Journal of Biophysical Chemistry
Vol.4 No.2(2013), Article ID:31772,8 pages DOI:10.4236/jbpc.2013.42010
Selection of highly efficient small interference RNA (SiRNA) targeting mammalian genes
![]()
Biophysics Department, Faculty of Science, Cairo University, Cairo, Egypt; *Corresponding Author: lakkani@netscape.net
Copyright © 2013 Ali El-Iakkani et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 21 January 2013; revised 25 February 2013; accepted 9 March 2013
Keywords: RNAi; SiRNA; SiRNA Design
ABSTRACT
RNAi is the method of silencing the expression of targeted genes. RNAi applications include gene function analysis and target validation. Designing highly efficient small interference RNA (siRNA) sequence with maximum target specificity for mammalian RNAi is one of important topics in recent years. In this work, a statistical analysis of the information for a large number (3734) of siRNA presented in the database available on the internet is done. This is to improve the design of efficient siRNA molecules. The (3734) siRNAs are classified according to their efficiency to three groups (high efficient, moderate efficient and low efficient). Thirteen properties (positional and thermodynamics) are identified in the high efficient group in the primary statistical study. In the final statistical study, the average weight of each identified property is calculated. A very good linear correlation was found between the average percentage efficiency and the weighted score of siRNA properties. It is found that the most important feature of highly efficient siRNA is the difference in binding energy between the 5’ end and the 3’ end of the anti-sense strand. The (RISC) activation step is a critical step in RNAi process where the efficiency of this process depends on the instability of the 5’ end of the anti-sense strand.
1. INTRODUCTION
RNA interference (RNAi) is the process through which double stranded RNA (dsRNA) molecules inhibit gene expression by mediating sequence-specific messenger RNA (mRNA) degradation. The process starts when dsRNA molecules are degraded into short interfering RNA (siRNA) molecules, about 21 - 23 nucleotides in length, by the RNase enzyme Dicer. These siRNAs are then incorporated into a ribonucleoprotein complex named RNA-induced silencing complex (RISC). The RISC helicase subunit unwinds a duplex siRNA into two singlestranded RNAs, namely the passenger strand (sense) and the guide strand (antisense). The passenger strand will be degraded, and the guide strand is incorporated into the RNA-induced silencing complex (RISC) and guides it to bind with the complementary mRNA. The mRNA can be inhibited or degraded, depending on the degree of pairmatching [1-3].
Multiple statistical and computational models have been proposed in recent years to design functional siRNA depending on a large number of “features”. These features can be roughly classified into three categories, the first category is sequence features, the second category includes features defined based on the thermodynamics of the siRNA and the third category of features are defined based on the target sites on the mRNA, including target location-related features [4].
In this work, statistical studies are done on a large number of data (3734 siRNA) available on internet: http://gesteland.genetics.utah.edu/members/olgaM/siRN A database September 2006.xls. In the primary statistical study, thirteen properties (positional and thermodynamics) are identified in the high efficient siRNA group. In the final statistical study, the average weight of each identified property is computed. The advantages of this study are the average weights obtained for the identified thermodynamics properties of siRNA and it is done on a large number of data.
2. MATERIAL AND METHODS
The database contain a large number of mammalian experimentally validated (siRNAs) targeting mRNA of different mammalian genes [5-7]. The database contains the sequences and the thermo-dynamics properties of the siRNA besides the experimentally determined of the percent of remaining level of the mRNA in cells.
2.1. Classification of the SiRNA
The siRNAs are classified according to their silencing percentage efficiency (calculated from the percentage of the remaining level of mRNA) to three groups: high-efficient siRNA group (experimental efficiency ≥ 80%), moderate-efficient group (efficiency < 80% and ≥ 50%), and low-efficient group (efficiency < 50%).
2.2. Statistical Studies
Positional properties of the anti-sense strand (AS) of siRNA, such as the presence of the nucleotide (A/U) or (G/C) in certain position are studied in each group. The thermodynamics properties including Gibbs free energy change ΔG of sense-antisense duplex, Gibbs free energy change ΔG of antisense oligo secondary structure, Gibbs free energy change ΔG of antisense oligo-oligo dimer and the binding energy difference dΔG or (ΔG1 - ΔG18) between the 5’end and the 3’end of (AS) strand are studied in each group. The average GC content in each group is also studied.
From these studies the important positional and thermodynamics properties present significantly in the high efficient group are deduced. The thirteen important chosen properties are:
1) A/U at position 1 of AS strand.
2) A/U at position 2 of AS strand.
3) A/U at position 7 of AS strand.
4) A/U at position 10 of AS strand.
5) A/U at position13 of AS strand.
6) A/U at position 14 of AS strand.
7) G/C at position 19 of AS strand.
8) G/C at position 18 of AS strand.
9) G/C content ranged between (6 - 10) nucleotides of 19 ones of AS strand.
10) ΔG of sense-antisense duplex (≥ −36.720 Kcal/mole).
11) ΔG of antisense oligo secondary structure (≥ −1.240 Kcal/mole).
12) ΔG antisense oligo-oligo dimer (≥ −8.414 Kcal/mole).
(13) The binding energy difference between the 5’end and the 3’end of (AS) strand > 0 Kcal/mole.
2.3. Determining the Weight of Each Property
The 3743 siRNAs in the database are sorted according to the type of properties found in each one (from the thirteen identified properties). If more than one siRNA have the same type of properties then the average corresponding efficiency for them is calculated. From these data the different weights of the thirteen properties are computed by using a developed computer program. The program obtains the best fit of the data to a second order polynomial function (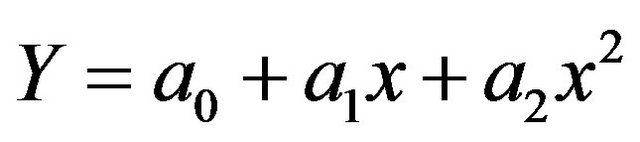 ). The best fit weights minimizing the sum squared error function were computed 200 times after randomizing the order in which the properties were considered. Then the average weight of each property was computed.
). The best fit weights minimizing the sum squared error function were computed 200 times after randomizing the order in which the properties were considered. Then the average weight of each property was computed.
3. RESULT AND DISCUSSION
Figure 1 shows the percent of siRNAs in the three groups (high-efficient, moderate-efficient and low-efficient) which contain A/U at positions from 1 to 19 of AS strand starting from 5’-AS end. It can be seen from Figure 1 that high percentage of siRNA has A/U at positions (1, 2, 7, 10, 13 and 14 of AS strand starting from the 5’end) are present in the high efficient group i.e. low internal stability in these positions. In contrast the low efficient group is enriched with siRNA molecules that have high internal stability (G/C) in these positions (see Figure 2). Khvorova et al. [8] found in their experimental study that the functional group of siRNA is enriched with siRNA molecules that have a low internal stability at the 5’-AS and an overall low internal stability profile, particularly in positions 9 - 14 100 (counted from 5’-AS end) and this is in contrast to the non-functional group.
Figure 2 shows the percent of siRNAs in the three groups (high-efficient, moderate-efficient and low-efficient) which contain G/C at positions from 1 to 19 of AS strand starting from 5’-AS end. From this figure, high percentage of siRNA in the high efficient group has G/C at positions (18, 19 of AS strand). Shabalina et al. [9] performed a comparative, thermodynamics and correlation analysis on a set of 653 siRNAs collected from the literature. From a t-test analysis they found that for efficient siRNA, the nucleotides C and U are preferred in position 18 and for position 19 nucleotides C and G are preferred in the AS strand.
Many studies [5,9,10] showed that the percentage of the GC content of the siRNA play an important role in its efficiency. In Figure 3 the average GC contents of the siRNA were calculated for the three groups (high efficient, moderate efficient and low efficient), which are equals (8.8, 10, 10.9 nt/19 nt of the AS strand) respectively. In another studies [11-15] it was found that in the high efficient siRNA the GC content are between 6 and 10 nt/19 nt of AS strand or 30% to 50% or 36% to 53% or 30% to 52%.
Duplex stability is very important for hybridization and anti-sense oligo-RNA interaction. It is hypothesized that oligo intraor intermolecular structures can compete with oligo-target duplex formation which may result in low hybridization efficiency. Extensive secondary structure of the siRNA can also limit this efficiency. It was found by Shabalina et al. [9] and also observed in the

Figure 1. The percentage of siRNAs which contain A/U at positions from (1 to 19 of AS strand starting from 5’end) in the three groups: ■high-efficient, ■moderate-efficient and ■low-efficient.
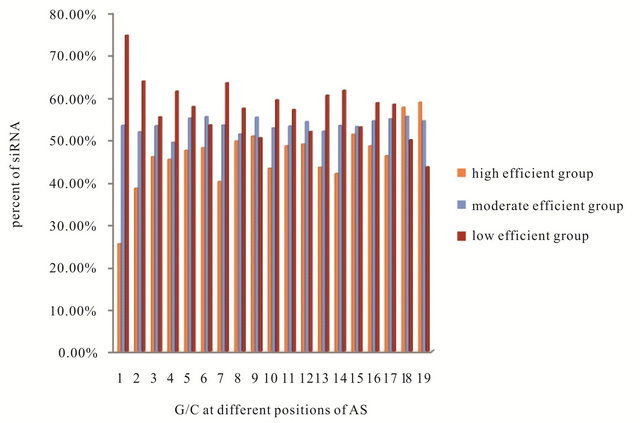
Figure 2. The percentage of siRNAs which contain G/C at positions from (1 to 19 of AS strand starting from 5’end) in the three groups (■high-efficient, ■moderate-efficient and ■low-efficient).
database that thermodynamic considerations improve the selection of efficient siRNA. The following thermodynamic features: Gibbs free energy change ΔG of sense-anti-sense duplex, Gibbs free energy change ΔG of anti-sense oligo secondary structure, Gibbs free energy change ΔG of antisense oligo-oligo dimer and the energy difference dΔG or (ΔG1-ΔG18) between the 5’end and the 3’end of (AS) strand in the high, moderate and low efficient siRNAs groups are studied. Figure 4 shows the average free energy change ΔG of the SS-AS duplex formation of the siRNAs in the three groups. It can be observed from this figure that as the SS-AS duplex is stable, the efficiency of the siRNA decreases. As the SS-AS duplex is stable, the (RISC) may be unable to separate SS-AS duplex to activate itself and complete the RNAi efficiently (Kcal/mole). The average of ΔG of AS oligo secondary structure formation in high-efficient, moderate-efficient and low-efficient groups are (−1.240, −1.69, −2.179 Kcal/ mole) respectively (Figure 5). The AS of the siRNAs in the high efficient group has the higher oligo secondary
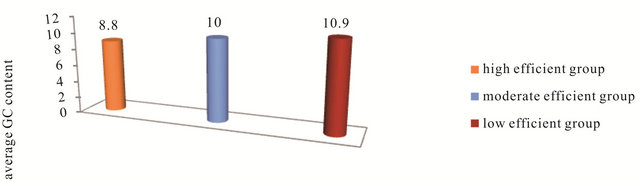
Figure 3. The average GC contents of the siRNA in the three groups (■high efficient, ■moderate efficient and ■low efficient).
Figure 4. The average free energy change ΔG of the SS-AS duplex formation for the siRNA in the three groups.
structure average free energy change. The high efficient siRNA should be unstable in oligo secondary structure of AS strand to be able to form duplex with the sense strand not secondary structure and complete RNAi process efficiently. The same action should be occur for oligo-oligo dimer reaction of AS strand which can reduce the efficiency of RNAi process by reacting AS strand with itself not its target of mRNA. So the high efficient siRNA has higher average ΔG of AS oligo-oligo dimer than moderate and low efficient AS strand. This can be observed in Figure 6. One of the most important steps in the RNAi process is the selection of the correct strand to bind with RISC. This step is depending on the binding energy difference between the antisense 5’ and 3’ends. The efficient RNAi occurs when the 5’end of the AS has lower binding energy than the 3’end. In this case the RISC will be able to unwinding the SS-AS duplex from the 5’end of AS and the correct strand (AS) will reaches the target (mRNA). The average of the biding energy difference dΔG or (ΔG1 - ΔG18) between 5’end and 3’end of (AS) strand of the siRNA in the three groups are shown in Figure 7. This figure shows that in the high efficient group the average dΔG or (ΔG1 - ΔG18) for the siRNA has a positive value 0.71 Kcal/mole while the siRNA in the low efficient group has a negative value −0.615 Kcal/ mole.
From the previous explanation, thirteen properties present in the high efficient group are selected as mentioned before. The 3743 siRNAs in the database are sorted according to the type of properties found in each one (from the thirteen identified properties) with the corresponding efficiency or average efficiency. This data are fitted to a second order polynomial 200 times after randomizing the order in which the properties were considered. One of these curves is shown in Figure 8. Each curve (from the 200 curves) gets a weight values for the

Figure 5. The average free energy change ΔG of the oligo secondary structure of the AS for the siRNAs in the three groups.
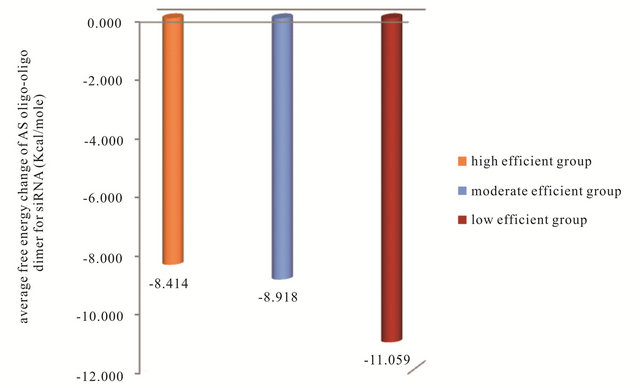
Figure 6. The average free energy change ΔG of the AS oligo-oligo dimer for siRNA in the three groups.
thirteen properties then the average weight for each property is calculated and tabulated in Table 1.
Finally, from the average weight determined for each property a score can be calculated for each siRNA in the databases according to the type of properties found in each (from the thirteen identified properties). scoresiRNA = Σ Wi Ai where Wi are the average weights obtained for each of the thirteen properties and Ai are the binary entries for each of them. Ai takes the value (1) if the property (i) is present and (0) if the property is not present. The 3734 siRNAs in the database are classified into 20 groups according to their scores and the average percentage efficiency and the standard deviation of the percentage efficiency are calculated for the siRNAs in each class. The correlation between the average score and the average experimental percentage efficiency value of the siRNA can be obtained from the final curve in Figure 9. The average percentage efficiency was computed for a score bin size of 5. The error bars are the standard deviation in the percentage efficiency values. The data was fitted using a second order polynomial, the fitted value for the coefficient ɑ2 of the square term is zero so a linear correlation of 0.97 was observed.
From the positive values of the weights in Table 1, the 13 properties are important properties which must be present in the siRNA for high efficient RNAi. This is
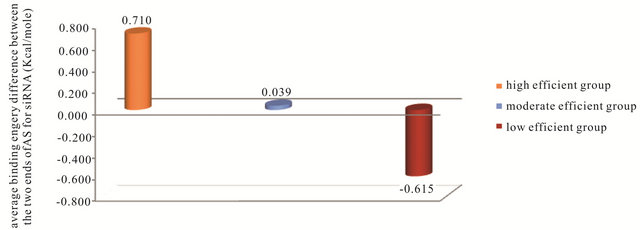
Figure 7. The average binding energy difference between 5’end and 3’end of (AS) for the siRNA in the three groups.
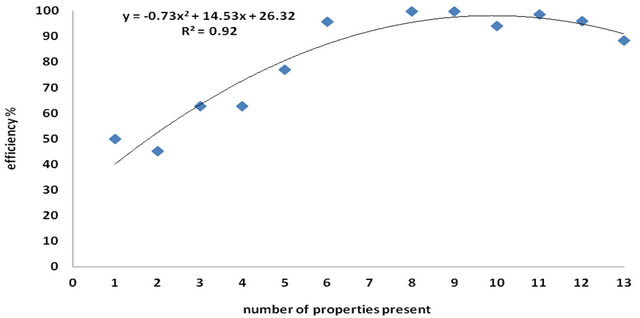
Figure 8. The number of properties present in the siRNA against the efficiency. The continuous line is the fitted second order polynomial.
because these properties are especially found in the high efficient group and not found in the low efficient group as mentioned in the primary statistical study. The properties 13 and 2 have the highest weights 0.081 and 0.080 respectively, next to them the properties 1 and 9 which have equal weights 0.072 and the remaining properties have weight values from 0.060 to 0.070. The three properties 1, 2 and 13 are related to the instability of the high efficient siRNA at the 5’end of the anti-sense strand. The properties 1 and 2 which mean the presence of A or U nucleotides at positions 1 and 2 starting from the 5’end of the anti-sense strand and also mean that the siRNA at the 5’end of the anti-sense strand is unstable are needed for the correct unwinding of SS and AS by RISC. The same reason for the property 13 which means that the difference in the binding energy at the two ends (5’-AS and 3’-AS) must be positive. The properties 1, 2 and 13 have a higher weight and may be related to each other, so from this study it appears that the instability of the siRNA at the 5’end with respect to the 3’end of the anti-sense strand is an important factor for the efficiency of RNAi. This needs that the 3’end of the anti-sense strand must be more stable or at lower binding energy than the 5’end. This is achieved by the presence of the nucleotides G or C in the positions 19 and 18 starting from the 5’end of the anti-sense strand. This are represented by properties 7 and 8 respectively.
SS-AS duplex should be flexible duplex and this is achieved by the presence of instability nucleotides such as A or U in different positions in the siRNA as the properties 3, 4, 5 and 6. These properties are the presence of A or U in the positions 7, 10, 13 and 14 starting from the 5’ end of the anti-sense strand respectively. Khvorova et al. [8] said that “the observed low overall internal stability of efficient siRNAs, especially the cleavage region (9 - 14 nt) may facilitate product release, thus allowing the RISC to find a second substrate.” Also property 10 is related to the flexibility of siRNA. Property 10 which is “the average free energy change of the SS-AS duplex formation should be greater or equal to −36.72 Kcal/ mole” has weight value (0.063). The instability of the siRNA depends also on the GC content represented by property 9 where the presence of more G or C nucleo-

Figure 9. The average score of the class against the average percentage efficiency of the siRNAs in the class.
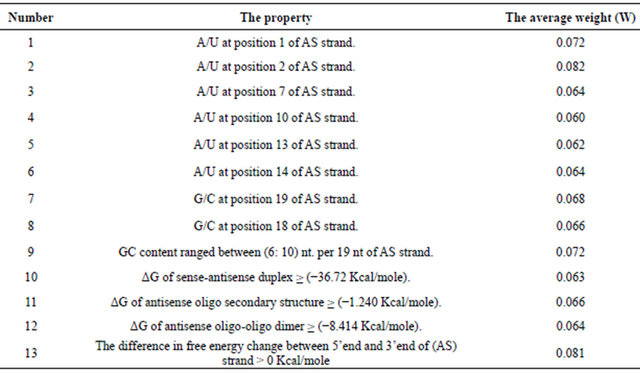
Table 1. The type of the property and its average weight.
tides gives more stability, while the presence of A or U gives instability which is needed in certain regions of siRNA for efficient RNAi as mentioned before. The property 9 is found to be of high weight.
Some reactions must be avoided as the oligo seconddary structure and oligo-oligo dimer of AS. These are represented by the properties 11 and 12 respectively.
4. CONCLUSION
The weighted scoring system developed in this search includes sequence as well as thermodynamics properties and can be used to determine the optimal sequence within a target mRNA and thus aids in the rational selection of siRNA sequences. The weighted scoring system provides better flexibility in designing an appropriate siRNA when compared to the un-weighted scoring system. This weighted scoring system shows a linear correlation with the silencing efficiency of siRNAs.
From the primary and the final statistical studies, it is obvious that the thirteen properties are important properties for efficient RNAi. If the siRNAs satisfy all the thirteen properties, the silencing efficiency can reach 88.4%. The remaining percentage efficiency may be related to the properties of targeted mRNA.
The most important feature of highly efficient siRNA is the difference in binding energy between the 5’end and the 3’end of the anti-sense strand which is related to the properties 1, 2, 7, 8 and 13. The (RISC) activation step is a critical step in RNAi process where the efficiency of this process depends on the instability of the 5’end of antisense strand. This agrees with the high weights of the properties (the difference in binding energy between the 5’end and the 3’end of the antisense strand) and the presence of the nucleotides A or U at positions 1 and 2 and the nucleotides G or C at positions 18 and 19 starting from 5’end of the antisense strand.
5. ACKNOWLEDGEMENTS
The many thanks for the authors of the database on the net, their names are mentioned in the paper.
REFERENCES
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. and Tuschl, T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494-498. doi:10.1038/35078107
- Martinez, J., Patkaniowska, A., Urlaub, H. Lührmann, R. and Tuschl, T. (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell, 110, 563-574. doi:10.1016/S0092-8674(02)00908-X
- Hannon, G.J. and Rossi, J.J. (2004) Unlocking the potential of the human genome with RNA interference. Nature, 431, 371-378. doi:10.1038/nature02870
- Hsieh, A.C., Bo, R., Manola, J., Vazquez, F., Bare, O., Khvorova, A., Scaringe, S. and Sellers, W.R. (2004) A library of siRNA duplexes targeting the phosphoinositide 3-kinase pathway: Determinants of gene silencing for use in cell-based screens. Nucleic Acids Research, 32, 893- 901. doi:10.1093/nar/gkh238
- Huesken, D., Lange, J., Mickanin, C., Weiler, J., Asselbergs, F., Warner, J., Meloon, B., Engel, S., Rosenberg, A., Cohen, D., Labow, M., Reinhardt, M., Natt, F. and Hall, J. (2005) Design of a genome-wide siRNA library using an artificial neural network. Nature Biotechnology, 23, 995- 1001. doi:10.1038/nbt1005-1315a
- Jagla, B., Aulner, N., Kelly, P.D., Song, D., Volchuk, A., Zatorski, A., Shum, D., Mayer, T., De Angelis, D.A., Ouerfelli, O., Rutishauser, U. and Rothman, J.E. (2005) Sequence characteristics of functional siRNAs. RNA, 11, 864-872. doi:10.1261/rna.7275905
- Katoh, T. and Suzuki, T. (2007) Specific residues at every third position of siRNA shape its efficient RNAi activity. Nucleic Acids Research, 35, 27-40. doi:10.1093/nar/gkl1120
- Khvorova, A., Reynolds, A. and Jayasena, S.D. (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell, 115, 209-216. doi:10.1016/S0092-8674(03)00801-8
- Shabalina, S. A., Spiridonov, A.N. and Ogurtsov, A.Y. (2006) Computational models with thermodynamic and composition features improve siRNA design. BMC Bioinformatics, 7, 65-80. doi:10.1186/1471-2105-7-65
- Shah, J.K., Garner, H.R., White, M.A., Shames, D.S. and Minna, J.D. (2007) sIR: siRNA information resource, a web-based tool for siRNA sequence design and analysis and an open access siRNA database. BMC Bioinformatics, 8, 178. doi:10.1186/1471-2105-8-178
- Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W.S. and Khvorova, A. (2004) Rational siRNA design for RNA interference. Nature Biotechnology, 22, 326-330. doi:10.1038/nbt936
- Ui-Tei, K., Naito, Y., Takahashi, F., Haraguchi, T., OhkiHamazaki, H., Juni, A., Ueda, R. and Saigo, K. (2004) Guidelines for the selection ofhighly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Research, 32, 936-948. doi:10.1093/nar/gkh247
- Chalk, A.M., Wahlestedt, C. and Sonnhammer, E.L. (2004) Improved and automated prediction of effective siRNA. Biochemicaland Biophysical Research Communications, 319, 264-274. doi:10.1016/j.bbrc.2004.04.181
- Matveeva, O., Nechipurenko, Y., Rossi, L., Moore, B., Saetrom, P., Ogurtsov, A.Y., Atkins, J.F. and Naito, Y., Yamada, T., Ui-Tei, K., Morishita, S. and Saigo, K. (2004) siDirect: Highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Research, 32, W124-W129. doi:10.1093/nar/gkh442
- Tilesi, F., Fradiani, P., Socci, V., Willems, D. and Ascenzioni, F. (2009) Design and validation of siRNAs and shRNAs. Molecular Therapeutics, 11, 156-164. http://ukpmc.ac.uk/abstract/MED/19330721

