, –9 and –7. These abnormalities were proposed to follow the major route of clonal evolution, whereas other changes evolving more rarely were suggested to follow the minor route [37]. In view of our present knowledge, it may be reasonable to expand the major evolutionary route to include all these aberrations, using 5% as a reasonable cut-off value. Our study shows that FISH analysis using directly labeled DNA probes specific for chromosome 8 is an excellent way to detect trisomy 8 cells in hematologic malignancies. iFISH could also be used to monitor the effects of treatment, in patients found to have trisomy 8 at the time of diagnosis, or to detect early relapse. We believe this study predicts that such analyses could be extended to other chromosomal trisomies and monosomies; although the latter will require even stricter attention to normal value studies.
In conclusion, our results confirmed that iFISH has become an invaluable tool in defining and monitoring acquired chromosome abnormalities associated with hematologic malignancies, and is a sensitive technique for the evaluation of response to treatment in patients with CML. Despite the high incidence of typical iFISH patterns of BCR/ABL gene rearrangements, atypical patterns are also found in BCR/ABL+ CML and AML. Moreover, our study suggests that FISH analysis using directly labeled DNA probes specific for chromosome 8 is an excellent way to detect trisomy 8 cells in hematologic malignancies, or it could be used to supplement conventional cytogenetic studies in patients with normal or uncertain results.
5. Acknowledgements
This study was supported by the Department of Medical Biology and Genetics, Faculty of Medicine, Çukurova University, Adana-Turkey.
REFERENCES
- E. S. Jaffe, N. L. Harris, H. Stein and J. W. Vardiman, “World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues,” IARC Press, Lyon, 2001.
- F. Mitelman, B. Johansson and F. Mertens, “The İmpact of Translocations and Gene Fusions on Cancer Causation,” Nature Reviews Cancer, Vol. 7, No. 4, 2007, pp. 233-245. doi:10.1038/nrc2091
- J. D. Rowley, “Chromosomal Translocations: Revisited yet Again”, Blood, Vol. 112, No. 6, 2008, p. 2183. doi:10.1182/blood-2008-04-097931
- B. Mohr, M. Bornhauser, U. Platzbecker, J. FreibergRichter, R. Naumann, G. Prange-Krex, J. Mohm, F. Kroschinsky, G. Ehninger and C. Thiede, “Problems with İnterphase Fluorescence in Situ Hybridization in Detecting BCRABL Positive Cells in Some Patients Using a Novel Technique with Extra Signals,” Cancer Genet Cytogenet, Vol. 127, No. 2, 2001, pp. 111-117. doi:10.1016/S0165-4608(00)00371-X
- G. W. Dewald, T. C. Smyrk, E. C. Thorland, R. R. McWilliams, D. L. van Dyke, J. G. Kefe, K. J. Belongie, S. A. Smoley, D. L. Knutson, S. R. Fink, A. E. Wiktor and G. M. Petersen, “Fluorescence in Situ Hybridization to Visualize Genetic Abnormalities in Interphase Cells of Acinar Cell Carcinoma, Ductal Adenocarcinoma, and Islet Cell Carcinoma of the Pancreas,” Mayo Clinic Proceedings, Vol. 84, No. 9, 2009, pp. 801-810. doi:10.4065/84.9.801
- W. G. Finn and L. C. Peterson, “Hematopathology in Oncology,” Kluwer Academic Publishers, New York, 2004. doi:10.1007/b113185
- A. Hagemeijer, A. Buijs, E. Smit, B. Janssen, G. J. Creemers, D. van der Plas and G. Grosveld, “Translocation of BCR to Chromosome 9: A New Cytogenetic Variant Detected by FISH in Two Ph-Negative, BCR-Positive Patients with Chronic Myeloid Leukemia,” Genes, Chromosomes and Cancer, Vol. 8, No. 4, 1993, pp. 237-245. doi:10.1002/gcc.2870080406
- M. J. Macera, L. J. Smith, E. Frankel, P. Szabo and R. S. Verma, “A Philadelphia Negative Chronic Myelogenous Leukemia with the Chimeric BCR/ABL Gene on Chromosome 9 and a B3-A2 Splice Junction,” Cancer, Genetics and Cytogenetics, Vol. 101, 1998, pp. 143-147. doi:10.1016/S0165-4608(97)00257-4
- W. T. Hsu, H. Preisler, K. Szego, R. Sprudzs and X. Z. Gao, “The ABL/BCR Fusion Gene on Chromosome 9 in Ph-Negative Chronic Myelogenous Leukemia: A Case for Vigilance in Fluorescence in Situ Hybridization İnterpretation,” Cancer, Genetics and Cytogenetics, Vol. 104, 1998, pp. 57-60. doi:10.1016/S0165-4608(97)00430-5
- E. Abruzzese, M. J. Pettenati, K. Cox, B. Jackle, R. G. Watts, A. J. Carroll, M. C. Steuterman and P. N. Rao, “Identification of BCR/ABL Fusion on Chromosome 9 by Fluorescence in Situ Hybridization in Two Chronic Myeloid Leukemia Cases,” Cancer, Genetics and Cytogenetics, Vol. 105, 1998, pp. 164-167. doi:10.1016/S0165-4608(98)00020-X
- K. S. Reddy and B. Grove, “A Philadelphia-Negative Chronic Myeloid Leukemia with a BCR/ABL Fusion Gene on Chromosome 9,” Cancer, Genetics and Cytogenetics, Vol. 107, No. 1, 1998, pp. 48-50. doi:10.1016/S0165-4608(98)00064-8
- H. R. Qiu, K. R. Miao, R. Wang, C. Qiao, J. F. Zhang, S. J. Zhang, S. X. Qian, W. Xu and J. Y. Li, “The Application of Fluorescence in Situ Hybridization in Detecting Chronic Myeloid Leukemia,” Chinese Journal of Medical Genetics, Vol. 26, No. 2, 2009, pp. 207-210.
- T. H. Lim, S. L. Tien, P. Lim and A. S. Lim, “The Incidence and Patterns of BCR/ABL Rearrangements in Chronic Myeloid Leukaemia (CML) Using Fluorescence In situ Hybridisation (FISH),” Annals Academy of Medicine Singapore, Vol. 34, No. 9, 2005, pp. 533-538.
- G. W. Dewald, C. R. Schad, E. R. Christensen, A. L. Tiede, A. R. Zinsmeister, S. L. Spurbeck, S. N. Thibodeau and S. M. Jalal, “The Application of Fluorescent in Situ Hybridization to Detect Mbcr/Abl Fusion in Variant Ph Chromosomes in CML and ALL,” Cancer, Genetics and Cytogenetics, Vol. 71, 1993, pp. 7-14. doi:10.1016/0165-4608(93)90196-S
- J. L. Diez-Martin, G. W. Dewald and R. V. Pierre, “Possible Cytogenetic Distinction between Lymphoid and Myeloid Blast Crisis in Chronic Granulocytic Leukemia”, American Journal of Hematology, Vol. 27, No. 1, 1988, pp. 194-203. doi:10.1002/ajh.2830270309
- P. B. Sinclair, E. P. Nacheva, M. Leversha, N. Telford, J. Chang, A. Reid, A. Bench, K. Champion, B. Huntly and A. R. Gren, “Large Deletions at the T(9; 22) Breakpoint are Common and may İdentify a Poor-Prognosis Subgroup of Patients with Chronic Myeloid Leukemia,” Blood, Vol. 95, 2000, pp. 738-743.
- C. Herens, F. Tassin, V. Lemaire, Y. Beguin, E. Collard, S. Lampertz, C. Croisiau, M. Lecomte, B. De Prijk, L. Longrée and L. Koulischer, “Deletion of the 5’-ABL Region: A Recurrent Anomaly Detected by Fluorescence in Situ Hybridization in About 10% of PhiladelphiaPositive Chronic Myeloid Leukaemia Patients,” British Journal of Haematology, Vol. 110, No. 1, 2000, pp. 214-216. doi:10.1046/j.1365-2141.2000.02142.x
- B. J. Huntly, A. G. Reid, A. J. Bench, L. J. Campbell, N. Telford, P. Shepherd, J. Szer, H. M. Prince, P. Turner, C. Grace, E. P. Nacheva and A. R. Gren, “Deletions of the Derivative Chromosome 9 Occur at the Time of the Philadelphia Translocation and Provide a Powerful and İndependent Prognostic İndicator in Chronic Myeloid Leukemia,” Blood, Vol. 98, No. 6, 2001, pp. 1732-1738. doi:10.1182/blood.V98.6.1732
- N. Cohen, G. Rozenfeld-Granot, I. Hardan, F. BrokSimoni, N. Amariglio, G. Rechavi and L. Trakhtenbrot, “Subgroup of Patients with Philadelphia-Positive Chronic Myelogenous Leukemias Characterized by a Deletion of 9q Proximal to ABL Gene: Expression Profiling, Resistance to İnterferon Therapy, and Poor Prognosis,” Cancer, Genetics and Cytogenetics, Vol. 128, No. 2, 2001, pp. 114-119. doi:10.1016/S0165-4608(01)00412-5
- A. G. Reid, B. J. Huntly, C. Grace, A. R. Green and E. P. Nacheva, “Survival İmplications of Molecular Heterogeneity in Variant Philadelphia-Positive Chronic Myeloid Leukaemia,” British Journal of Haematology, Vol. 121, No. 3, 2003, pp. 419-427. doi:10.1046/j.1365-2141.2003.04291.x
- E. Nacheva, T. Holloway, K. Brown, D. Bloxham and A. R. Gren, “Philadelphianegative Chronic Myeloid Leukemia: Detection by FISH Of BCR-ABL Fusion Gene Localized Either to Chromosome 9 or Chromosome 22,” British Journal of Haematology, Vol. 87, No. 2, 1994, pp. 409-412. doi:10.1111/j.1365-2141.1994.tb04933.x
- A. N. Mohamed, F. Koppitch, M. Varterasian, C. Karanes, K. L. Yao and F. H. Sarkar, “BCR/ABL Fusion Located on Chromosome 9 in Chronic Myeloid Leukemia with a Masked Ph Chromosome,” Genes Chromosome Cancer, Vol. 13, No. 2, 1995, pp. 133-137. doi:10.1002/gcc.2870130210
- D. Primo, M. D. Tabernero, A. Rasillo, J. M. Sayagues, A. B. Espinosa, M. C. Chillon, R. Garcia-Sanz, N. Gutierrez, M. Giralt, A. Hagemeijer, J. F. San Miguel and A. Orfao, “Patterns of BCR/ABL Rearrangements by İnterphase Fluorescence in Situ Hybridization (FISH) in BCR/ABL+ Leukemias: İncidence and Underlying Genetic Abnormalities,” Leukemia, Vol. 17, 2003, pp. 1124-1129. doi:10.1038/sj.leu.2402963
- H. Lawce, C. Durum, N. Unsworth, S. Olson and R. E. Magenis, “BCR-ABL FISH: Probes, Patterns and Prognosis,” Association of Genetic Technologists, Vol. 28, 2002, pp. 40-46.
- B. J. Huntly, A. J. Bench, E. Delabesse, A. G. Reid, J. Li, M. A. Scott, L. Campbell, J. Byrne, E. Pinto, A. Brizard, D. Niedermeiser, E. P. Nacheva, F. Guilhot, M. Deininger and A. R. Gren, “Derivative Chromosome 9 Deletions in Chronic Myeloid Leukemia: Poor Prognosis is not Associated with Loss of ABL-BCR Expression, Elevated BCR-ABL Levels, or Karyotypic İnstability,” Blood, Vol. 99, No. 12, 2002, pp. 4547-4553. doi:10.1182/blood.V99.12.4547
- B. J. Huntly, F. Guilhot, A. G. Reid, G. Vassiliou, E. Hennig, C. Franke, J. Byrne, A. Brizard, D. Niederwieser, J. Freeman-Edward, G. Cuthbert, N. Bown, R. E. Clark, E. P. Nacheva, A. R. Green and M. W. Deininger, “Imatinib İmproves but May not Fully Reverse the Poor Prognosis of CML Patients with Derivative Chromosome 9 Deletions,” Blood, Vol. 102, No. 6, 2003, pp. 2205-2215. doi:10.1182/blood-2002-09-2763
- M. B.-L. Coniat, K. F. Nguyen, M. T. Daniel, O. A. Bernard and R. Berger, “Chromosome 21 Abnormalities with AML1 Amplification in Acute Lymphoblastic Leukemia,” Genes Chromosome Cancer, Vol. 32, No. 3, 2001, pp. 244–249. doi:10.1002/gcc.1188
- F. Morel, A. Herry, M.-J. Le Bris, N. Douet-Guilbert, G. Le Calvez, V. Marion, C. Berthou and M. De Braekeeler, “AML1 Amplification in a Case of Childhood Acute Lymphoblastic Leukemia,” Cancer, Genetics and Cytogenetics, Vol. 137, No. 2, 2002, pp. 142-145. doi:10.1016/S0165-4608(02)00566-6
- D. Penther, C. Preudhomme, P. Talmant, C. Roumier, A. Godon, F. Mechinaud, N. Milpied, R. Bataille and H. Avet-Loiseau, “Amplification of AML1 Gene is Present in Childhood Acute Lymphoblastic Leukemia but not in Adult, and is not Associated with AML1 Gene Mutation,” Leukemia, Vol. 16, No. 6, 2002, pp. 1131-1134. doi:10.1038/sj.leu.2402479
- C. Preudhomme, D. Warot-Loze, C. Roumier, N. Grardel-Duflos, R. Garand, J. L. Lai, N. Dastugue, E. Macintyre, C. Denis, F. Bauters, J. P. Kerckaert, A. Cosson and P. Fenaux, “High İncidence of Biallelic Point Mutations in the Runt Domain of the AML1/PEBP2 Alpha B Gene in Mo Acute Myeloid Leukemia and in Myeloid Malignancies with Acquired Trisomy 21,” Blood, Vol. 96, 2000, pp. 2862-2869.
- G. W. Dewald, W. A. Wyatt, A. L. Juneau, R. O. Carlson, A. R. Zinsmeister, S. M. Jalal, J. L. Spurbeck and R. T. Silver, “Highly Sensitive Fluorescence in Situ HybridiZation Method to Detect Double BCR/ABL Fusion and Monitor Response to Therapy in Chronic Myeloid Leukemia,” Blood, Vol. 91, No. 9, 1998, pp. 3357-3365.
- D. C. van der Plas, A. B. Hermans, D. Soekarman, E. M. Smit, A. de Klein, N. Smadja, G. Alimena, R. Goudsmit, G. Grosveld and A. Hagemeijer, “Cytogenetic and Molecular Analysis in Philadelphia Negative CML,” Blood, Vol. 73, No. 4, 1989, pp. 1038-1044.
- F. Mitelman, “Recurrent Chromosome Aberrations in Cancer,” Mutat Research, Vol. 462, 2000, pp. 247-253. doi:10.1016/S1383-5742(00)00006-5
- W. Xu, X. Cao, Q. Liu, L. Fan, K. R. Mio, H. R. Qiu, D. X. Zhu, H. X. Qiu and J. Y. Li, “Trisomy 8 in Two Newly Diagnosed Chinese Patients with Chronic Lymphocytic Leukemia,” Cancer, Genetics and Cytogenetics, Vol. 192, No. 2, 2009, pp. 79-81. doi:10.1016/j.cancergencyto.2009.04.007
- B. Johansson, T. Fioretos and F. Mitelman, “Cytogenetic and Molecular Genetic Evolution of Chronic Myeloid Leukemia,” Acta Haematologica, Vol. 107, No. 2, 2002, pp. 76-94. doi:10.1159/000046636
- J. Anastasi, J. Feng, M. M. Le Beau, L. A. Larson, J. Rowley and J. W. Vardiman, “Cytogenetic Clonality in Myelodysplastic Syndromes Studied with Fluorescemnce in Situ Hybridization: Lineage, Response to Growth Factor Therapy, and Clone Expansion,” Blood, Vol. 81, No. 6, 1993, pp. 1580-1585.

