Journal of Environmental Protection
Vol.07 No.07(2016), Article ID:67389,8 pages
10.4236/jep.2016.77091
Characterizations and Analysis of the Mold from Animal Specimens in Shenzhen Museum
Rui Zhang1*#, Kang Wang1*, Meirong Sun2, Bo Zhang3, Honglian Zhang1, Sidong Li1, Hui Du1, Yanlin Lu3, Yang Ye3, Xiaoqiang Yu4
1Center of Biology of Experimental Teaching Department, Guangdong Ocean University, Zhanjiang, China
2Shenzhen Guibao Science & Technology Company, Ltd., Shenzhen, China
3Shenzhen Museum, Shenzhen, China
4School of Biological Sciences, University of Missouri-Kansas City (UMKC), Kansas City, USA

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 17 March 2016; accepted 13 June 2016; published 16 June 2016
ABSTRACT
Biological specimens play an important role in cultural exchange, science popularization, scientific research and economic window, but the preparation and preservation technology system of biological specimens is relatively unsafe and inefficient. Mold grows seriously on animal specimens, which is not only harmful to human beings’ health and environment, but also is one of the factors that restricts the development of the natural history museums where these specimens are kept. This paper identified the mold species of animal specimens by PCR with ITS primers, bio-micro- scopic observation, sequencing and phylogenetic tree analysis. The results showed the mold of animal specimens mainly belonged to Aspergillus and Neurospora. This study established the foundations of controlling and restoring the mold that infected animal specimens and guided a new methodology of preparation and environmental friendly exhibition for animal specimens.
Keywords:
Animal Specimens, Mold Identification, PCR, Sequencing, Phylogenetic Tree Analysis
1. Introduction
1.1. Beautiful Appearance but Problems inside
Biological specimens display beautiful appearances [Figure 1], but there are serious potential problems hidden
Figure 1. Biological specimens display the very beautiful appearance.
inside, especially in south China. Exposure to hot and humid environments has damaged specimens. This environment has been especially hard on animals that have been taxidermied because they have external skin, fur, internal fat, protein, connective tissue, etc. Infection by mold spores and hyphae is an acute problem to animal specimens, which have been associated with many human beings’ serious health problems and environmental safety problems. It would also lead to great loss of specimens in natural history collections. Because animal specimens are more precious than any other forms of biological specimens such as plants and other specimens. They are very expensive, so it would be a huge economic lost if they were damaged. The climate of South China is hot and humid which lasts for about 8 - 10 months in one year. In Guangdong province, the annual average relative humidity is 65% - 85%. Especially during the mold rainy season, the relative humidity often reached more than 90%, sometimes even was 100%.
At present, there are many reports on the ancient cultural relics such as paper, ancient lacquer painting, silk cotton fabric, leather, ivory, bamboo, jade, pottery and porcelain, and glass [1] - [4] . Recently there have been a few reports on the mold that infected animal specimens.
So, there are great challenges and difficulties in the most important techniques for sample collection, preparation and storage of animal specimens.
1.2. Current Situation: The Harmfulness of Chemical Mold Proof Reagents Used in Preparation of Animal Specimens
At present, there are many poisonous reagents used in preparation of animal specimens, such as arsenic trioxide (As2O3), Camphor, Phenol, Oxymethylene, and so on. It is harmful to people’s health and the environment of exhibition, such as conservation workers, and the general public. Mold is the most important role to damage the animal specimens [Figure 2].
1.3. Mold Damage: The Harmfulness of Animal Specimens Mold to Human Beings
The incidence and prevalence of serious mycoses continue to be a public health problem.
Figure 2. There are many poisonous reagents used in preparation of animal specimens.
Thousands of species of microscopic MOLD are common in the environment, these mold feed on dead organic matter, including natural history specimens. The species involved Aspergillus, Penicillium, Geotrichum, Alternaria, Cladosporium, Mucor, Rhizopus, Trichoderma, Fusarium which are associated mostly with allergic response of different types. At first, mold is harmful to human beings. Microbiological contamination with mold, It not only can pose a significant health hazard to those working in archives or museums [5] , but also could lead to many serious illness. The nose and paranasal sinuses almost always harbor numerous mold species. Amongst the species recovered, many have the potential to produce mycotoxins including Aspergillus (flavus, niger, fumigatus, versicolor), Chaetomium, Fusarium, Penicillium and Trichoderma. This group also found “mold elements (hyphae, destroyed hyphae, conidiae and spores)”. Mold and eosinophilic mucin are the markers of sinus involvement in the CRS patients [6] . Chronic Rhinosinusitis (CRS), A primary concern was that exposure to high concentrations of Aspergillus spores may cause respiratory illness, particularly in individuals that are immunocompromised, it also led to endocrinopathy with growth hormone deficiency; Dennis-Robertson syndrome [7] - [9] . Molds and mycotoxins cause neurobehavioral and pulmonary impairment as well as adverse health effects especially with allergies and asthma [10] - [19] .
So, it is very important to characterize and analyze the mold of animal specimens.
Shenzhen Museum was established in 17th Oct, 1981. The area of structure is 32,000 square meter. By the end of 2015. There are collected more than 35,000 different kinds of cultural relics, including about 25,000 of marine animals and wild animal specimens and fossils collected. The specimens of biological fossils dated from 100 million years, ancient history and art treasures, showing 5000 years of civilization of China and important historic materials about the development history of modern and contemporary Shenzhen.
2. Materials and Methods
2.1. Experimental Materials
The mold samples were collected from animal specimens of Shenzhen museum during December 2013-March 2014. These specimens include porcupine, birds, turtles, antelope and deer. Because of the mould rainy day, there are plenty of mold exposed on them (Figure 3).
2.2. Methods
2.2.1. Mold Cultured and Isolated and Microscopes Observation
PDA medium was used to culture and isolate the collected mold. Identification of the mold was confirmed by culturing swabs from infected specimens and examining mold hyphae with both light bio-microscopes.
The swabs were immersed and homogenized in a sterile physiological solution and serial dilutions were made. Each dilution was inoculated (0.1 ml) on MEA. After 2 days of cultivation at 25˚C, identification of the mold was performed. After 3 days of fostering, seemingly different mold colony grew on 12 plates. A small sample from the middle of the colony was dug and transferred to new plate to culture the different colonies. Strains that were previous activated had a small amount of sterile water with the pipettor added. The mold was then extracted from the solution and inoculated on a slant medium. This was repeated.
Figure 3. The mold samples was collected from animal specimens.
Then slightly digged a small piece of colony from the center of the same kind. Mold were transferred to a new plate one by one until separating into single and stable colonies plates. Take strains previously activated and kept in the AGAR slant tube, added some amount (bacteriostatic ring according to the experimental plate number) of sterile water with the pipettor on the clean work platform, then moved the pipettor under the liquid level, absorbed a little liquid and gently hit out, beat the mold on the inclined medium, let more mold into the suspension, make of the mold suspension.
The mold was observed through bio-microscopic observation. A small amount mold hyphae was dropped onto the water slide to make a suspension. At this point, the class slide was turned 45 degrees slowly to cover the mold to make observations of the bubbles. The slide was put on the staged and to observe the structure of the mold. Computer software was then used to characterize the mold.
Cultural and micromorphological characteristics of mold colonies observation and identification was performed through using identification ways.
The collection consisted of deteriorated animal specimens, including 12 species of animal specimens, 1) Porcupine, 2) wood, 3) bird, 4) turtle, 5) antelope, 6) deer 1, 7) deer 2, 8) wale, 9) Chimpanzee, 10) Raja porosa.
2.2.2. PCR Detection
The isolated single colonies and ITS primers were used to do PCR (S1000 Thermal Cycler BioRad, USA). Primers were ITS1 and ITS4. The mold DNA was amplified using the highly conserved fungal rRNA gene primers (ITS1F and ITS4) as White and Gardes previously described [20] - [22] . The sequence of ITS1F and ITS4 are ITS1F 5’-CTTGGTCATTTAGAGGAAGTAA-3’, ITS4 5’-TCCTCCGCTTATTGATATGC-3’.
PCR reaction conditions was 94˚C predenatured for 5 min, 98˚C denatured for 30 s, 58˚C annealed for 30s, 68˚C extended for 2 - 3 min. After the total 27 cycles. They were detect by agarose gel electrophoresis.
2.2.3. Sequencing and Phylogenetic Tree Analysis
The isolated single colonies were sent to Shanghai Shenggong Company, Ltd to be sequenced. And the results were analyzed. Distance-matrix sequence alignment methods used Clustal W. Maximum Likelihood (ML) phylogenetic tree for the fungal Operational Taxonomic Units (OTUs) constructed based on fungal ITS gene sequences obtained using fungal specific primer set ITS1F/ITS4. These sequences resulted in 8 fungal Operational Taxonomic Units (OTUs) with ITS and 19 OTUs with 18S rDNA primer sets respectively by taking into account the 2% sequence divergence cut-off for species delineation. Topology was built using Mega v.5.03 from a ClustalW 1.83 alignment. Numbers below branches indicate bootstrap values (>50%) from 1000 replicates. New sequence types are marked with triangle. B: Proportional distribution of different fungal taxa in the clone library constructed with fungal specific primer set ITS1F/ITS4.
3. Results and Discussion
3.1. Mold Microscopic Observation Results
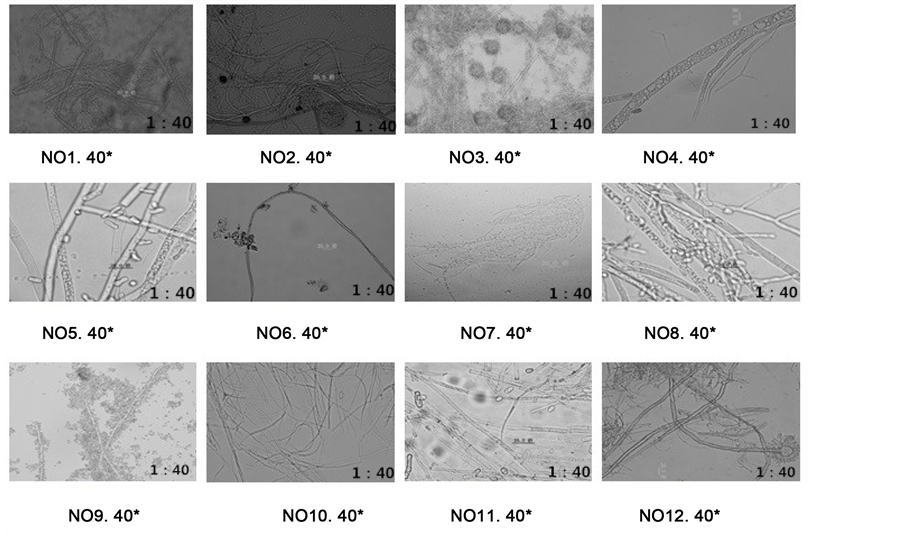
Identification of the mold was confirmed by culturing swabs from infected specimens and examining mold hyphae both light bio-microscopes. Mold microscopic observation of 12 mold infected species were identified
The examined animal specimens in Shen Zhen museum showed clear signs of biodeterioration. Superficial colonies of mold were clearly visible and abundant. A total of 12 mold species were identified from all analyzed samples with 12 taxa identified (Figure 4).
The mold hyphae was detected used 40 fold light bio-microscopes. It need to improve the fold of bio-micro- scopes. It would have better observation results.
Figure 4. Mold hyphae microscopic observation that isolated from 12 mold that infected animal specimens.
3.2. Single Colony-PCR Detection
Because the mold collected from the museum was mixed mold samples, they were still unable to determine the type and the single degree, so PCR was used for species identification. The isolated single colonies were detected by PCR primers ITS1 and ITS4. The results showed that the DNA bands were right (Figure 5).
Twelve animal species mold samples DNA were amplified through colony PCR amplification. Then it were done the species identification. The results proved that the colony PCR experiments could be successfully used for species identification.
Mold genetic materials were amplified in colony PCR expansion test and species identification was performed. The results proved that the colony PCR experiments could be successfully used for species identification.
Figure 5 showed the electrophoresis bands after the PCR amplification of 1 - 12 strains. It could be seen the bright and clear 12 bands from the figure, Colony PCR experiment successfully copied the genetic material of 12 strains.
Colony PCR constructed based on fungal ITS gene sequences obtained using fungal specific primer set ITS1F/ITS4. But the general primers sometimes had some limited. It would have better PCR results after screening advanced specific primers [23] .
3.3. Sequenced and Analyzed, Phylogenetic Tree Analysis
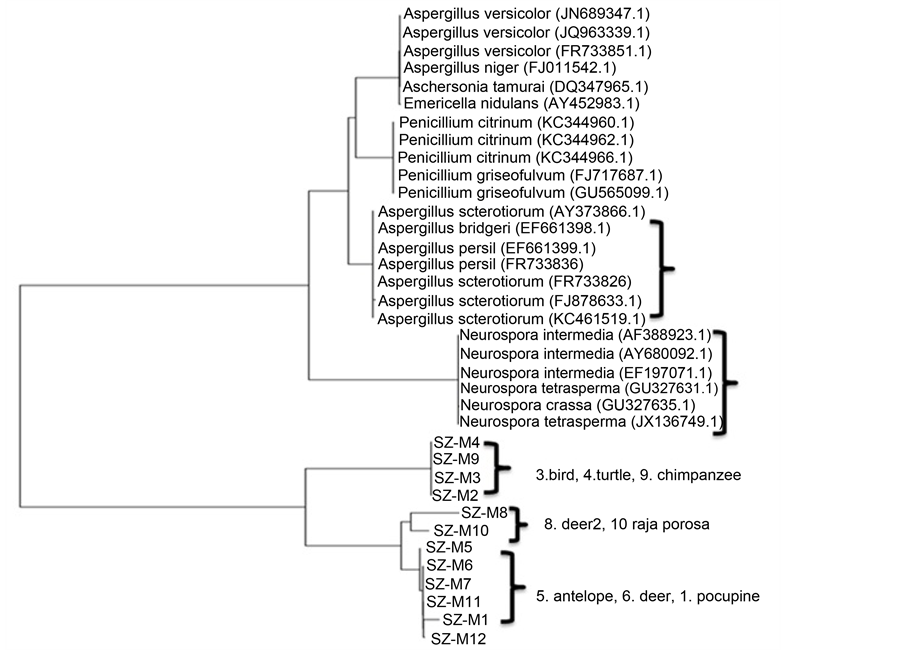
The single isolated colony was sent to Shanghai Shenggong Company, Ltd to be sequenced. The results were analyzed. Distance-matrix sequence alignment methods used Clustal W (Figure 6). The results showed the mold from animal specimens mainly belong to Aspergillus and Neurospora. The results were benefit for different novel mold inhibitor screening according to the different types of mold that molecular species identification.
Phylogenetic tree for the fungal Operational Taxonomic Units (OTUs) constructed based on fungal ITS gene sequences obtained using fungal specific primer set ITS1F/ITS4. These sequences resulted just in fungal Operational Taxonomic Units (OTUs) with ITS and 19 OTUs with 18S rDNA primers that with different fungal taxa in the clone library constructed with fungal specific primer set ITS1F/ITS4. But fungal strains used for specificity test of new 28S rDNA primers would be more significant results [24] - [26] .
Figure 5. Colony-PCR, ITS primer be used to identify of mold samples. 1-12 mold samples collected from 12 spieces. 1) Porcupine, 2) wood, 3) bird, 4) turtle, 5) antelope, 6) deer 1, 7) deer 2, 8) wall, 9) chimpanzee, 10) Raja porosa. M: Marker, Del2000. 1-12 was the mold samples PCR that was collected from animal specimens. This figure was the electrophoresis stripes after the PCR amplification of 1-12 strains, could see the bright and clear 12 stripes from the figure, colony PCR experiment successfully copied the genetic material of 12 strains.
Figure 6. Mold genetic sequenced and analysed, phylogenetic tree analysis. The results showed the prevailing mold species isolated from animal specimens was Aspergillus niger Tiegh (62.5%), followed by Neurospora crassa Shear & B. O. Dodge (25%) The signal colony that isolated sent to Shanghai Shenggong Company, Ltd. to be sequenced. And the results was analysed. Distance-matrix sequence alignment methods was used Clustal W.
4. Conclusion
All in all, after identifying the mold species from animal specimens, by PCR with ITS primer, bio-microscopic observation and sequencing and phylogenetic tree analysis, the results showed the mold from animal specimens mainly belonged to Aspergillus and Neurospora. This study established the foundations of controlling and restoring the mold that infected animal specimens and guided a new methodology of preparation and environmental friendly exhibition for animal specimens. It was benefit for screening different novel mold inhibitor according to the different types of mold.
Acknowledgements
This research was supported by the Science Technology and Innovation Committee of Shenzhen Municipality (the Grant No. JCYJ20130325105212218), and supported by “300701-A14025” and Guangdong Science and Technology Department of public welfare project research and capacity building special funds, 2015A020219008 (K15272) It was also supported by CSC (China Scholarship Council ) funds 201508440158.
Cite this paper
Rui Zhang,Kang Wang,Meirong Sun,Bo Zhang,Honglian Zhang,Sidong Li,Hui Du,Yanlin Lu,Yang Ye,Xiaoqiang Yu, (2016) Characterizations and Analysis of the Mold from Animal Specimens in Shenzhen Museum. Journal of Environmental Protection,07,1033-1040. doi: 10.4236/jep.2016.77091
References
- 1. Kolomodin-Hedman, B., Blomquist, G. and Sikstrom, E. (1986) Mould Exposure in Museum Personnel. International Archives of Occupational and Environmental Health, 57, 321-323. http://dx.doi.org/10.1007/BF00406187
- 2. Krake, A.M., Worthington, K.A., Wallingford, K.M. and Martinez, K.F. (1999) Evaluation of Microbiological Contamination in a Museum. Applied Occupational and Environmental Hygiene, 14, 499-509.http://dx.doi.org/10.1080/104732299302468
- 3. Krysińska-Traczyk, E. (1994) Contamination of Archives by Filamentous Fungi and Their Evaluation for Potential Pathogenicity. Medycyna Pracy, 45, 495-500.
- 4. Lugauskas, A. and Krikstaponis, A. (2004) Microscopic Fungi Found in the Libraries of Vilnius and Factors Affecting Their Development. Indoor and Built Environment, 13, 169-182. http://dx.doi.org/10.1177/1420326X04045274
- 5. Zielińska-Jank, K., Kozajda, A., Piotrowska, M. and Szadkowska-Stanczyk, I. (2008) Microbiological Contamination with Mould in Work Environment in Libraries and Archive Storage Facilities. Annals of Agricultural and Environmental Medicine, 15, 71-78.
- 6. Brewer, J.H., Thrasher, J.D., Straus, D.C., Madison, R.A. and Hooper, D. (2013) Detection of Mycotoxins in Patients with Chronic Fatigue Syndrome. Toxins, 5, 605-617. http://dx.doi.org/10.3390/toxins5040605
- 7. Dennis, D.P. (2003) Chronic Defective T-Cells Responding to Superantigens, Treated by Reduction of Fungi in the Nose and Air. Archives of Environmental Health, 58, 433-451.
- 8. Dennis, D.P., Roberson, D. and Curtis, L. (2009) Fungal Exposure Endocrinopathy with Growth Hormone Deficiency; Dennis-Robertson Syndrome. Toxicology and Industrial Health, 25, 669-680. http://dx.doi.org/10.1177/0748233709348266
- 9. Rea, W.J., Didriksen, N., Simon, T.R. and Griffiths, G. (2003) Effects of Toxic Exposure to Mold and Mycotoxins in Building-Related Illnesses. Archives of Environmental Health, 58, 399-405.
- 10. Campbell, A., Thrasher, J.D., Gray, M.R. and Vojdani, A. (2004) Mold and Mycotoxins: Effects on the Neurological and Immune Systems in Humans. Advances in Applied Microbiology, 55, 375-398. http://dx.doi.org/10.1016/S0065-2164(04)55015-3
- 11. Gray, M.R., Thrasher, J.D. and Crago, R. (2003) Mixed Mold Mycotoxicosis: Immunological Changes in Humans Following Exposure to Water Damaged Buildings. Archives of Environmental Health, 58, 410-420.
- 12. Kilburn, K.H. (2009) Neurobehavioral and Pulmonary Impairment in 105 Adults with Indoor Exposure to Molds Compared to 100 Exposed to Chemicals. Toxicology and Industrial Health, 35, 681-692. http://dx.doi.org/10.1177/0748233709348390
- 13. Fisk, W.J., Eliseeva, E.A. and Mendell, M.J. (2010) Association of Residential Dampness and Mold with Respiratory Tract Infections and Bronchitis: A Meta-Analysis. Environmental Health, 9, 72-84. http://dx.doi.org/10.1186/1476-069X-9-72
- 14. El-Morsy, S.M., Khafagy, Y.W., El-Naggar, M.M. and Beih, A.A. (2010) Allergic Fungal Rhinosinusitis: Detection of Fungal DNA in Sinus Aspirate Using Polymerase Chain Reaction. Journal of Laryngology & Otology, 124, 152-160. http://dx.doi.org/10.1017/S0022215109991204
- 15. Vennewald, I. and Wollina, U. (2005) Cutaneous Infections Due to Opportunistic Molds: Uncommon Presentations. Clinics in Dermatology, 23, 565-571. http://dx.doi.org/10.1016/j.clindermatol.2005.01.003
- 16. Lionakis, M.S. and Kontoyiannis, D.P. (2004) Fusarium Infections in Critically Ill Patients. Seminars in Respiratory and Critical Care Medicine, 25, 159-169. http://dx.doi.org/10.1055/s-2004-824900
- 17. De Paoli, P. (2005) Bio-Banking in Microbiology: From Sample Collection to Epidemiology, Diagnosis and Research. FEMS Microbiology Reviews, 29, 897-910.
- 18. Ljaljevic-Grbic, M., Stupar, M., Vukojevic, J., Maricic, I. and Bungur, N. (2013) Molds in Museum Environments: Biodeterioration of Art Photographs and Wooden Sculptures. Archives of Biological Sciences, 65, 955-962. http://dx.doi.org/10.2298/ABS1303955G
- 19. Bush and Portnoy (2001) The Presence of Fungal Propagules in Indoor Air Causes Adverse Health Effects, Especially Allergies and Asthma. Allergy, 2, 59-64.
- 20. White, T.J., Bruns, T.D., Lee, S.B. and Taylor, J.W. (1990) Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. and White, T.J. Eds., PCR Protocols—A Guide to Methods and Applications, Academic Press, San Diego, 315-322. http://dx.doi.org/10.1016/b978-0-12-372180-8.50042-1
- 21. Gardes, M. and Bruns, T.D. (1993) ITS Primers with Enhanced Specificity for Basidiomycetes—Application to the Identification of Mycorrhizae and Rusts. Molecular Ecology, 2, 113-118. http://dx.doi.org/10.1111/j.1365-294X.1993.tb00005.x
- 22. Lai, X., Cao, L., Tan, H., Fang, S., Huang, Y. and Zhou, S. (2007) Fungal Communities from Methane Hydrate-Bear- ing Deep-Sea Marine Sediments in South China Sea. The ISME Journal, 1, 756-762. http://dx.doi.org/10.1038/ismej.2007.51
- 23. Daniel, K.M. and Jorge, M.V. (2007) Use of the ITS Primers, ITS1F and ITS4, to Characterize Fungal Abundance and Diversity in Mixed-Template Samples by qPCR and Length Heterogeneity Analysis. Journal of Microbiological Methods, 71, 7-14. http://dx.doi.org/10.1016/j.mimet.2007.06.016
- 24. Purnima, S., Chandralata, R., Pankaj, V. and Yogesh, S. (2012) Assessment of Fungal Diversity in Deep-Sea Sediments by Multiple Primer Approach. World Journal of Microbiology and Biotechnology, 28, 659-667. http://dx.doi.org/10.1007/s11274-011-0859-3
- 25. Mueller, R.C., Gallegos-Graves, L., Zak, D.R. and Kuske, C.R. (2016) Assembly of Active Bacterial and Fungal Communities along a Natural Environmental Gradient. Microbial Ecology, 71, 57-67. http://dx.doi.org/10.1007/s00248-015-0655-y
- 26. Dirk Krüger, D.K., Christiane, F.R. and Daniel, T.W. (2012) Diversity Measures in Environmental Sequences Are Highly Dependent on Alignment Quality—Data from ITS and New LSU Primers Targeting Basidiomycetes. PLoS ONE, 7, e32139. http://dx.doi.org/10.1371/journal.pone.0032139
NOTES
*These authors contributed equally to this work.
#Corresponding author.