Advances in Bioscience and Biotechnology
Vol.4 No.3A(2013), Article ID:29284,8 pages DOI:10.4236/abb.2013.43A059
Into the life and death: RecA a WISE factor working to integrate survival and evolution in Escherichia coli
![]()
1Museo de la Naturaleza de Cantabria, Plaza Don Pedro Ygareda, Cantabria, Spain
2OAS-BioAstronomy Group, Observatorio Astronómico de Segurilla, Camino Valparaiso S/N Segurilla, Toledo, Spain
Email: chemaseg@yahoo.es
Received 5 January 2013; revised 20 February 2013; accepted 28 February 2013
Keywords: Life & Death; E. coli RecA; RecA Nucleofilament; RecA*; RecA Anfi-Funcionality; SOS Response; Apoptosis-Like Death; Swarming Motility; Multicellularity; Sociability; Cheaters; Cooperation; WISE Factor
ABSTRACT
Escherichia coli RecA has been considered traditionally a cellular protein with multiple vital functions working to ensure the maintenance of integrity of genome in each individual bacterial cell as well as promoting swarming migration in collectivity. On the contrary, recently it has been described that RecA promotes cellular apoptotic-like death (ALD), a pathway of programmed cellular death (PCD). In fact, RecA has been dubbed as the major apoptotic executor in E. coli. From these studies, RecA emerges as a prototypical Gin/Gan protein that despite of their intrinsic vital and lethal anfi-funcionality becomes in a WISE factor: a Worker to Integrate Survival and Evolution in E. coli evolving populations living in community. Here, I provide a review of recent experimental and conceptual advances trying to understand these RecA’s antagonistic roles in appearance contradictory under a unified biological vision.
1. INTRODUCTION
Evolution is the principal issue in Biology [1]. Darwinian evolution depends of two interrelate fundamental processes: emergence of genetic diversity in a population and Natural Selection of the fittest (adapted) organisms in each environmental setting [1]. A genetic level the diversity is created by two fundamental processes mutation and recombination [1]. Under vegetative conditions of growth, the mutation results from inevitable random errors introduced by the Replisome apparatus due to their limited fidelity during the replication of hereditary material [1]. On the other hand, genetic recombination is the outcome of homologous recombination (HR) process, a housekeeping mechanism involved in the maintenance of genome integrity and generation of genetic variability through the rearrangement of (shuffling) genes within and between chromosomes [2]. HR has a crucial role to all forms of life and involves the exchange (i.e., breakage and reunion) of DNA sequences between two DNA homologous molecules for the repair of a variety of DNA damage of exogenous and endogenous origin [2]. Such molecular exchange provides an evolutionary force that contributes to promote genetic diversity and to conserve genetic integrity [2]. In addition, homologous recombination is also used in horizontal gene transfer (HTG) to exchange genetic material between different strains and species of bacteria and viruses contributing to the genetic diversification of bacterial genomes [3].
Homologous recombination depends mainly of existence of DNA recombinases [4]. Bacterial E. coli RecA protein is prototypical member of a class of recombinases proteins with homologs in all domains of life: RadA in Archaea, and Rad51 and Dmc1 in Eukarya [5]. The functional form of RecA protein in these processes is the nucleoprotein filament, a scaffold structure formed by assembly of RecA protein on DNA, generally singlestranded DNA (see later) [5]. During homologous recombination, the RecA nucleoprotein filament catalyzes the pairing and exchange of complementary DNA strands between homologous regions of DNA. This DNA strand exchange reaction is used during the recombinational DNA repair [5]. In addition, this RecA nucleofilament is required to induction of the SOS response by promotion of the autocatalytic cleavage of the LexA repressor, activation of UmuD’ by mediating autocatalytic cleavage of UmuD, and direct participation in SOS mutagenesis by activation of DNA polymerase V (see next section) [6].
Additionally to this RecA traditional function other functions have been discovered in the last years. Thus, RecA is required to control of swarming behaviour [7]
(see fourth section), and participates directly and indirectly in the control of programmed cellular death (PCD) phenomenon [8] (see third section). These RecA functions related to HR and DNA repair have been covered in much more detail in recent reviews [9,10]. However, firstly I carry out a short review of the different RecA activities as background for discussion. At my knowledge this is the first review that trying to integrate the different RecA activities under the same unitary conceptual framework having as background referential an important evolutionary question:
What effective control (i.e., management) have the cells both individually and collectively on the different processes underlying adaptive changes that driving the bacterial evolution?
2. RECA STAR (RECA*) FOR LIFE
Bacteria must cope many stress situations in nature. Several programmed (encoded genetically) responses are induced in different environmental stressful conditions [11]. Several kind of intracellular signals inform to cell of their environmental situation [11]. For stress situations that lead to DNA damage (e.g., UV radiation) historically the term RecA* was used to described the intracellular signal inducer of SOS response [6]. After of identification the RecA* as a RecA nucleofilaments, RecA* was defined as molecular entity that could promote of SOS induction and SOS mutagenesis [6]. Recently, E. coli RecA* has been defined as active RecA filaments bound to DNA in the presence of ATP or an ATP analog, filaments that are active for the complete range of activities normally associated with RecA [12]. This is the molecular entity that we will have in mind when RecA* is mentioned.
The formation of RecA nucleofilament in E. coli is a complex orchestrated process at the molecular level [6]. Their formation has been studied in deep in a singleDNA molecule level with RecA molecules tagged fluorescently [13].
In a nutshell, the RecA filament formation begin with a RecA dimer nucleating on ssDNA in a dynamical molecular competition with SSB (single stranded-ssDDNAbinding) proteins, after this, the RecA monomers are added in both direction 3’ to 5’and 5’ to 3’ but the filament (extension) has a net growth in 5’ to 3’ direction. In their competition with SSB proteins for binding on ssDNA, RecA is aided with the “mediator” proteins RecF and RecOR [13]. Others proteins such as DinI and RecX modulate the assembly and disassembly of RecA filaments [14].
RecA* thus defined participate in several functions:
2.1. HR and Recombinational Repair
As recombinase RecA promote a DNA strand exchange reaction. Mechanistically, the active nucleoprotein filament (RecA*) once formed aligns the bound single strand with homologous sequences in a different duplex DNA then promotes a strand exchange reaction in which one strand of the duplex is transferred to the bound single strand to create a new duplex. The second strand from the original duplex is displaced [6]. This exchange process is central to RecA function in recombinational DNA repair (e.g., double strand break-DSB-repair) [6] and rescue of stalled replication fork [15].
2.2. SOS Induction
The SOS response to DNA-damage in Escherichia coli is a paradigmatic example of a genetic programmed bacterial stress response involves the coordinated expression of at least 43 genes, the SOS regulon [6]. Under non inducing conditions, the LexA (a transcriptional regulatory protein) binds to the operator (the SOS box) of each of the SOS-inducible genes, and to its own operator, repressing transcription [6,16]. Different kinds of stressors are able to triggers SOS response. Thus, SOS is induced under DNA-damage conditions (e.g., UV radiation) [6] or cell-wall inhibition by β-lactam antibiotics [17,18] or conditions that block DNA replication [6]. For example, once DNA is damaged and replication is halted at the sites of lesions, persisting regions of single-stranded DNA (ssDNA) near the replication blockage allow the assembly of activated RecA nucleoprotein (RecA*). Derepression of SOS genes results from the autocatalytic selfcleavage of LexA, a process mediated by RecA* that acts a coprotease ring the LexA self-cleavage (rendering it unable to bind to the operators of SOS-inducible genes) triggers the expression of the SOS genes that previously were repressed by LexA. The induction of gene SOS is a timed response. Several error-free reparation systems are induced in the first minutes of inductions in SOS cells [6].
The first genes SOS to be induced are uvrA, uvrB, and uvrD. These proteins, together with the endonuclease UvrC, form the UvrABC multiprotein complex that catalyze nucleotide excision repair (NER), a reaction that excises the damaged nucleotides from double-stranded DNA. RecA-mediated HR allows the repair of lesions that occur on ssDNA regions at replication forks by rendering that double-stranded dsDNA (and hence a substrate for NER). The production of division inhibitor SfiA mediated by inhibition of the activity of FtsZ division protein delay the division to give the bacterium time to complete the repairs [6]. Finally, if the damage was not fully repaired by NER and HR the mutagenic DNA repair polymerase PolV (encoded from umuC and umuD genes) is induced (see next section [6]). Others genes encoding the alternative SOS DNA polymerases PolII
(DinA), PolIV (DinB) are also SOS induced and contribute to maintaining genome integrity in different stressful situations [19].
The SOS induction is switch off when genomic damages are repaired. This shutoff is a reverting process that is possible because there is a disappearance of the intracellular SOS inducer: The RecA nucleofilament that allows that the newly synthesized LexA molecules can to bind to SOS gene promoters silencing their expression.
However, despite the intensive work in this system, the role of the products of many SOS induced genes related to the different aspects of the E. coli SOS response remain to be unveiled yet and are waiting to be discovered.
2.3. PolV Mutasome Formation
The PolV complex active in SOS mutagenesis has been dubbed PolV mutasome [20]. In the mutasome formation the product of SOS induced umuD and umuC genes are involved. Their formation begins when RecA* promote the cleavage of UmuD protein to generate UmuD’ polypeptide, which associates with UmuC to form UmuD’2UmuC complex, now known as DNA polymerase V (PolV). Additionally, a direct molecular way RecA* participates actively in convert Pol V in a polymerase that synthesizes DNA through of replicationblocking lesions in a process known as translesion synthesis (TLS), i.e., PolV mutasome is a lesion bypass DNA polymerase, to synthesize DNA at otherwise irreparable lesions. Mutasome TLS activity allows that various kinds of DNA lesions, including abasic sites, photodimers (e.g., pyrimidine dimers produced by UV irradiation), and a wide variety of damaged bases, can be by passed although with low fidelity and often producing mutations. It is considered that the SOS mutagenesis phenomenon is a by-product of mutasome TLS DNA synthesis activity [21]. The PolV mutasome formation is a last-ditch response of the cell to repair the genome damage (mainly double strand break-DSB) but to expense of introducing mutations in the genome. In this way mutasome is a mechanism to tolerate DNA damage to expense of increase the mutation rate under stressful conditions. The implication for bacterial evolution has been discussed in previous reviews [12,21].
The cell has a last resort when conditions are too stressful and the accumulation of mutations increases the risk of death without having achieved a complete genome repair: dying to help the rest of the population. When food is scarce, the death of part of the bacterial population may provide nutrients for the surviving cells.
3. RECA* FOR DEATH
Life and Death are the two side of a same vital coin. Through of genetic programming life is able to control Death [22]. This control is essential in many developmental processes in multicellular metazoan organism. In fact, the programmed cellular death (PDC) was initially discovered in this organism. Generally, PCD is required for the elimination of superfluous or potentially harmful cells [23]. The PCD phenomenon has also been described in bacteria that traditionally had been considered unicellular organisms [24]. In fact, it has been recently suggested that PCD is another manifestation bacterial multicellularity [25]. It has been argued that the presence of a regulated suicide network in unicellular bacteria might—in analogy to PCD of eukaryotic microorganisms—increase the fitness of populations by facilitating the elimination of unwanted cells (see next section).
When E. coli is challenges with bactericidal antibiotic stress two different types of “PCD subroutines” have been described to be acting: The toxin–antitoxin system module mazEF mediated cell death (a nonapoptotic PCD subroutine, RecA independent) and the apotosis-like death (ALD) with RecA playing a critical role in this pathway having dubbed as the “main apoptotic executor” of the cell contributing to the antibiotic-triggered apoptotic demise of bacteria [8]. In ALD E. coli exhibit several hallmarks of apoptosis that accompany cell death. These hallmarks include DNA fragmentation and condensation, and membrane alterations including PS exposure, decreased membrane potential, and cell division arrest (SulA-induced filamentation) [8]. Both the mazEF and apoptotic pathways permit survival of a small fraction of the bacterial population responding to external stress.
Importantly, the mazEF-mediated death required of the production and extracellular liberation of the EDF (Extracellular Death Factor), a linear pentapeptide (NNWNN) which acts as a Quorum Sensing factor [26] and induces the endoribonucleolytic (killer) activities of MazF and ChpBK [27]. The Clp protease complex ClpXP is known to be involved in the synthesis of the quorum sensing signalling factor EDF. RecA participate directly and indirectly in the ALD pathway. Indirectly by that the ALD induction required of inactivation of the repressor LexA by RecA*: i.e., ALD is part of phenomenology that characterize the SOS response [28]. Furthermore, it believed that RecA has direct and central role in the ALD supposedly with a caspase-like activity that modified target (still unknown proteins) that lead in last term to the phenotypical apoptotic manifestations that are associated with this death pathway [8]. However, this direct role in ALD needs more experimental work for a complete molecular clarification.
The SOS response in addition appears to play a downstream unidentified role in this process of ADL. Interestingly, both pathways are interwoven through the proteolytic activity of ClpXP, and via reduction (mazEFmediated) of recA mRNA levels [28], although the precise mechanism of this control remains still unknown. Thus the intensity of the SOS response in ALD would be modulated indirectly through the EDF mediated quorum sensing. Significantly, this would be the first glimpse of a collectively modulation, although indirect, of recA gene expression, a feature to my knowledge that has not previously been indicated.
Therefore, two cellular agents RecA, ClpXP, and the SOS response act in a concerted manner to alter cellular behaviour when cellular stress is persistent through of modulation of the function of target proteins (unknown) involved in the appearance of apoptotic phenotypes and pushing the cell toward its death [29].
4. A RECA STARLESS FUCTION FOR LIFE COLLECTIVE MIGRATION
Escherichia coli use flagella for their motility [30]. In liquid medium individual planktonic cells swimming searching foods guided by their chemotactic system [30], but when the viscosity of the medium increase and sugars are available (e.g., glucose) E. coli is able to simultaneously grow and spreading rapidly over a semisolid surface in a coordinated collectively manner [31]. This groupal movement is called “swarming” [32]. Swarming as swimming is propelled by rotating flagella [32]. Swarming motility is view as a complex multicellular and social behaviour by which bacteria can rapidly migrate on moist surfaces in search of new resources. This moistness environment is generated when bacteria extract water from the underlying agar, producing a thin lubrication layer, known as well as a viscous slime layer [32] keeping the cells in a moist environment [33]. Swarming motility normally requires that population of cells to reach a certain cell number before the process is initiated, i.e., in many bacterial species swarming is a quorum sensing controlled behaviour [33]. This kind of the social motility provides ecological advantages to the bacteria because as a motile populations can rapidly reach novel niches, which they can colonize.
Swarming motility demands a lot of energy and requires the integration of many environmental cues, which leads to the physiological and morphological differentiation of the bacteria into swarmer cells. These are often elongated as a result of the suppression of cell division. In E. coli unlike to swimming motility chemotaxis is not required for swarming [34]. The environmental cues (chemical and physical signals), nutritional requirements, the physiological signalling networks (e.g., c-diGMP second messenger [35]) and genes and cellular structures (e.g., lipopolysaccharide, the enterobacterial common antigen, and Type 1 fimbriae [36] required for E. coli swarming motility has been studied intensively in the last years [35,36]. Despite this work, nevertheless, a clear picture of the different steps of developmental process that changes an E. coli swimmer cell in a swarmer cell has not been still articulated. At this respect by example, it is a conventional notion that E. coli swarmer cells are multinucleated and multiflagellated when swarming on Eiken-agar semisolid surfaces [37], but it has also been described that swarmer E. coli cells are able to migrate on semisolid prepared with Difco-agar that neither are hyperflagellated and not multinucleated [38].
RecA protein had been discovered as an unexpected and unanticipated cellular player for the control of E. coli swarming motility [7]. To difference of the others RecA activities commented in previous paragraphs that dependent critically of the RecA nucleofilament formation (RecA*), RecA promotion of swarming migration is a RecA function that apparently does not require the formation of a canonical RecA* molecular scaffold [7]. As it is shown in Figure 1, the molecular mechanism(s) through RecA modulate E. coli swarming motility are not know yet, although a tentative molecular mechanism was proposed [7]. Thus, it was postulate that a direct interaction between the CheW protein (involved in the chemosensory signal transduction) and RecA would be the mediator link [7]. Remarkably, it has recently been experimental data obtained in Salmonella enterica that support this model. Thus, it has been proposed exists between the intracellular concentrations of RecA and CheW an equilibrium that is necessary for the swarming motility in S. enterica [39]. Additional experimental work is required to clarify these issues in a molecular level.
The evolutionary implications of the RecA control of swarming behaviour in E. coli has been discussed previously [7], being this control an intriguing feature of this multifunctional protein that need to be explored in deep. To this respect, for example, it is interesting to speculate with the possibility that RecA modulates an evasive motile response under stress situation to increase the survival whereas the repair of genome occurs.
5. UNDERSTANDING THE BIOLOGICAL MEANIG OF GIN/GAN RECA “ANFI-FUNCIONALITY”
Several important questions have been raised by these not anticipate new RecA functions: Why RecA has these different antagonistic roles? How RecA protein integrates these diverse functions?
The diverse roles the RecA only can to be understood when they are put in the appropriate conceptual background under two perspectives unicelullarity versus multicellularity. It is important to remember here the principal ideas of this complementary conceptual approach.
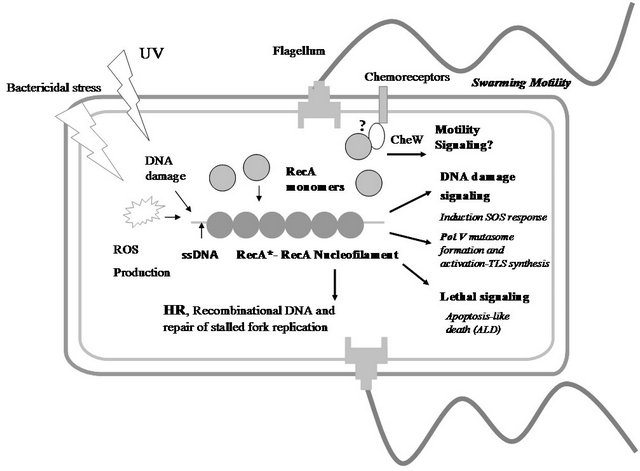
Figure 1. RecA as a WISE factor. The multiples functions of RecA protein in Escherichia coli bacteria are schematically depicted in this figure. Different stressors (bactericidal antibiotics, UV, etc...) lead lately to the intracellular formation of a RecA nucleofilament (RecA*) that has different signalling and direct activities: Induction SOS response, formation and activation of PolV Mutasome (a TLS polymerase), and activation of apoptosis-like death (ALD) pathway. On the other hand, a RecA function independent of the canonical RecA* formation is the promotion of swarming motility. This diagram shows a hypothetical interaction between RecA monomers and the CheW protein that would mediate in the RecA’s control of swarming migration [7]. The understanding how RecA is able to coordinate and to integrate these activities is a challenge for future experimental inquiries. Abbreviation: HR, homologous recombination; ROS, reactive oxygen species; ssDNA, single-stranded DNA. UV, ultraviolet radiation; TLS, translesion synthesis.
Bacteria were traditionally considered single-celled microorganisms that have an individualistic lifestyle [40]. Increasing experimental evidence indicate that bacteria seldom behave as isolated organisms, but in natural environment they more often live in communities capable of intercellular communication and concerted social behaviour bearing a most active and interesting intercomnected social life that in many ways resemble that of multicellular organisms [41]. The Sociomicrobiology is the new scientific discipline that studies these social aspects of microbial life [42]. All social activities need communication, bacteria engage in communicative activities to achieve multicellular and social behaviour. Chemical communication is the key process to entangle bacterial activities [43].
Bacteria are engaged in cell-cell communication through the production, extracellular release, and detection by the community of diffusible small signalling molecules called autoinducers [44]. These substances accumulate in a cell density-dependent manner, reaching a threshold concentration that triggers a synchronous and concerted response of bacterial populations that have achieved an effective quorum [44]. Therefore, this communicative process is most often referred by the general term of “quorum sensing” (QS), because it requires the concerted action of numerous cells (quorum) to carry out a productive group behaviour [45]. QS is the bacterial communicative phenomenon most intensively studied [45]. Interestingly, a bacterial paracrine communication process has been recently described, giving more support to the notion of bacteria as multicellular entities [46]. Thus, Multicellularity and sociability make the two key concepts of the new scientific paradigm to understand bacterial life.
With this background in mid I will now analyze conceptually the RecA anfi-functionality.
In unicellularity, in the level single cell, RecA works to safeguard the genome integrity maintaining the gene information. As shown in Figure 1, the RecA functions in a unique cell can be divided qualitatively in different modes: intracellular signalling and RecA mediated directly transactions. For example, RecA* has a double role in DNA repair: participate indirectly as SOS inducer distress signal and directly in the activation of PolV mutasome activity.
On the other hand, in under extreme (terminal) stress situation the RecA* work mainly to ensure the population survival. In this case, RecA* participate indirectly to activate SOS response required to the acquisition of a complete apoptotic phenotype [8] and modulating directly the functionality of proteins still unknown in ADL pathway [8].
RecA Promotes ADL Death to Avoid Cheaters?
Multicellularity requires cooperative behaviour. But this biological organization has a weak point their Achilles’s ankle: it is susceptible to apparition of cheaters individuals who reap the benefits of collective behaviour but contribute less or nil to the communitarian effort [47]. It has been assumed that cooperation persists in evolutive longterm time, despite this weakness because individuals tend to interact with their relatives, and reducing the fitness of relatives by cheating indirectly reduces an individual’s own fitness [48]. This is, however, a controversial issue [49]. Mechanisms that prevent, mitigate or eliminate social conflict among interacting individuals are required for cooperation or multicellularity to succeed [50]. Even simple organisms such as bacteria can evolve to suppress social cheaters [51].
It has been proposed that when, for example, DNA repair systems fail to overcome excessive damage to the chromosome, mazEF-mediated cell death might be activated eliminating cells that carry genomic defects and mutations [28]. In this way the mazEF system might contribute to the maintenance of genomic stability of the whole population [8,28]. To this respect the cell death mediated by mazEF may also be important in the response of bacteria to severe nutritional stress. It has been suggested that this is a mechanism of “altruistic” cell death among bacterial populations [26].
Because the mazFZ death pathway is an EDF-mediated quorum signalling pathway, the altruistness of mazEF is susceptible to be cheating [8]. In agreement with previous suggestion that the ALD pathway would to be the last resource programmed in each individual cell to suppress in last term the emergence of cheaters. Therefore, the existence of ADL-PCD pathway could provide a safeguard mechanism for the survival of bacterial population by forcing of cheaters to die under chronic stress.
In addition, these cheaters ADL mediated demise would be modulated by the entanglement between the mazFZ pathway and ALD pathway mediated by the ClpXP protease and the mazFZ mediated decrease of recA messenger [8].
6. CONCLUSIONS AND FUTURE DIRECTIONS
Metaphorically and abstractly, the RecA double-side functionality would be viewed as a möbius strip with RecA in the role of an “anfi-functional or Gin/Gan” protein that is capable of integrates both cellular functions in a unique biological function: the driving of bacterial survival and evolution in multicellularity and sociality.
In view of all the previous discussion I want to suggest here that E. coli RecA is configured as the principal bacterial WISE-Factor (for Worker for Integrate the Survival and Evolution) (see Figure 1) because controlling many aspects of the bacteria physiology related in all with survival under stressful conditions in an individualistic way (SOS response and mutasome PolV formation and action) as in a communitarian setting (ALD). In addition, RecA controls a collective social phenomenon as the swarming migration under non stress situation.
In summary, RecA is a great biological manager promoting LIFE in unicellularity and population survival in multicellularity through the activation in this case of DEATH. RecA converts the Stochasticity (Chance) in Necessity by harnessing the inherent randomness of the genetic change. In this way, RecA is one of the fundamental factors that contributes to promote the “transformation” of bacterial genomes in evolomes [52].
Mechanistically a molecular level, however, many issues still need to be answered:
Thus, it remains elusive whether RecA’s vital and lethal functions might be dissociated from each other. This could be explored, for instance by generating specific point mutants that would preferentially affect one or the other function.
How the activities of RecA in swarming motility are integrated when the cells additionally undergoes a stress situation? How that function of RecA in DNA repair and recombination and motility are coupled and coordinated?
REFERENCES
- Koonin, E.V. (2012) The logic of change: The nature and origin of biological evolution. Pearson Education, Inc., Publishing as FT Press Science, Upper Saddle River.
- Hiom, K. (2000) Homologous recombination. Current Biology, 10, R359-R361. doi:10.1016/S0960-9822(00)00500-5
- Rocha, E.P.C, Cornet, E. and Michel, B. (2005) Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genetics, 1, e15. doi:10.1371/journal.pgen.0010015
- Bianco, P.R., Tracy, R.B. and Kowalczykowski, S.C. (1998) DNA strand exchange proteins: A biochemical and physical comparison. Frontiers in Biosciences, 3, D570-D603.
- Galletto, R. and Kowalczykowski, S.C. (2007) RecA. Current Biology, 17, R395-R397. doi:10.1016/j.cub.2007.03.009
- Friedberg, E.C., et al. (2006) DNA repair and mutagenesis. ASM Press, Washington DC.
- Gómez-Gómez, J.M., Manfredi, C., Alonso, J.C. and Blázquez, J. (2007) A novel role for RecA under non-stress: Promotion of swarming motility in Escherichia coli. BMC Biology, 5, 14. doi:10.1186/1741-7007-5-14
- Dwyer, D.J., Camacho, D.M., Kohanski, M.A., Callura, J.M. and Collins, J.J. (2012) Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Molecular Cell, 46, 561-572. doi:10.1016/j.molcel.2012.04.027
- West, S.C. (2003) Molecular views of recombination proteins and their control. Nature Reviews of Molecular Cell Biology, 4, 435-445. doi:10.1038/nrm1127
- Michael, M.C. (2007) Motoring along with the bacterial RecA protein. Nature Reviews of Molecular Cell Biology, 8, 127-137. doi:10.1038/nrm2099
- Storz, G. and Hengge, R. (2011) Bacterial stress responses. 2nd Edition, ASM Press, Washington DC.
- Patel, M., Jiang, A., Woodgate, R., Cox, M.M. and Goodman, M.F. (2010) A new model for SOS-induced mutagenesis: How RecA protein activates DNA polymerase V. Critical Reviews in Biochemistry and Molecular Biology, 45, 171-184. doi:10.3109/10409238.2010.480968
- Lovett, S.T. (2012) A glimpse of molecular competition. Nature, 491, 199-200. doi:10.1038/nature11639
- Cox, M.M. (2007) Regulation of bacterial RecA protein function. Critical Reviews in Biochemistry and Molecular Biology, 42, 41-63. doi:10.1080/10409230701260258
- Cox, M.M., Goodman, M.F., Kreuzer, K.N., Sherratt D.J., Sandler, S.J. and Marians, K.J. (2000) The importance of repairing stalled replication forks. Nature, 404, 37-41. doi:10.1038/35003501
- Blázquez, J. and Gómez-Gómez, J.M. (2008) Evolution of antibiotic resistance by hypermutation. In: Baquero, F., Nombela, C., Cassell, G.H. and Gutiérrez-Fuentes, J.A., Eds., Evolutionary Biology of Bacterial and Fungal Pathogens. American Society for Microbiology (ASM) Press, Washington DC, 319-331.
- Miller, C., Thomsen, L.E., Gaggero, C., Mosseri, R., Ingmer, H. and Cohen, S.N. (2004) SOS response induction by β-lactams and bacterial defense against antibiotic lethality. Science, 305, 1629-1631. doi:10.1126/science.1101630
- Pérez-Capilla, T., Baquero, M.R., Gómez-Gómez, J.M., Ionel, A., Martin, S. and Blázquez, J. (2005) SOS-independent induction of dinB transcription by beta-lactammediated inhibition of cell wall synthesis in Escherichia coli. Journal of Bacteriology, 187, 1515-1518. doi:10.1128/JB.187.4.1515-1518.2005
- Rosenberg, S.M., Chandan, S.R., Frisch, L. and Hastings, P.J. (2012) Stress-induced mutation via DNA breaks in Escherichia coli: A molecular mechanism with implications for evolution and medicine. Bioessays, 34, 885-892. doi:10.1002/bies.201200050
- Schalcter, K., Leslie, K., Wyman, C., Woodgate, R., Cox, M.M. and Goodman, M.F. (2005) DNA polymerase V and RecA, a minimal mutasome. Molecular Cell, 17, 561- 572. doi:10.1016/j.molcel.2005.01.006
- Schlacher, K. and Goodman, M.F. (2007) Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nature Reviews in Molecular Cell Biology, 8, 587-594. doi:10.1038/nrm2198
- Jacobson, M.D., Weil, M. and Raff, M.C. (1997) Programmed cell death in animal development. Cell, 88, 347- 354. doi:10.1016/S0092-8674(00)81873-5
- Lewin, K. (2000) Programmed death in bacteria. Microbiology and Molecular Biology Reviews, 64, 503-514. doi:10.1128/MMBR.64.3.503-514.2000
- Rosenberg, S.M. (2009) Life, death, differentiation, and the multicellularity of bacteria. PLoS Genetics, 5, Article ID: e1000418. doi:10.1371/journal.pgen.1000418
- Engelberg-Kulka, H., Amitai, S., Kolodkin-Gal, I. and Hazan, R. (2006) Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genetics, 2, Article ID: e135. doi:10.1371/journal.pgen.0020135
- Kolodkin-Gal, I., Hazan, R., Gaathon, A., Carmeli, S. and Engelberg-Kulka, H. (2007) Linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science, 318, 652-655. doi:10.1126/science.1147248
- Belitsky, M., et al. (2011) The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpB. Molecular Cell, 41, 625-635. doi:10.1016/j.molcel.2011.02.023
- Erental, A., Sharon, I. and Engelberg-Kulka, H. (2012) Two programmed cell death systems in Escherichia coli: An apoptotic-like death is inhibited by the mazEF-mediated death pathway. PLoS Biology, 10, Article ID: e1001281. doi:10.1371/journal.pbio.1001281
- Carmona-Gutíerrez, D., Kroemer, G. and Madeo F. (2012) When death was young: An ancestral apoptotic network in bacteria. Molecular Cell, 46, 552-554. doi:10.1016/j.molcel.2012.05.032
- Berg, H.C. (2004) E. coli in motion. Springer, New York.
- Kearns D.B. (2010) A field guide to bacterial swarming motility. Nature Reviews in Microbiology, 8, 634-644. doi:10.1038/nrmicro2405
- Copeland, M.F. and Weibel, D.B. (2009) Bacterial swarming: A model system for studying dynamic self-assembly. Soft Matter, 5, 1174-1187. doi:10.1039/b812146j
- Daniels, R., Vanderleyden, J. and Michiels, J. (2004) Quorum sensing and swarming migration in bacteria. FEMS Microbiology Review, 28, 261-289.
- Burkart, M., Toguchi, A. and Harshey, R.M. (1998) The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proceeding of the National Academy of Sciences USA, 95, 2568-2573. doi:10.1073/pnas.95.5.2568
- Girgis, H.S., Liu, Y., Ryu, W.S. and Tavazoie, S. (2007) A Comprehensive genetic characterization of bacterial motility. PLoS Genetics, 3, Article ID: e154. doi:10.1371/journal.pgen.0030154
- Inoue, T., Shingaki, R., Hirose, S., Waki, K., Mori, I. and Fukui, K. (2007) Genome-Wide screening of genes required for swarming motility in Escherichia coli K-12. Journal of Bacteriology, 189, 950-957. doi:10.1128/JB.01294-06
- Harshey, R.M. and Matsuyama, T. (1994) Dimorphic transition in Escherichia coli and Salmonella typhimurium: Surface-induced differentiation into hyperflagellate swarmer cells. Proceedings of the National Academy of Sciences USA, 91, 8631-8635. doi:10.1073/pnas.91.18.8631
- Zorzano, M.-P., Cuevas, M.-T., Hochberg, D. and GómezGómez, J.-M. (2005) Reaction-diffusion model for pattern formation in E. coli swarming colonies with slime. Physical Review E, 71, Article ID: 031908. doi:10.1103/PhysRevE.71.031908
- Medina-Ruiz, L., Campoy, S., Latasa, C., Cárdenas, P., Alonso, J.C. and Barbé, J. (2010) Overexpression of the recA gene decreases oral but not intraperitoneal fitness of Salmonella enterica. Infection and Immunity, 78, 3217- 3225. doi:10.1128/IAI.01321-09
- Shapiro, J.A. and Dworkin, M. (1997) Bacteria as multicellular organisms. Oxford University Press, New York.
- Shapiro, J.A. (1998) Thinking about bacterial populations as multicellular organisms. Annual Review in Microbiology, 52, 81-104. doi:10.1146/annurev.micro.52.1.81
- Parsek, M.R. and Greenberg, E.P. (2005) Sociomicrobiology: The connections between quorum sensing and biofilms. Trends in Microbiology, 13, 27-33. doi:10.1016/j.tim.2004.11.007
- Schauder, S. and Bassler, B.L. (2001) The languages of bacteria. Genes & Development, 15, 1468-1480. doi:10.1101/gad.899601
- Bassler, B.L. and Losick, R. (2006) Bacterially speaking. Cell, 125, 237-246. doi:10.1016/j.cell.2006.04.001
- López, D., Vlamakis, H., Losick, R. and Kolter, R. (2009) Paracrine signalling in a bacterium. Genes & Development, 23, 1631-1638. doi:10.1101/gad.1813709
- West, S.A., Griffin, A.S., Gardner, A. and Diggle, S.P. (2006) Social evolution theory for microorganisms. Nature Review in Microbiology, 4, 597-607.
- Strassmann, J.E. (2000) Evolution: Bacterial cheaters. Nature, 404, 555-556.
- Carmona-Fontaine, C. and Joao, B.X. (2012) Altruistic cell death and collective drug resistance. Molecular System Biology, 8, 627. doi:10.1038/msb.2012.60
- Joao, B.X. (2011) Social interaction in synthetic and natural microbial communities. Molecular System Biology, 7, 483. doi:10.1038/msb.2011.16
- Travisano, M. and Velicer, G.J. (2004) Strategies of microbial cheater control. Trends in Microbiology, 12, 72- 78. doi:10.1016/j.tim.2003.12.009
- West, S.A., Diggle, P.D., Buckling, A., Gardner, A. and Griffin, A.S. (2007) The social lives of microbes. Annual Review of Ecology and Evolution Systematic, 38, 53-77. doi:10.1146/annurev.ecolsys.38.091206.095740
- Gómez-Gómez, J.M. (2010) Aging in bacteria, immortality or not a critical review. Current Aging Science, 3, 198- 218. doi:10.2174/1874609811003030198
NOTES
*To Marta Saínz de la Maza from el Museo de la Naturaleza Cantabria for your constant support.

