Advances in Bioscience and Biotechnology
Vol.2 No.3(2011), Article ID:5045,7 pages DOI:10.4236/abb.2011.23023
Genetic diversity among Indian phytopathogenic isolates of Fusarium semitectum Berkeley and Ravenel
![]()
Department of Biotechnology, Sant Gadge Baba Amravati University, Amravati, India.
Email: mkrai123@rediffmail.com; pmkrai@hotmail.com
Received 7 February 2011; revised 11 April 2011; accepted 12 April 2011.
Keywords: Fusarium; Genetic Diversity; RAPD; Macroconidia; UPGMA
ABSTRACT
We report total ten isolates of F. semitectum recovered from different hosts. Identity of these isolates was determined by morphological and cultural characteristics and confirmed by RAPD-PCR analysis using forty random primers. Morphologically all the ten isolates showed similarity, but based on RAPDPCR analysis, these isolates can be categorized in three groups depending upon similarity co-efficient. Genetic similarity coefficients between pair wise isolates varied from 0.00 to 1.95 based on an unweighted paired group method of arithmetic average (UPGMA) cluster analysis. RAPD-PCR technique can be used as an important tool for the genetic differentiation among isolates of F. semitectum.
1. INTRODUCTION
Fusarium is one of the most important genus of plant pathogenic fungi [1,2]. It causes infection in plants, animals and human beings [3-6]. Among the different Fusarium species, F. semitectum was found to be responsible for causing diseases like wilts, blights, root rots, and cankers in coffee, pine trees, wheat, corn, rice, cereals, carnations and grasses [7]. Knight and colleagues [8] reported the crown-rot disease of bananas caused by F. semitectum. Recently, Hawa et al. [9] found F. semitectum associated associated with red fleshed dragon fruit disease (Hylocereus polyrhizus) in Malaysia.
Apart from parasitic nature, F. semitectum is also used as biocontrol agent. Mikunthan and Manjunatha [10] reported the use of F. semitectum as a potential mycopathogen against thrips and mites in chilli. In India, chilli (Capsicum annuum L.) suffered with a characteristic leaf curl symptoms due to the attack of mite, (Polyphagotarsonemus latus) (Banks) and thrips (Scirtothrips dorsalis) or both. In such cases F. semitectum was found to be active against these mites and thrips. In another study, Manjunatha and group [11] studied the biocontrol nature of F. semitectum and found that F. semitectum showed the significant reduction in the attack of tobacco aphid Myzus persicae.
Different keys are available for the identification of Fusarium species on the basis of morphological and cultural characteristics. Seifert [12] developed the “Fuskey” for the identification of Fusarium. She carried out the morphological study on potato dextrose agar only. While Leslie and Summerell [13] mentioned the detailed characteristics features of many Fusarium species on different media like potato dextrose agar and carnation leaf agar in their book “The Fusarium laboratory manual”.
Different molecular markers are available for the differentiation and clarification of fungal taxa [2]. Out of these Random amplified polymorphic DNA (RAPD) [14] and amplified fragment length polymorphisms (AFLP) [15] are specific PCR-based molecular markers. RAPD has the advantage of being technically simple and rapidly facilitated. It includes use of random primers and thus there is no need to have genetic information of test organism. This technique has been used since a long time for fungal genetics and phylogenetic studies [14, 15].
A number of approaches to differentiate Fusarium species and formae speciales or races of F. oxysporum have been reported. Feng et al. [16] used RAPD markers to assess genetic diversity among three races (races 3, 7, 8) of F. oxysporum f. sp. vasinfectum in China. Smith et al. [17] characterized two types of F. oxysporum f. sp. vasinfectum, a high sporing type (HS) and a low sporing type (LS) by RAPD analysis and pathogenicity tests. Similarly, genetic variation in F. oxysporum f. sp. fragariae causing wilt disease in strawberry was studied using different molecular markers including RAPD [18]. Gupta and co-workers [14] reported the genetic polymorphism in six isolates of F. solani causing wilt disease in Guava, isolated from different places of in India.
In the present study, ten isolates of F. semitectum isolated from different infected plant materials including fruits, vegetables and foodgrains were identified on the basis of morphological and cultural characteristics. Also, the intra-species genetic variations among these isolates were studied by using RAPD-PCR method.
2. EXPERIMENTAL PROCEDURES
2.1. Collection of Infected Plant Materials
Infected plant materials like fruit; (papaya) foodgains (ground nut, mung bean soybean, arhar and cotton) and vegetables (chilly, cauliflower, ladies finger and snake gourd) were collected from different locations of Akola and Amravati cities of Maharashtra, India (Table 1).
2.2. Isolation of Fusarium semitectum
Different isolates of F. semitectum were recovered from the infected plant materials on potato dextrose agar (PDA). For the isolation, infected materials were surface sterilized for 1min with 1% sodium hypochlorite solution, rinsed twice in sterile distilled water, dried in a laminar air flow cabinet and inoculated on to solidified PDA plates at 25 ± 2˚C for 3 days. The cultures were further purified by growing single-spore on the PDA. All the isolates were maintained at 4˚C in a refrigerator.
2.3. Identification on the Basis of Morphology and Microscopic Characteristics
All the auxenic cultures of different isolates of Fusarium were identified on the basis of morphological and cultural characters like morphology of colony (colour of the colony, pigmentation, texture of mycelia, etc.), morphology of macroconidia, microconidia and chlamydospores (presence or absence of conidia, shape and size of conidia, etc.) on PDA. For species identification, Seifert’s Fusarium key and Laboratory manual for Fusa rium were used.
2.4. DNA Extraction
Each isolate of F. semitectum was grown on PDA at 25 ± 2˚C for 3 days. The mycelia were harvested and total DNA was extracted using fungal genomic DNA isolation kit from Chromous Biotech Pvt. Ltd, Bangalore, India according to manufacturer’s instructions.
2.5. RAPD Analysis
Forty random decamer primers (OPC:1-20 and OPD: 1-20) (Table 2) from Operon primer “C” and “D” kits (Operon Technology Inc, USA) were tested for PCR amplification of ten F. semitectum DNA. Preliminary amplifications determined the optimal concentration of the component in PCR reaction mixture and amplification conditions. Amplifications were performed in a total volume of 25 µl containing 12.5 µl PCR master mix (2X) (Fermentas Life Sciences, Canada) 5 µl of template DNA (20 ng), 1.5 µl MgCl2 (25 mM), 0.3 µl Taq DNA polymerase (Genaxy, 5U/µl), 1 µl each of primer (Operon, USA) and 4.7 µl nuclease free distilled water (supplied with Fermentas PCR master mix).
PCR was carried out on gradient PCR machine (PalmCycler from Corbett Research, Australia). The programme included an initial denaturation at 94˚C for 2 min, 35 cycles with denaturation at 94˚C for 30 sec, annealing at 38˚C for 1 min, extension at 72˚C for 2 min and final extension at 72˚C for 5 min with holding temperature at 4˚C for 10 min. Negative control (without template DNA) was maintained for each set of experiment to test for the presence of non-specific banding. All experiments were repeated for three times. PCR products were electrophoresed on 1.5% agarose by using 1X TAE buffer (Fermentas Life Sciences, Canada), stained with ethidium bromide, visualised in a UV-transilluminator and the gel were photographed using Gel Doc (AlphaImager, Gel documentation system, USA), system.
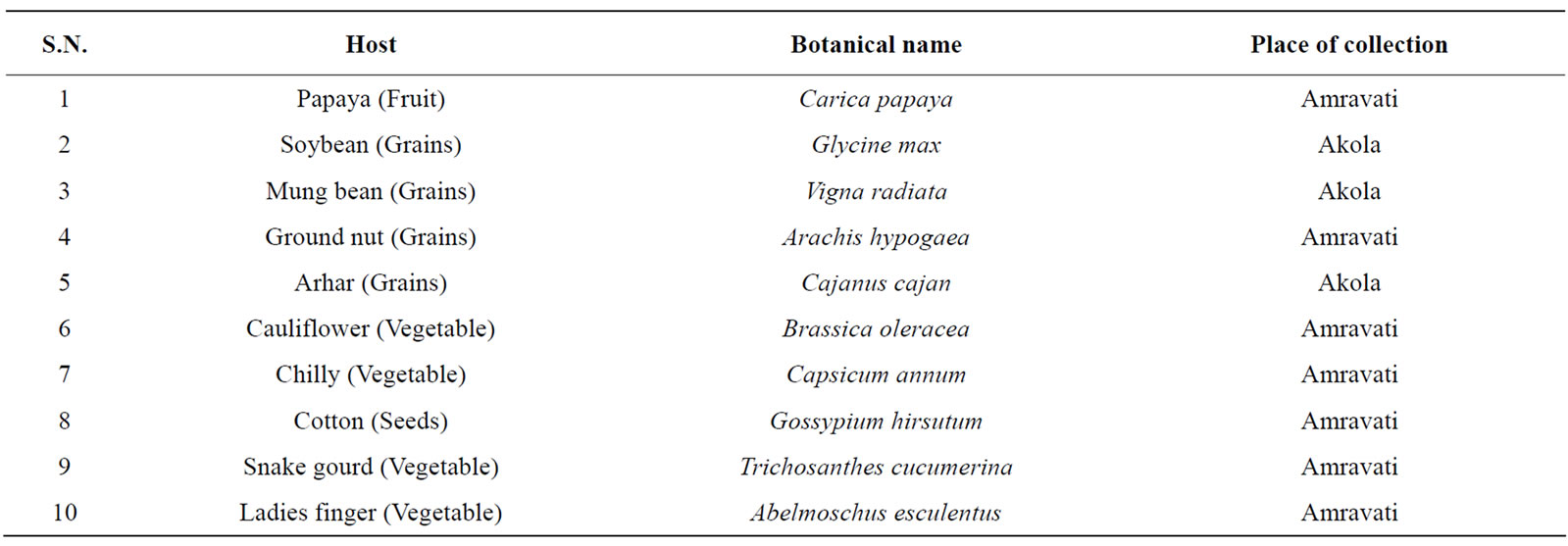
Table 1. Fusarium semitectum isolated from different plant hosts and locality.

Table 2. Primer and their sequences used in RAPD-PCR test.
3. RESULTS AND DISCUSSION
3.1. Morphological and Cultural Studies
All the ten isolates of F. semitectum grew rapidly with dense aerial mycelia on potato dextrose agar. Table 3 showed the detailed morphological study including macroscopic and microscopic characteristics.
3.2. A. Macroscopic Characteristics
Ten isolates of F. semitectum were recovered from different infected hosts (Table 1) on Potato dextrose agar (PDA) and cultured in pure form from a single spore culture. All the ten isolates were identified on the basis of morphological and cultural characteristics. F. semitectum isolates showed the growth of dense aerial mycelia, initially with white/cream and later brown colour. Cream to brown colour pigmentation on reverse of the colony was also observed (Figure 1). All the isolates attained similar growth rate of 8 - 8.5 cm after 6 days incubation at 28˚C, except F. semitectum isolated from cotton seed, which showed slower growth (5.5 cm) than other nine isolates. The morphological characters of different isolates of F. semitectum species in the present study showed the resemblance with morphological characters reported by many researchers in past [12,19-21]. These characters confirmed the identity of F. semitectum isolates.
3.3. B. Microscopic Characteristics
Microscopic characteristics also revealed the similar characters of F. semitectum. Microscopic examination showed the presence of septate and hyaline hyphae, and also presence of microconidia and chlamydospores. Macroconidia observed were abundant and borne in the aerial mycelium were mostly straight or slightly curved, generally 3 - 5 septate, and measured about 23.52 to 62.10 µm in length (Figure 2 (a)). The apical shape of macroconidia was tapering or curved while the basal shape was foot or notched shaped (Figure 2 (b)). Microconidia were also observed in all the isolates of F. semitectum these were 1 - 2 septate (Figure 2 (c)). Two types of chlamydospores were reported in the present microscopic study. In one case chlamydospores were found in the hyphae singly and sometimes in chains (Figure 2 (d)), while in another case, these were found singly in the macroconidia (Figure 2 (e)) which is a specific characteristic of F. semitectum. It provides the strength for the identification of all these isolates on the basis of microscopic characteristics. Hawa et al. [9] reported the similar microscopic characters for the F. semitectum isolated from red fleshed dragon fruit disease in Malaysia. They reported the formation of macroconidia in abundant and absence of chlamydospores in the culture. The macroconidia produced were 3 - 5 septate with average size range of 35.5 × 4.8 µm.
3.4. RAPD Analysis
Genomic DNA isolated from ten isolates of F. semitectum was subjected to RAPD-PCR analysis with forty random decamer primers from Operon “C” and “D” kits (Table 2). In the preliminary experiments, out of 40 primers, 19 primers (11-OPC primers, viz. OPC-5, OPC- 7, OPC-8, OPC-9, OPC-10, OPC-11, OPC-12, OPC-13, OPC-18, OPC-19 and OPC-20 and 8-OPD primers, viz. OPD-2, OPD-3, OPD-5, OPD-7, OPD-8, OPD-11, OPD- 12 and OPD-17) produced distinct and reproducible band profile and polymorphisms in all isolates of F. semitectum. Out of these 19 primers four primers, in cluding OPC-7 (5’-GTCCCGACGA-3’), OPC-18 (5’- TGAGTGGGTG-3’), OPD-3 (5’-GTCGCCGTCA-3’) and OPD-8 (5’-GTGTGCCCCA-3’) were used for comparative analysis of isolates of F. semitectum (Figure 3).
All the amplified fragments were ranged from 0.5 kb to 3.0 kb. RAPD assays of all ten isolates with four above mentioned primers yielded 236 bands which were found to be polymorphic. Above data showed that RAPD is a convenient method for distinguishing the isolates of F. semitectum and also revealed a significant genetic variation among these isolates. Other most studied Fusarium species viz. F. oxysporum also showed the genetic variation and differentiation among its isolates [18]. Assigbetse et al. [22] differentiated races of F. oxysporum f.

Table 3. Morphological characteristics of different isolates of F. semitectum recovered from different hosts.
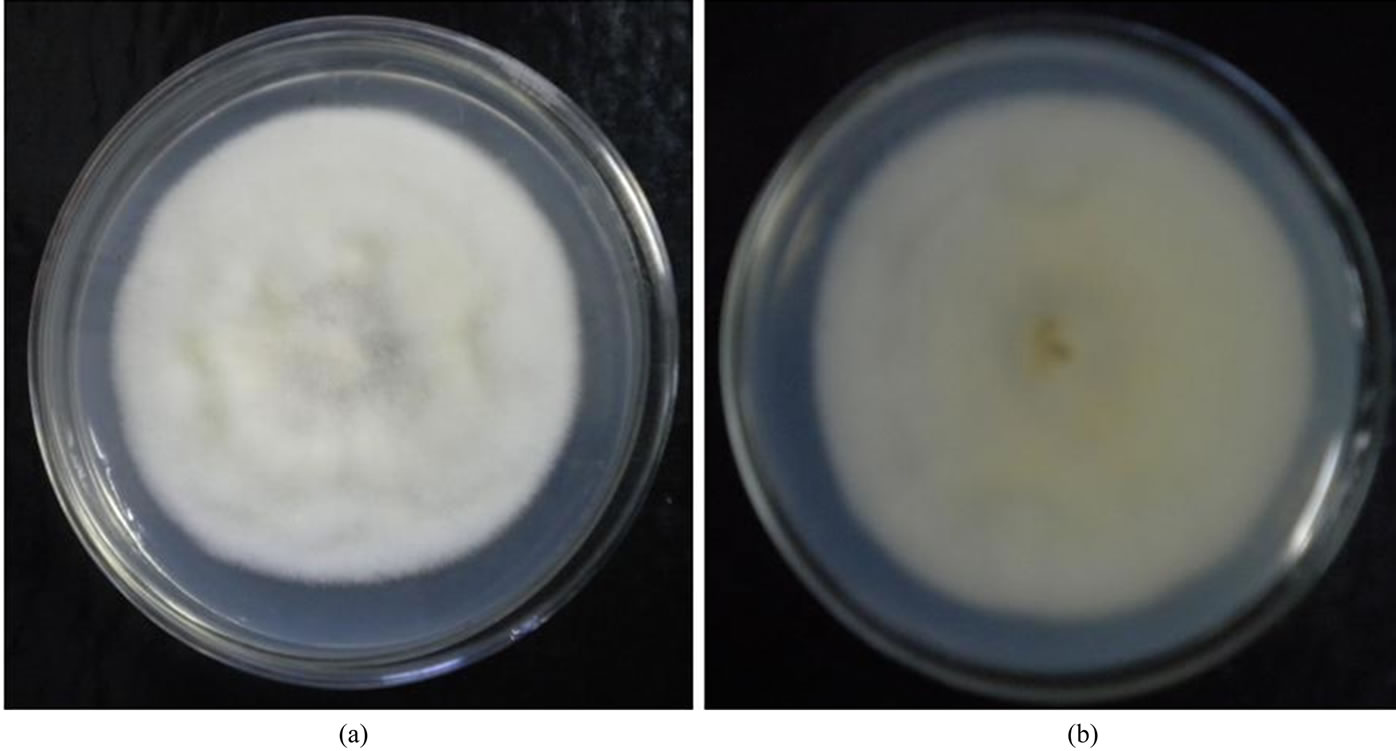
Figure 1. Growth of F. semitectum on PDA medium. (a) Ventral view; (b) Dorsal view isolated from soybean.
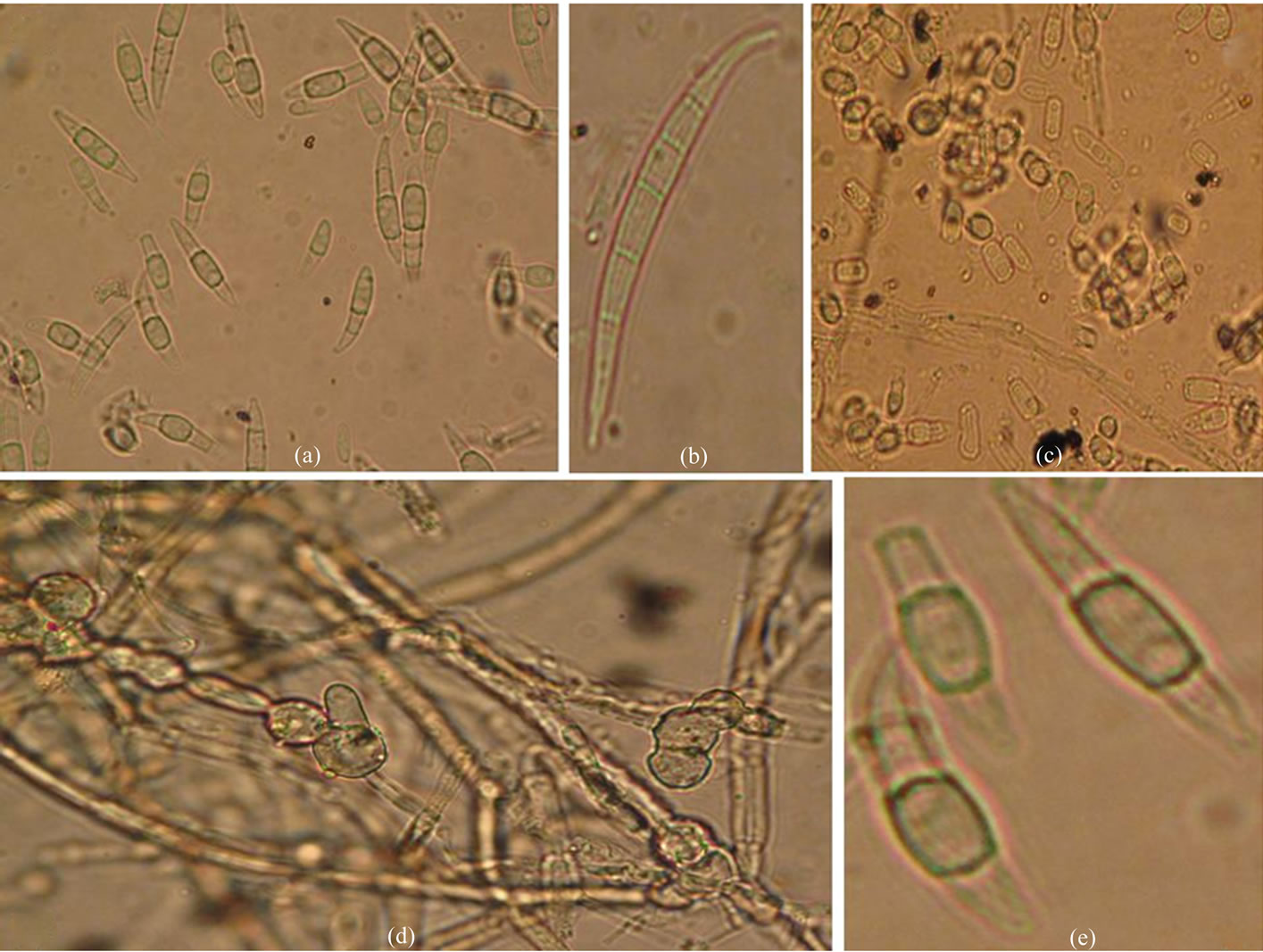
Figure 2. Microscopic characteristics of F. semitectum. (a) Macroconidia; (b) Single Macroconidia (5 septate) showing tapering apical and notched basal shape; (c) Microconidia (1-2 septate); (d) Chlamydospore in chains in hyphae; (e) Chlamydospore singly in conidia.
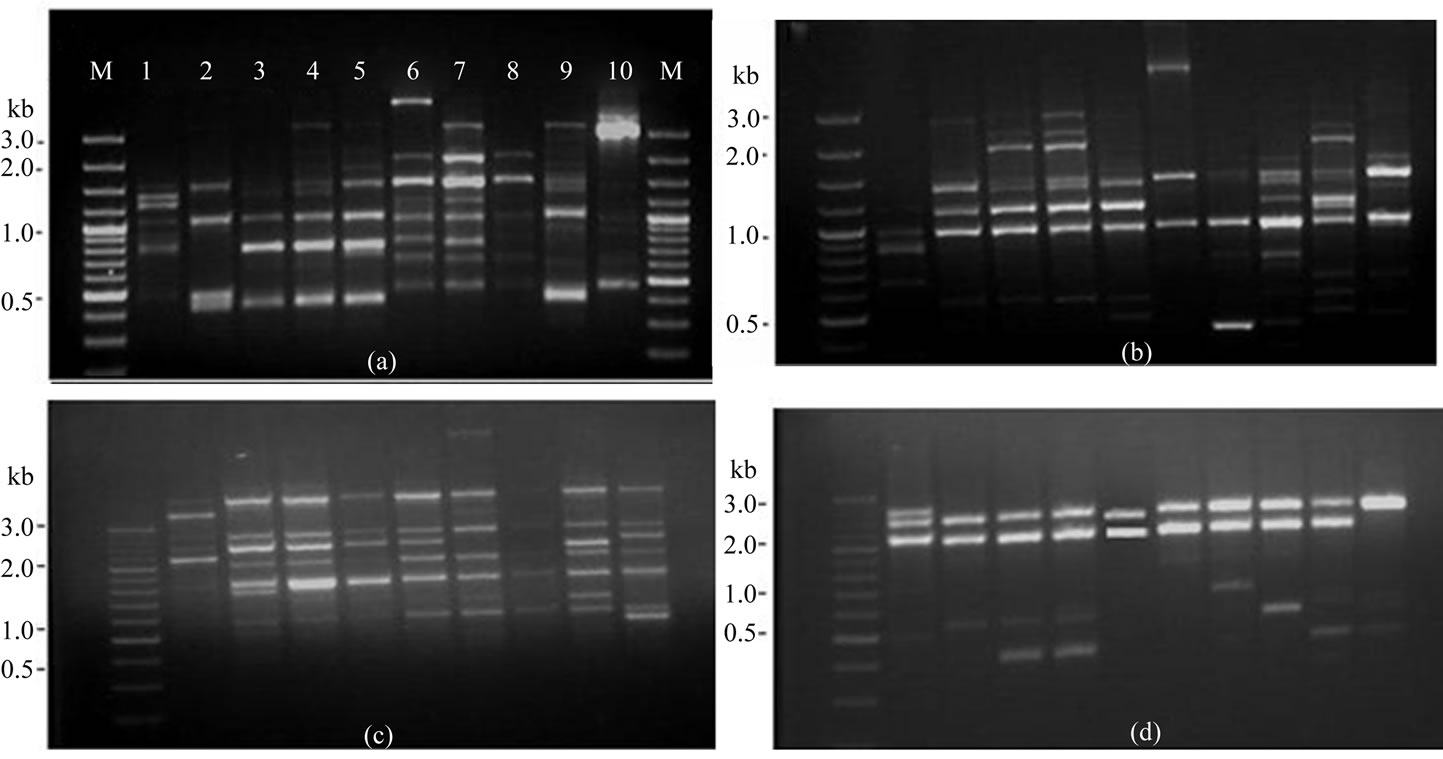
Figure 3. RAPD patterns on 1.5% agarose gel of amplified fragments generated from 10 isolates of F. semitectum with random primers. (a) OPC-7; (b) OPC-18; (c) OPD-3; (d) OPD-8, Lane M, DNA marker (100 bp), lane 1- isolate-1, lane 2- isolate-2, lane 3- isolate-3, lane 4- isolate-4, lane 5- isolate-5, lane 6- isolate-6, lane 7- isolate-7, lane 8- isolate-8, lane 9- isolate-9, lane 10- isolate-10.
sp. vasinfectum on cotton by using RAPD as molecular tool. They reported a significant genetic variation in these isolates of F. oxysporum. In another study carried out by Edel and colleagues [23], it was observed that the isolates of F. oxysporum isolated from different soil samples in France showed genetic diversity. While in another study, Nagarajan and group [18] studied the genetic variation in F. oxysporum f. sp. fragariae population causing wilt in Strawberry using RAPD and RFLP analysis. The work carried out by Leslie and co-workers [24] supports the findings of present study. They observed the interand intra specific genetic variation in different Fusarium species. RAPD-PCR technique as a suitable method was used for rapid identification and differentiation of Fusarium species [15,25]. Gupta et al. [14] reported the genetic polymorphism and diversity in isolates of F. solani isolated from wilt disease of Guava in India. Recently, Hawa et al. [9] reported the genetic variation among the different isolates of F. semitectum using different molecular markers like RAPD-PCR and AFLP. They observed that both the methods were convenient to determine the intra-species genetic variation in the isolates of same species.
3.5. Unweighted Pair Group Method with Arithmetic Mean Analysis (UPGMA)
A similarity matrix on simple matching co-efficients was calculated from the data based on the RAPD of all ten isolates. The matrix was used to construct a dendrogram using UPGMA tool of NTSYS for establishing to analyze the level of relatedness among the ten isolates. The dendrogram obtained from the data showed that hierarchical clustering separated the isolates into three groups according to their similarity co-efficients (Figure 4). The similarity co-efficients among the all isolates were found
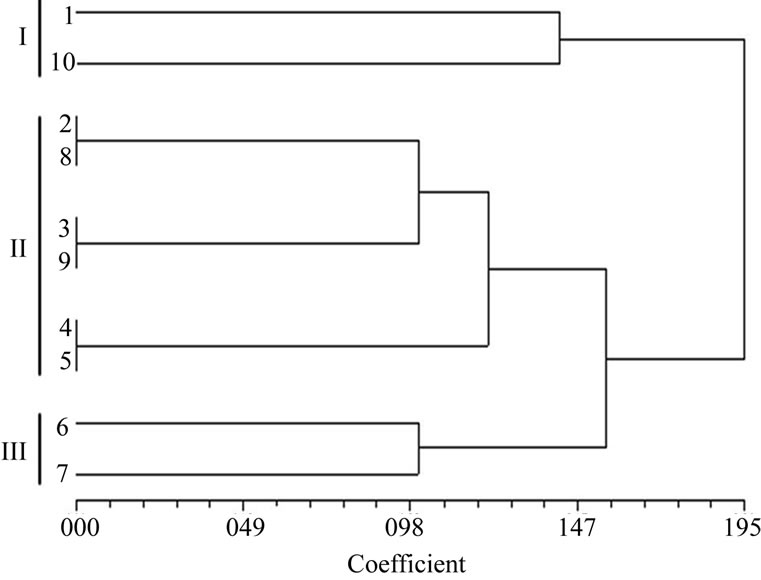
Figure 4. Dendrogram showing genetic relationship among the ten isolates of F. semitectum.
in between 0.00 to 1.95. UPGMA analysis thus carried out in the present study showed the genetic variation in these isolates of F. semitectum.
UPGMA is an important bioinformatics tool used to calculate the distance matrix and similarity co-efficient among isolates of same species. The results obtained in the present study are noteworthy and showed the similarity with the observations of Gupta et al. [14] and Nagarajan et al. [18]. In their studies on isolates of F. solani and F. oxysporum respectively, they used data generated from RAPD banding pattern for the UPGMA analysis and found that there was genetic variations in different isolates of same Fusarium. Not only other species of Fusarium but isolates of F. semitectum also showed genetic variation and it was proved by the UPGMA analysis carried out from data obtained from RAPD and AFLP [21].
Although the ten isolates of F. semitectum recovered from different hosts showed the similar morphological characteristics, RAPD analysis suggest a significant genetic variation among the isolates of F. semitectum. It seems that the fungal species showed the host dependant genetic variation.
4. CONCLUSIONS
F. semitectum is an important species of the genus Fusarium. It causes disease in plants and also produces mycotoxin, which is hazardous to animals and human beings. It can be concluded that morphologically similar strains of the same fungal species may show genetic variations. Finally, we suggest that RAPD marker may be used as a quick and reliable tool for the determination of genetic variations among the Fusarium species. However, further in-depth study is required before reaching to a final conclusion.
REFERENCES
- Ingle, A., Karwa, A., Rai, M.K. and Gherbawy, Y. (2009) Fusarium: Molecular detection, mycotoxins and biocontrol. In: Gherbawy, Y., Mach, R. and Rai, M. Eds., Current Advances in Molecular Mycology, Science Publishers Inc., Enfield, 85-106.
- Steinkellner, S., Mammerler R. and Vierheilig, H. (2008) Germination of Fusarium oxysporum in root exudates from tomato plants challenged with different Fusarium oxysporum strains. European Journal Plant Pathology, 122, 395-401. doi:10.1007/s10658-008-9306-1
- Godoy, P., et al. (2004) Onychomycosis caused by Fusarium solani and Fusarium oxysporum in São Paulo, Brazil. Mycopathologia, 157, 287-290. doi:10.1023/B:MYCO.0000024186.32367.d4
- Mansoory, D., Roozbahany, N.A., Mazinany, H. and Samimagam, A. (2003) Chronic Fusarium infection in an adult patient with undiagnosed chronic granulomatous disease. Clinical Infectious Diseases, 37, 107-108. doi:10.1086/377608
- Nucci, M. and Anaissie, E. (2007) Fusarium infections in immunocompromised patients. Clinical Microbiology Review, 20, 695-704. doi:10.1128/CMR.00014-07
- Sultana, N. and Hanif, N.Q.N. (2009) Mycotoxin contamination in cattle feed and feed ingredients. Pakistan Veterinary Journal, 29, 211-213.
- Girish, G.K. and Goyal, R.K. (1986) Prevention and control of mycotoxins in food grains in India. Bull Grain Technology, 24, 157-177.
- Knight, C., Cutts, D.F. and Colhoun, J. (2008) The role of Fusarium semitectum in causing crown rot of banana. Journal Phytopathology, 89, 170-176. doi:10.1111/j.1439-0434.1977.tb02853.x
- Hawa, M.M., Salleh, B. and Latiffah, Z. (2010) Characterization and intraspecific variation of Fusarium semitectum (Berkeley and Ravenel) associated with red-fleshed dragon fruit (Hylocereus polyrhizus [Weber] Britton and Rose) in Malaysia. African Journal Biotechnology, 9, 273-284.
- Mikunthan, G. and Manjunatha, M. (2006) Fusarium semitectum, a potential mycopathogen against thrips and mites in chilli, Capsicum annuum. Communications in Agricultural and Applied Biological Sciences, 71, 449- 463.
- Manjunatha, A.M., et al. (2009) Evaluation of fungal pathogen, Fusarium semitectum Berk and Ravenel against tobacco aphid under laboratory and greenhouse conditions. Karnataka Journal of Agriculture Science, 22, 495-498.
- Seifert, K. (1996) Fusarium interactive key. Agriculture and Agri food Canada, 1-30.
- Leslie, J.F. and Summerell, B.A. (2006) The Fusarium laboratory manual. 3rd Ed., Blackwell Publishing Professional, Ames.
- Gupta, V.K., et al. (2009) Studies of genetic polymorphism in the isolates of Fusarium solani. Australian Journal Crop Science, 3, 101-106.
- Niessen, L. (2007) PCR based diagnosis and quantification of mycotoxin producing fungi. International Journal Food Microbiology, 119, 38-46. doi:10.1016/j.ijfoodmicro.2007.07.023
- Feng, J.I.E., et al. (2000) RAPD analysis of physiologic races of Fusarium oxysporum f. sp. vasinfectum in China. Mycosystema, 19, 45-50.
- Smith, S.N., et al. (2001) Soil-borne populations of Fusarium oxysporum f. sp. vasinfectum, a cotton wilt fungus in California fields. Mycologia, 93, 737-743. doi:10.2307/3761828
- Nagarajan, G., et al. (2004) Genetic variation in Fusarium oxysporum f. sp. fragariae populations based on RAPD and rDNA RFLP analyses. Plant Pathology Journal, 20, 264-270. doi:10.5423/PPJ.2004.20.4.264
- Nelson, P.E., Toussoun, T.A and Marasas, W.F.O. (1983) Fusarium species: An illustrated manual for identification. Pennsylvania State University Press, University Park.
- Sutton, D.A., Fothergill, A.W. and Rinaldi, M.G. (1998) Guide to clinically significant fungi, 1st Ed., Williams & Wilkins, Baltimore.
- Abd-Elsalam, K.A., Schnieder, F., et al. (2003) Intra- -species genomic groups in Fusarium semitectum and their correlation with origin and cultural characteristics. Journal Plant Disease Protection, 110, 409-418.
- Assigbetse, K.B., et al. (1994) Differentiation of Fusarium oxysporum f. sp. vasinfectum races on cotton by Random amplified polymorphic DNA (RAPD) analysis. Phytopathology, 84, 622-626. doi:10.1094/Phyto-84-622
- Edel, V., et al. (2001) Genetic diversity of Fusarium oxysporum populations isolated from different soils in France. FEMS Microbiology and Ecology, 36, 61-71. doi:10.1111/j.1574-6941.2001.tb00826.x
- Leslie, J.F., et al. (2007) Interand intra specific genetic variation in Fusarium. International Journal Food Microbiology, 119, 25-32. doi:10.1016/j.ijfoodmicro.2007.07.059
- El-Fadly, G.B., et al. (2008) Identification of some Fusarium spp. using RAPD-PCR technique. Egyptian Journal Phytopathology, 36, 71-80.

