World Journal of Vaccines
Vol.4 No.1(2014), Article ID:43089,11 pages DOI:10.4236/wjv.2014.41006
Human Enterovirus 71 DNA Vaccine Constructs Containing 5’UTR with Complete Internal Ribosome Entry Site Sequence Stimulated Improved Anti-Human Enterovirus 71 Neutralizing Immune Responses
1Tropical Infectious Diseases Research and Education Center (TIDREC), Department of Medical Microbiology, Faculty of Medicine, University Malaya, Kuala Lumpur, Malaysia; 2Virology Unit, Institute for Medical Research, Kuala Lumpur, Malaysia.
Email: sazaly@um.edu.my
Copyright (c) 2014 Nor-Aziyah Mat-Rahim, Sazaly AbuBakar. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights (c) 2014 are reserved for SCIRP and the owner of the intellectual property Nor-Aziyah Mat-Rahim, Sazaly AbuBakar. All Copyright (c) 2014 are guarded by law and by SCIRP as a guardian.
Received December 27th, 2013; revised January 27th, 2014; accepted February 7th, 2014
Keywords:Human Enterovirus 71; 5’Untranslated Region (5’UTR); Internal Ribosome Entry Site (IRES); DNA Vaccine; Neutralizing Antibodies
ABSTRACT
Recent improvement in the technologies for efficient delivery of DNA vaccines has renewed interest in the DNA-based vaccines. Several DNA-based vaccines against human enterovirus 71 (EV71), the causative agent for hand, foot and mouth disease (HFMD) have been developed. Here we examined the potential of improving the vaccines by inserting the EV71 5’ untranslated region (5’ UTR) containing the full length internal ribosome entry site (IRES) sequence to the EV71 VP1-based DNA vaccine constructs. Four vaccine constructs designated as 5’ UTR-VP1/EGFP, VP1/EGFP, 5’ UTR-VP1/pVAX and VP1/pVAX, were designed using the pEGFP-N1 and pVAX-1 expression vectors, respectively. Transfection of Vero cells with the vaccine constructs with the 5’-UTR (5’-UTR-VP1/EGFP and 5’ UTR-VP1/pVAX) resulted in higher percentages of cells expressing the recombinant protein in comparison to cells transfected with vectors without the 5’-UTR (67% and 57%, respectively). Higher IgG responses (29%) were obtained from mice immunized with the DNA vaccine construct with the full length 5’ UTR. The same group of mice when challenged with life EV71 produced significantly higher neutralizing anti-body (NAb) titers (>5-fold). These results suggest that insertion of the EV71 5’ UTR sequence consisting of the full length IRES to the EV71 DNA vaccine constructs improved the efficacy of the constructs with enhanced elicitation of the neutralizing antibody responses.
1. Introduction
Hand, foot and mouth disease (HFMD) caused by enterovirus 71 (EV71) has been estimated to affect millions of young children worldwide [1]. The disease is usually mild and most are self-limiting [2,3]. In recent years, EV71-associated brainstem encephalitis cases with high fatalities have become more frequent, especially in several Asian countries [3-9].
There are no commercially available vaccine to prevent the infection, however, various strategies have been employed to develop candidate vaccines against EV71 [6, 8,10-14]. The success of vaccination in eradicating poliovirus, a virus belonging to the same family as EV71, strongly suggests that vaccination is likely among the most effective means to prevent and contain EV71 infection. Up to date, few EV71 inactivated vaccines are being evaluated in the clinical trial stage [8,15,16]. While it is possible that the conventional live-attenuated EV71 vaccine could prove to be the most effective in stimulating protective immune responses, there are concerns of possible reversion to the infectious form [4,17], similar to that reported for poliovirus vaccine [18-20]. A DNA-based vaccine, hence, is proposed as possible alternative as it could mimic the effects conferred by live-attenuated vaccines without the usage of infectious agents [21-23]. DNA vaccines have been demonstrated to stimulate both Class I major histocompatibility complex (MHC-I) and Class II major histocompatibility complex (MHC-II) responses that allows for the induction of the CD8+ and CD4+ T cells, respectively [24]. It is expected note peculiarities. For example, the head margin in this template measures that the ability to stimulate the T cell responses would accord long-lasting immunity.
One of the major hurdles of DNA-based vaccine is the inability to induce robust immune responses in large animals and human, and the inefficiency in inducing humoral immune responses [25]. The low cellular uptake efficiency of the DNA-based vaccine and low level of expression from the vector may cause the inefficiency in inducing strong antigen-specific immune responses [26]. Approaches to enhance the efficacy of DNA-based vaccine therefore, are very much needed. The advents of latest technology for the delivery of DNA-based vaccines [27-30] warrant relooking at possible DNA-based vaccines for EV71.
We had previously described the potential of VP1- based DNA vaccines to elicit immune responses against EV71 [23]. The immune responses however, elicited only low level of neutralizing antibody titers. In this study, the EV71 DNA vaccine has been improved by incorporation of complete sequence of EV71 internal ribosome entry site (IRES). The IRES, especially picornaviruses’ are widely used in biotechnology applications [31-33]. Numerous studies on IRES of encephalomyocarditis (EMCV), foot-and-mouth virus (FMDV), poliovirus and hepatitis C virus (HCV) have been performed by various groups and these reports showed that the IRESes have helped in enhancing the expression of gene located upstream of the IRES [32,34-39]. However, not much is known about IRES of EV71 except for the report by Lee et al. [32] who reported that the EV71 IRES mediated protein expression up to 10- to 15-fold higher than the expression mediated by commonly used EMCV IRES. In this study, we explored the possibility of improving the EV71 DNA vaccine constructs ability to stimulate the neutralizing antibody responses by incorporating the EV71 IREScontaining sequences within the vaccine constructs.
2. Methods
2.1. Virus
Enterovirus 71 strain EV71/9/97/SHA89 [GenBank:
AJ586873] maintained strictly in Vero cells was used for this study. Virus inoculum was prepared by infecting cells and incubating the cells at 37˚C until ~70% of the cells showed cytopathic effects (CPE). The supernatant was clarified by centrifugation and used as virus inoculum. Viral RNA was extracted from the inoculum using Tri Reagent (Molecular Research Center, USA) following the manufacturer’s protocol. Protein lysate was extracted from the EV71-infected cell pellet and used as antigen for enzyme-linked immunosorbent assay (ELISA).
2.2. Construction of DNA Vaccine Constructs
Four different candidate DNA vaccine constructs were designed. Initially, the VP1 gene region of EV71 isolate S2861/SAR/00 [GenBank:AF376085] was amplified from VP1/pCR® Blunt (courtesy of Prof Mary Jane Cardosa, Universiti Malaysia Sarawak) using VP1F and VP1R primer (Table 1) with NheI and BamHI restriction enzyme (RE) sequences included in the forward and reverse primer, respectively. Following amplification, the VP1 gene fragment was digested with the respective REs and ligated into pEGFP-N1 (VP1/EGFP). For cloning into pVAX-1 expression vector, the VP1 gene was amplified using VP1HindF and VP1R (Table 1) with HindIII and BamHI RE sequences included in the forward and reverse primer, respectively. The VP1 gene fragment was digested using HindIII and BamHI REs (Promega, USA) and ligated with the pVAX-1 vector (VP1/pVAX). The 5’ UTR gene region of EV71 isolate EV71/9/97/ SHA89 consisting of full IRES sequence was amplified directly from the viral RNA using a two-step RT-PCR. The cDNA was generated using EVR944 primer (Table 1) followed by amplification using IRESF and IRESR primer set (NheI RE sequence included at both forward and reverse primer), to generate 5’ UTR fragment, and IRESF2 and IRESR2 primer set (HindIII RE sequences included at both forward and reverse primer), to generate 5’ UTR 2 (Table 1). Both 5’ UTR and 5’ UTR2 fragments were digested with NheI and HindIII RE, respectively. Concurrently the VP1/EGFP and VP1/pVAX was linearized using NheI and HindIII RE, respectively. The 5’ UTR fragment was ligated into linearized VP1/EGFP, while 5’ UTR2 fragment was ligated into linearized VP1/ pVAX, both ligated at the 5’ proximal end of VP1 gene region. The plasmids consisting of 5’ UTR and VP1 gene regions were designated as 5’ UTR-VP1/EGFP and 5’ UTR-VP1/pVAX, respectively. All plasmids were transformed into competent Top10F’ E.coli (Invitrogen, USA), grown and prepared using Endotoxin-free Plasmid Maxi Kit (Qiagen, Germany). The integrity of all the DNA vaccine constructs was checked by DNA sequencing.

Table 1. Oligonucleotide primers used for amplification of EV71 5’ UTR and VP1 gene regions. The sequences underlined represent the sequence for the restriction endonucleases. The bold letters represent the codon inserted in the sequence. The underlined and bold sequences represent the inserted start codon.
2.3. In Vitro Expression
Protein expression from the DNA vaccine constructs (5’ UTR-VP1/EGFP, VP1/EGFP, 5’ UTR-VP1/pVAX and VP1/pVAX) was verified by transfecting Vero cells using Tfx™-20 transfection reagent (Promega, USA). At 48 hours post-transfection, cells transfected with pEGFPN1 and pVAX-1 harboring gene regions were fixed with 4% paraformaldehyde followed by permeabilization using 1% Nonidet P-40 (NP-40). Cells transfected with pEGFP-N1 consisting of the VP1 and 5’ UTR-VP1 were counterstained with propidium iodide, while cells tranfected with constructs cloned into pVAX-1 expression vector were probed with EV71 VP1 specific monoclonal antibody (Chemicon International, USA). Cells transfected with pEGFP-N1 vector backbone and EV71 RNA (full length genomic RNA) were included as positive controls for the transfection. Expression of constructs cloned into pEGFP-N1 was detected by observing the fluorescent EGFP protein under a microscope with ultraviolet (UV) light source, while cells expressing the recombinant proteins from DNA vaccine constructs with pVAX-1 backbone were detected by immunoperoxidase staining and viewed under a light microscope. The numbers of fluorescent and immunoperoxidase-stained cells were determined. The percentage of cells expressing the recombinant proteins was determined as described by Shafee and AbuBakar [40].
2.4. Mice Inoculation
All studies involving the use of mice received Institutional Research Board approval (No: MP/10/04/2007/ SAB(R)). Groups of female BALB/c mice were inoculated twice intramuscularly (IM) with the DNA vaccine constructs (100 μg) at 2-week intervals. Mice sera were collected by tail vein bleeding prior to any inoculation. All sera were kept at −20˚C until needed. Group of mice inoculated with only the expression vector pEGFP-N1 or pVAX-1 (100 µg) and heat-inactivated EV71 (10 µg) were used as controls. Five weeks following the final inoculation, mice were challenged intraperitoneally with 100 TCID50 of live EV71 virus (EV71/9/97/SHA89). Sera were collected at selected intervals post-challenge.
2.5. Assessment of IgG Responses
Antibody responses to the DNA vaccine constructs were determined by performing indirect ELISA using EV71- infected Vero cell lysate as antigen. Initially, ELISA plate was coated with the EV71 antigen diluted in carbonated buffer, pH 9.6, for 16 hours at 4˚C. Following the incubation, the plate was washed with 1X PBS containing 0.05% Tween-20 (PBS-T). Mice sera from the same treatment group were pooled, diluted (1:50) and added to the antigen-coated 96-well plate (100 µl). The plates were incubated for 2 hours followed by washing with PBS-T. Following the washing, alkaline phosphatase-conjugated anti-mouse IgG (Promega, USA) was added and incubated at 37˚C for 1 hour. After the incubation, phosphatase substrate was added and the optical density (O.D.) was read at 405nm with 650nm reference wavelength. The O.D. values were reported after they were normalized against the background O.D. value (value obtained from pre-immunized sera).
2.6. Enzyme-Linked Immunosorbent-Based Microneutralization Assay
Microneutralization assay was performed using sera of mice harvested at week 5 after immunization (referred as day −1), day 5 and day 10 after inoculation with live EV71 (strain EV71/9/97/SHA89). The neutralization test was performed using ELISA-based microneutralization assay [41]. Briefly, 108 FFU/ml of virus in 100 µl was added to 2-fold diluted pooled mice sera (according to their treatment group) in equal volume and incubated for 2 hours at 37˚C. The virus-serum mixture was transferred to Vero cells plated in 96-well plates. After 3 days of incubation, the supernatant was removed and cells were fixed with 4% paraformaldehyde followed by incubation at 4˚C for 30 minutes or overnight. The cells were then washed with PBS-T, permeabilized for 15 minutes at room temperature with 1% NP-40, sequentially added with rabbit-raised EV71 hyperimmune sera followed by addition of alkaline phosphatase-conjugated anti-rabbit IgG (Chemicon, USA). Alkaline phosphatase substrate, p-nitrophenyl-phosphate (PNP; KPL, USA) was then added. The optical density (O.D.) was read at 405 nm wavelength with 630 nm reference wavelength and the neutralization titer was calculated according to the method of Vorndam and Beltran [41].
2.7. Statistical Analyses
Where appropriate data from multiple datasets were expressed as the mean ± standard error mean (S.E.M). Data from the in vitro recombinant gene expression and analyses of the immune responses were transformed to log10 value prior to analysis using paired t-test with Prism 4 software (GraphPad Software, USA). Differences between groups were considered significant when p < 0.05.
3. Results
3.1. Construction of DNA Vaccine Constructs
The presence of nucleotide sequence consisting of the EV71 IRES and VP1 gene region within the DNA vaccine constructs were verified by PCR amplification followed by DNA sequencing. Amplification of DNA fragment of ~1600 base pair (bp) was obtained from the bacterial transformants harboring the 5’ UTR-VP1/EGFP (Figure 1(A)) and 5’ UTR-VP1/pVAX recombinant plasmid (Figure 1(B)). DNA fragment of ~900 bp was obtained from bacterial transformants harboring the VP1/ EGFP (Figure 1(A)) and VP1/pVAX (Figure 1(B)). No mutation or frameshifts was observed within the 5’ UTR and VP1 sequences in all DNA vaccine constructs in comparison to the original sequence (data not shown). The schematic of the DNA vaccine constructs were shown in Figure 2.
3.2. In Vitro Expression
Successful expression of constructs cloned into pEGFPN1 (5’ UTR-VP1/EGFP and VP1/EGFP) in Vero cells were confirmed by the detection of fluorescent EGFP under a microscope with ultraviolet (UV) light source (Figure 3(i)). The expression of constructs cloned into pVAX-1 (5’ UTR-VP1/pVAX and VP1/pVAX) were determined by observing the immunoperoxidase stained cells under light microscope, which appeared brownish following the staining (Figure 3(ii)). The percentage of cell transfected with 5’ UTR-VP1/EGFP was significantly
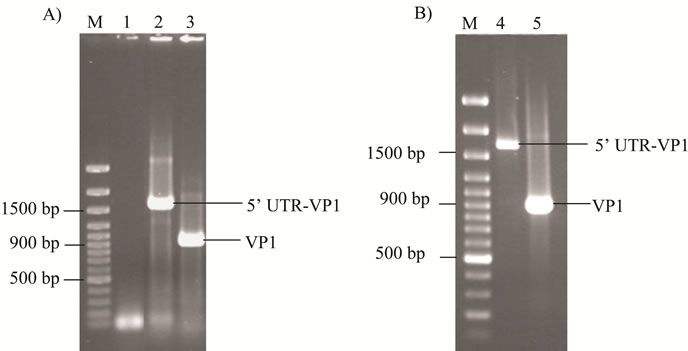
Figure 1. Amplification of 5’ UTR-VP1 and VP1 gene regions cloned into pEGFP-N1 and pVAX-1 expression vectors. (A) Lane 1 denotes amplification from bacterial transformant carrying the pEGFP-N1 vector without 5’ UTRVP1 or VP1 gene region insert. Lanes 2 and 3 indicate the amplified 5’ UTR-VP1 and VP1 gene regions from 5’ UTR-VP1/EGFP and VP1/EGFP constructs, respectively. (B) Lanes 4 and 5 denote amplified 5’ UTR-VP1 and VP1 fragments from constructs cloned into pVAX-1, respectively. Lane M denotes molecular marker.
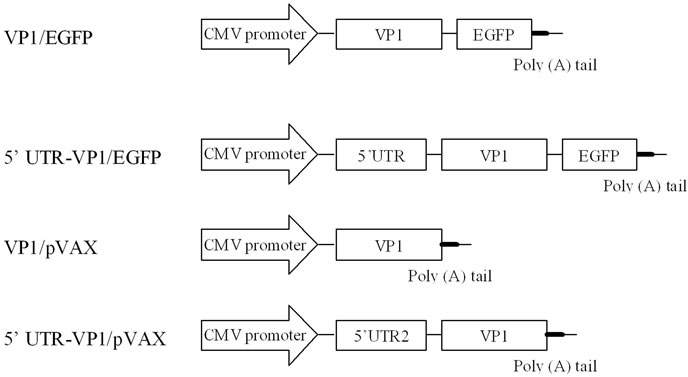
Figure 2. Schematic diagrams of the DNA vaccine constructs. For the DNA vaccine constructs using the pEGFPN1, the enhanced green fluorescent protein (EGFP) gene was fused to the 3’ proximal end of the VP1 gene region in the construct cloned into pEGFP-N1. The CMV promoter represents the human cytomegalovirus (CMV) immediate early gene promoter, 5’ UTR indicates the 5’ untranslated region and VP1 represents the viral protein 1 gene region. The constructs without 5’ UTR were expressed by the conventional cap-dependent mechanism, while the constructs with the 5’ UTR were expressed via the cap-independent mechanism using the IRES-mediated translation.
higher in comparison to VP1/EGFP at all four plasmid concentrations; 0.25 µg/ml (p = 0.0137), 0.5 µg/ml (p = 0.0488), 0.75 µg/ml (p = 0.0098) and 1.0 µg/ml (p = 0.0039) (Figure 4(i)). Cells transfected with DNA vaccine constructs cloned into pVAX-1 showed higher percentage (20% - 40%) of cells expressing the recombinant proteins in comparison to those cloned into pEGFP-N1. The 5’UTR-VP1/pVAX transfected cells showed higher percentage of transfected cells in comparison to that transfected with VP1/pVAX, however, the differences in the percentage of cells expressing the recombinant pro-
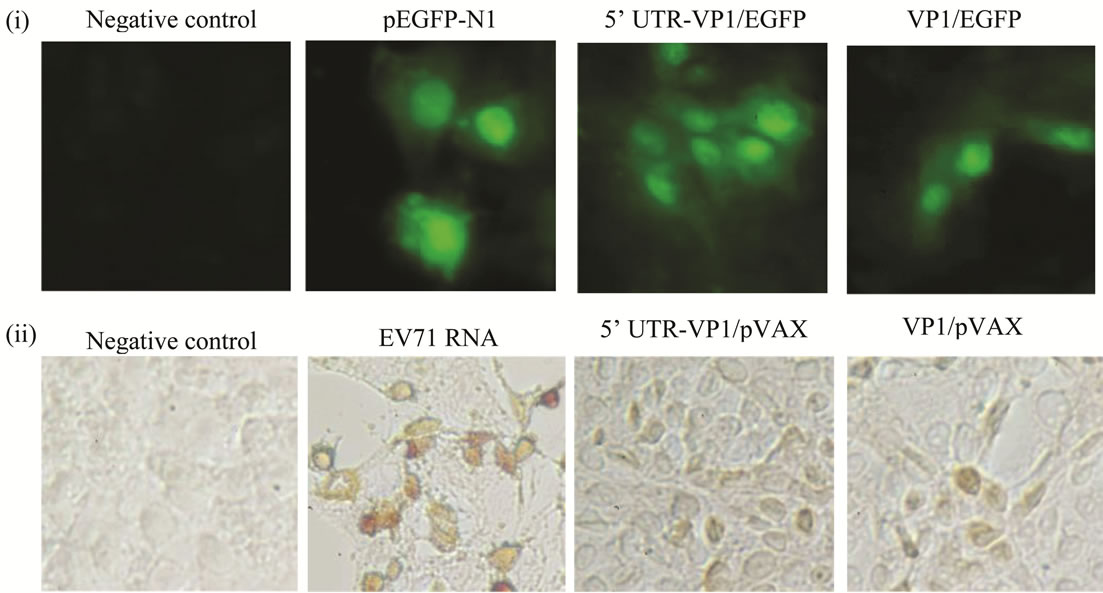
Figure 3. Expression of recombinant proteins from DNA vaccine constructs cloned into pEGFP-N1 vector and pVAX-1 expression vectors. Vero cells expressing the recombinant fusion protein from DNA vaccine constructs cloned into pEGFP-N1 (i) and pVAX-1 (ii) expression vectors. The DNA vaccine constructs (1.0 μg/ml) were transfected into Vero cells. Cells were fixed and viewed at 48 hours post-transfection. Cells transfected with pEGPF-N1 vector backbone and EV71 RNA (full length genomic RNA) were included as positive controls. Arrow indicates cells expressing the recombinant protein (in cells transfected with EV71 RNA and constructs cloned in pVAX-1, respectively).
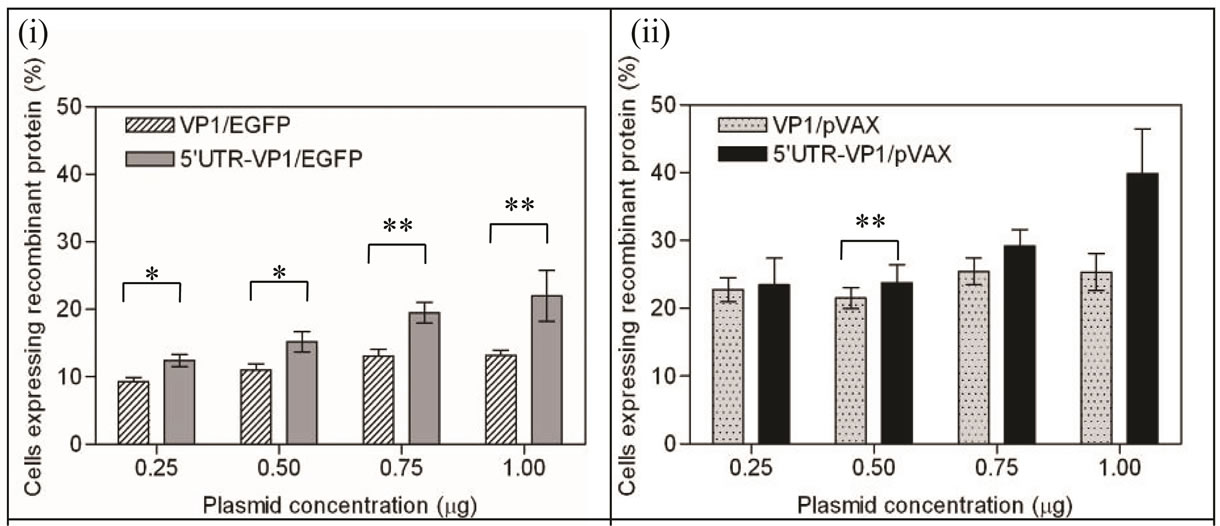
Figure 4. Percentage of cells expressing the EV71 VP1 recombinant protein. The percentage of cells expressing the recombinant protein from the DNA vaccine constructs VP1/EGPF and 5’ UTR-VP1/EGFP (i) and VP1/pVAX and 5’ UTR-VP1/ pVAX (ii) were compared. Cells were transfected with the plasmids 5’ UTR-VP1/EGFP, VP1/EGFP, 5’ UTR-VP1/pVAX and VP1/pVAX, respectively. The percentages of cells expressing the recombinant protein were calculated based on 10 different transfection experiments. On average 67 cells could be observed per microscopic field. p < 0.05 is indicated with * and p < 0.01 is indicated with **.
teins were not statistically significant (Figure 4(ii)). The percentage of cells expressing the recombinant proteins increased proportionally to the concentration of transfected plasmid in cells transfected with 5’ UTR-VP1/ EGFP (12% per μg) and 5’ UTR-VP1/pVAX (21% per μg). It is observed that the percentage of cells expressing the recombinant protein did not show linear increment over plasmid DNA concentration in those transfected with DNA constructs without the 5’UTR.
3.3. Immunogenicity of DNA Vaccine Constructs in Mice
The DNA vaccine constructs were used to immunize the BALB/c mice. The IgG responses to the EV71 VP1 in mice inoculated with the DNA vaccine constructs, the vector backbone and heat-inactivated EV71 was compared. At 2 weeks after the first immunization, the IgG levels of mice immunized with the construct 5’UTRVP1/EGFP and VP1/EGFP increased to ~27-fold (p < 0.001) and ~19-fold (p < 0.01), respectively, in comparison to the group immunized with only pEGFP-N1 (Table 2). The mice group immunized with 5’ UTR-VP1/pVAX and VP1/pVAX showed 11-fold (p < 0.05) and 19-fold (p < 0.01) increased of IgG levels, respectively, in comparison to the group immunized with only pVAX-1. At 4 week, no significant increase of IgG levels was obtained following the booster immunization in all mice group (Figure 5). In our study, we showed that the heat-inactivated EV71 stimulated less than 1-fold increase in IgG level in comparison to those immunized with the DNA vaccine constructs (p > 0.05), suggesting that the DNA vaccine is equally immunogenic.
3.4. Neutralization of EV71 by Sera from Vaccinated Mice after Challenged with Live EV71
The serum microneutralization test performed using the ELISA-based microneutralization assay showed that at day-1 PI, the neutralizing antibody (NAb) titer in group of mice inoculated with only pEGFP-N1 and pVAX-1, and the group immunized with heat-inactivated EV71 had a baseline NAb titer of <1:20. The group of mice immunized with the DNA vaccine constructs showed higher NAb up to 1:160 (Figure 6). Groups of mice immunized with VP1/EGFP and VP1/pVAX showed NAb titer of 1:40, whereas mice immunized with 5’ UTRVP1/EGFP and 5’ UTR-VP1/pVAX showed NAb titer of 1:160. At day 5 PI, the mice immunized with 5’ UTRVP1/EGFP and 5’ UTR-VP1/pVAX showed NAb titer of 1:640, ~7-fold higher than the negative control groups and the group of mice immunized with heat-inactivated EV71 (Figure 6). The NAb titer elicited in groups of mice immunized with VP1/EGFP and VP1/pVAX were
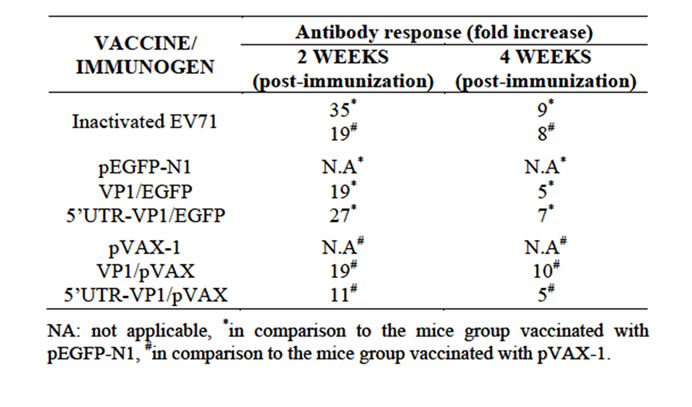
Table 2. Fold increases of the immune response in vaccinated mice were compared to the control mice group. Mice sera were collected 2 weeks after each inoculation with constructed DNA vaccine. Antibody level was determined by indirect enzyme-linked immunosorbent assay (ELISA) in 4 independent assays with triplicate for each assay.
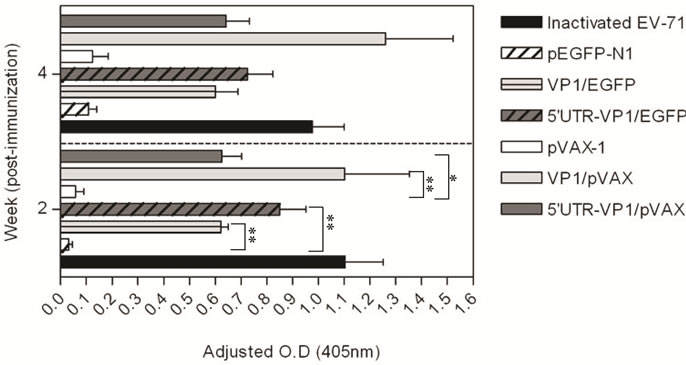
Figure 5. IgG antibody responses induced by immunization with DNA vaccine constructs. Groups of mice were immunized either with VP1/EGFP, 5’ UTR-VP1/EGFP, VP1/ pVAX or 5’ UTR-VP1/pVAX on days 0 and 14 by intramuscular injection without any adjuvant. Groups of mice immunized with pEGFP-N1, pVAX-1 or heat-inactivated EV71 were used as controls. Sera were collected after 2 weeks of each immunization. ELISA was performed using EV71-infected cell lysate as antigen. The O.D values were normalized against the O.D values of the pre-immunized sera. Arrow indicates the time of immunization and booster with DNA vaccine constructs. p < 0.05 is indicated with * and p < 0.01 is indicated with ** in comparison to the negative control groups (pEGFP-N1 or pVAX-1, respectively).
1:320 and 1:160, respectively. At day 10 PI, no significant increase in NAb were observed in mice immunized with pVAX-1 and pEGFP-N1 when compared to those immunized with the DNA vaccine constructs. The NAb titers decreased in all immunized mice groups, except in group of mice immunized with heat-inactivated EV71, which showed ~1-fold increase in the neutralizing titer. The NAb at day 10 PI were at 1:320 for mice immunized with 5’ UTR-VP1/EGFP and 5’ UTR-VP1/pVAX in comparison to the NAb titer for other groups of immunized mice with NAb titer of <1:40 (Figure 6).
4. Discussion
In the present study, we aimed to improve the EV71 VP1 DNA-based vaccine. The EV71 5’ untranslated region (UTR) containing the IRES was inserted into previously described DNA-based vaccine constructs [23]. Expression from the constructs was verified in vitro by transfection in Vero cells. Cells expressing the recombinant protein from the pEGFP-N1 vector showed diffuse fluorescence throughout the cytoplasm and nucleus suggesting successful expression of the recombinant protein. The intranuclear localization of the VP1-EGFP protein observed in this study is consistent with other earlier study, which suggests passive entry of the highly soluble protein into the nucleus [42].
In this study, the 5’ UTR-VP1 and VP1 gene regions were cloned upstream of the cytomegalovirus immediate early (CMV IE) gene promoter in both pEGFP-N1 and
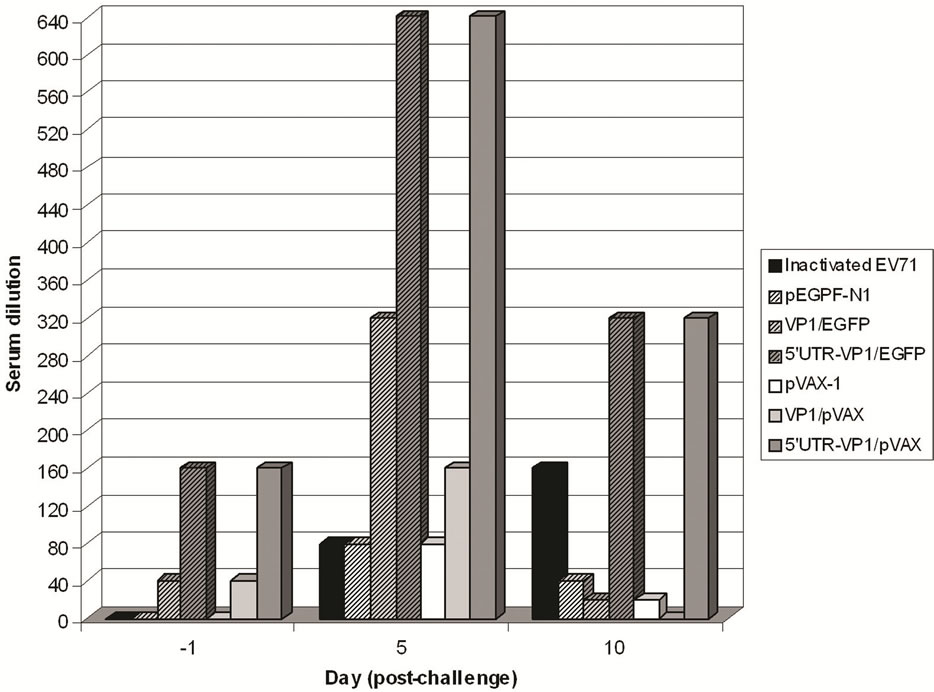
Figure 6. Neutralizing antibody titers of vaccinated mice after challenged with live EV71. Neutralizing antibody titers of mice groups immunized with the DNA vaccine constructs and heat-inactivated EV71 were evaluated at day -1, days 5 and 10 after the inoculation with live EV71.
pVAX-1 vector backbone. Transcription of the construct consisting of the 5’ UTR-VP1 gene region from the CMV IE gene promoter resulted in mRNA containing a highly structured (hairpin-like structure) 5’ region upstream of the VP1 gene region. The presence of the hairpin structure has been reported to block the cap-dependent ribosome scanning, which led to the inhibition of cap-dependant translation [43]. The IRES-mediated cap-independent translation effects may be observed in cells transfected with DNA constructs with 5’ UTR, where the percent of cells expressing the recombinant proteins showed proportionate increment over the concentration of transfected DNA constructs. Similar effect was not observed in the cells transfected with constructs without the 5’ UTR. This could be caused by the low efficiency of translation mediated through the 5’-cap due to limited eukaryotic initiation factors (eIFs) or elongation factors (EF) required for cap-dependent. In contrast, the IRESmediated cap-independent translation was not affected by the level of eIFs or EF, thus that limitation was not observed in cells transfected with constructs carrying the 5’ UTR [44,45]. Our results are in agreement with the report by Borman et al. [46] which described a linear relationship between the concentration of transfected plasmid DNA and translation efficiency in picornaviral IRESdriven expression.
In vivo evaluation showed that mice vaccinated with DNA-based vaccine construct carrying the 5’ UTR-VP1 and VP1 genes successfully elicited higher IgG level compared to the mice immunized with the vector backbone. However, the booster shot given 2 weeks after the initial immunization did not lead to further increase of the IgG levels in any of the mice group. This is consistent with previous studies that reported the second booster shot could actually lead to the decline of the IgG levels [23,47]. The decline in IgG level could be due to the shift in the immune response towards cellular immune responses [26,47]. It is noted that the immune responses profile elicited by the 5’ UTR-VP1/EGFP and 5’ UTRVP1/pVAX constructs were different, although both constructs carrying the 5’ UTR. This could be contributed by the difference in plasmid backbone used. Unlike 5’ UTRVP1/EGFP, no enhancement in the IgG level in mice immunized with 5’ UTR-VP1/pVAX at 2 weeks following the booster compared to those immunized with VP1/ pVAX. The reason for this observation is unclear, however, possible explanation could be that the 5’ UTRVP1/pVAX is better in inducing memory rather than inducing effector responses, which is reflected by high elicitation of IgG following the virus challenge (data not shown).
Elevation of antibody levels following immunization suggests successful stimulation of the host humoral immune response. However, it does not necessarily reflect the efficacy in providing protective responses [11]. The protective antibody response against enteroviruses infections is characterized by the presence of neutralizing antibodies [48]. In this study, the neutralizing antibody titer in challenged mice was measured using the ELISA-based microneutralization assay. This method is more accurate in comparison to the conventional neutralizing titer determination, since the cut-off point and the presence of neutralizing antibody titer were measured optically [41]. We noted that after challenged with live virus, groups of mice vaccinated with DNA vaccine constructs containing the 5’ UTR showed higher neutralizing antibody titer in comparison to the other mice groups, including those immunized with heat-inactivated EV71. Immunization with heat-inactivation did not lead to high elicitation of neutralizing antibody titer perhaps because no adjuvant was used in the present study. Adjuvant was not included in the vaccination regime simply because our main interest is to observe the ability of incorporated EV71 IRES within DNA-based vaccine construct in eliciting immune response. It is believed that combination of the constructed 5’ UTR-VP1 DNA vaccine with suitable adjuvant, such as ISCOMATRIX™ adjuvant using DNA prime-protein boost vaccination strategy could effectively generate protective immunity against EV71, comprising both humoral and cell-mediated immune responses [49].
It is noted that the mice immunized with heat-inactivated EV71, VP1/EGFP and VP1/pVAX showed slight increment in the neutralizing antibody titer at day 5 PI. The increase is most probably due to the presence of EV71 specific IgM elicited as a primary immune response to the challenge with live EV71, since microneutralization assay is unable to discriminate between the primary from secondary immune responses, unless the IgM was inactivated prior to the microneutralization assay. The high IgG responses in mice immunized with heat-inactivated EV71, VP1/EGFP and VP1/pVAX observed in this study were not corroborated with high neutralizing antibody titer. This could be that the immunogen possessed epitopes that elicited a broad anti-EV71 antibody response that is non-neutralizing [50,51].
In this study, the evaluation of immune responses elicited by the DNA vaccine constructs was determined by in vitro system, which may have overlooked other pathways of immune responses that could also been enhanced, such as activation of the complement cascades and antibody-dependent cytotoxicity [22]. With the availability of transgenic mouse model that demonstrate similar pathological features of EV71 infection as in human, the in vivo evaluation of protection confers by the constructed vaccine would be possible, which will give better reflection of elicited immune responses [52]. The exact mechanism of how the presence of the IRES sequence helped to improve the efficacy of the DNA vaccine is presently unknown.
In conclusion, improved efficiency of VP1 expression in the constructs with 5’ UTR was obtained. The higher efficiency of translation was mediated through a capindependent mechanism by the IRES could result in an over abundant presence of the recombinant protein per cell. This in turn could improve the presentation of specific EV71 VP1 neutralizing epitope either through presentation via the MHC molecules or direct stimulation of the B-cells, however, further subsequent studies would be required to address these possibilities.
Acknowledgements
We thank Prof. Mary Jane Cardosa, Universiti Malaysia Sarawak (UNIMAS) for providing us with the VP1 gene region of EV71 isolate S2/86/1 cloned in pCR® Blunt Vector (Zero Blunt™ PCR Cloning Kit, Invitrogen) and Prof Rozita Rosli, Universiti Putra Malaysia (UPM) for the pVAX-1 Vector (Invitrogen, USA). This project is funded in part by Ministry of Science, Technology and Innovation, Malaysia 06-02-09-001 BTK/TD/002 and Geran Sanjungan Penyelidikan (GSP) MO 006-2013.
REFERENCES
- J. Qiu, “Enterovirus 71 Infection: A New Threat of Global Public Health?” The Lancet Neurology, Vol. 7, No. 10, 2008, pp. 868-869. http://dx.doi.org/10.1016/S1474-4422(08)70207-2
- S. AbuBakar, H. Y. Chee, N. Shafee, K. B. Chua and S. K. Lam, “Molecular Detection of Enteroviruses from an Outbreak of Hand, Foot and Mouth Disease in Malaysia in 1997,” Scandinavian Journal of Infectious Diseases, Vol. 31, No. 4, 1999, pp. 331-335. http://dx.doi.org/10.1080/00365549950163734
- P. C. McMinn, “An Overview of the Evolution of Enterovirus 71 and Its Clinical and Public Health Significance,” FEMS Microbiology Reviews, Vol. 26, No. 1, 2002, pp. 91-107. http://dx.doi.org/10.1111/j.1574-6976.2002.tb00601.x
- Y. T. Lin, L. Y. Chang, S. H. Hsia, Y. C. Huang, C. H. Chiu, C. Hsueh, S. R. Shih, C. C Liu and M. H. Wu, “The 1998 Enterovirus 71 Outbreak in Taiwan: Pathogenesis and Management,” Clinical Infectious Diseases, Vol. 34, Suppl. 2, 2002, pp. S52-S57. http://dx.doi.org/10.1086/338819
- L. J. Herrero, C. S. Lee, R. J. Hurrelbrink, B. H. Chua, K. B. Chua and P. C. McMinn, “Molecular Epidemiology of Enterovirus 71 in Peninsular Malaysia, 1997-2000,” Archives of Virology, Vol. 148, No. 7, 2003, pp. 1369-1385. http://dx.doi.org/10.1007/s00705-003-0100-2
- H. F. Chen, M. H. Chan, B. L. Chiang and T. S Jeng, “Oral Immunization of Mice Using Transgenic Tomato Fruit Expressing VP1 Protein From Enterovirus 71,” Vaccine, Vol. 24, No. 15, 2006, pp. 2944-2951. http://dx.doi.org/10.1016/j.vaccine.2005.12.047
- S. AbuBakar, I. C. Sam, J. Mohd Yusof, M. K. Lim, S. Misbah, N. A. MatRahim and P. S. Hooi, “Enterovirus 71 Outbreak, Brunei,” Emerging Infectious Disease, Vol. 15, No. 1, 2009, pp. 79-82. http://dx.doi.org/10.3201/eid1501.080264
- P. C. McMinn, “Recent Advances in The Molecular Epidemiology and Control of Human Enterovirus 71 Infection,” Current Opinion in Virology, Vol. 2, No. 2, 2012, pp. 199-205. http://dx.doi.org/10.1016/j.coviro.2012.02.009
- M. A. Pallansch and M. S. Oberste, “Enterovirus 71 Encephalitis: A New Vaccine on the Horizon?” The Lancet, Vol. 381, No. 9871, 2013, pp. 976-977. http://dx.doi.org/10.1016/S0140-6736(13)60286-X
- C. H. Chiu, C. Chu, C. C. He and T. Y. Lin, “Protection of Neonatal Mice From Lethal Enterovirus 71 Infection by Maternal Immunization With Attenuated Salmonella Enterica Serovar Typhimurium Expressing VP1 of Enterovirus 71,” Microbes and Infections, Vol. 8, No. 7, 2006, pp. 1671-1678. http://dx.doi.org/10.1016/j.micinf.2006.01.021
- D. G. W. Foo, S. Alonso, V. T. K. Chow and C. L. Poh, “Passive Protection Against Lethal Enterovirus 71 Infection in Newborn Mice by Neutralizing Antibodies Elicited by a Synthetic Peptide,” Microbes and Infections, Vol. 9, No.11, 2007, pp. 1299-1306. http://dx.doi.org/10.1016/j.micinf.2007.06.002
- X. Li, C. Mao, S. Ma, X. Wang, Z. Sun, Y. Yi, M. Guo, X. Shen, L. Sun and S. Bi, “Generation of Neutralizing Monoclonal Antibodies Against Enterovirus 71 Using Synthetic Peptides,” Biochemical and Biophysical Research Communication, Vol. 390, No. 4, 2009, pp. 1126- 1128. http://dx.doi.org/10.1016/j.bbrc.2009.09.103
- X. Ye, Z. Ku, Q. Liu, X. Wang, J. Shi, Y. Zhang, L. Kong, Y. Cong and Z. Huang, “Chimeric Virus-Like Particle Vaccines Displaying Conserved Enterovirus 71 Epitopes Elicit Protective Neutralizing Antibodies in Mice Through Divergent Mechanisms,” Journal of Virology, Vol. 88, No. 1, 2014, pp. 72-81. http://dx.doi.org/10.1128/JVI.01848-13
- H. Y. Li, J. F. Han, C. F. Qin and R. Chen, “Virus-Like Particles for Enterovirus 71 Produced From Saccharomyces cerevisiae Potently Elicits Protective Immune Responses in Mice,” Vaccine, Vol. 31, No. 32, 2013, pp. 3281-3287. http://dx.doi.org/10.1016/j.vaccine.2013.05.019
- C. W. Chen, Y. P. Lee, W. F. Wang and C. K. Yu, “Formaldehyde-Inactivated Human Enterovirus 71 Vaccine is Compatible for Co-Immunization With a Commercial Pentavalent Vaccine,” Vaccine, Vol. 29, No. 15, 2011, pp. 2772-2776. http://dx.doi.org/10.1016/j.vaccine.2011.01.094
- A. Cheng, C. P. Fung, C. C. Liu, Y. T. Lin, H. Y. Tsai, S. C. Chang, A. H. Chou, J. Y. Chang, R. H. Jiang, Y. C. Hsieh, I. J. Su, P. C. H. Chong and S. M. Hsieh, “A Phase I, Randomized, Open-Label Study to Evaluate The Safety and Immunogenicity of an Enterovirus 71 Vaccine,” Vaccine, Vol. 31, No. 20, 2013, pp. 2471-2476. http://dx.doi.org/10.1016/j.vaccine.2013.03.015
- R. W. Ellis, “Technologies for the Design, Discovery, Formulation and Administration of Vaccines,” Vaccine, Vol. 19, No.17-19, 2001, pp. 2681-2687. http://dx.doi.org/10.1016/S0264-410X(00)00504-1
- H. Shimizu, B. Thorley, F. J. Paladin, K. A. Brussen, V. Stambos, L. Yuen, A. Utama, Y. Tano, M. Arita, H. Yoshida, T. Yoneyama, A. Benegas, S. Roesel, M. Pallansch, O. Kew and T. Miyamura, “Circulation of Type 1 Vaccine-Derived Poliovirus in the Philippines in 2001,” Journal of Virology, Vol. 78, No. 24, 2004, pp. 13512-13521. http://dx.doi.org/10.1128/JVI.78.24.13512-13521.2004
- O. M. Kew, R. W. Sutter, B. M. de Gourville, W. R. Dowdle and M. A. Pallansch, “Vaccine Derived PolioViruses and the End Game Strategy for Global Polio Eradication,” Annual Review of Microbiology, Vol. 59, 2005, pp. 587-635. http://dx.doi.org/10.1146/annurev.micro.58.030603.123625
- S. Mamishi, S. Shahmahmoudi, H. Tabatabaie, S. Teimourian, B. Pourakbari, Y. Gherisari, M. Yeganeh, A. Salavati, A. R. Esteghamati, M. M. Gooya, R. Nategh and N. Parvaneh, “Novel BTK Mutation Presenting With Vaccine-Associated Paralytic Poliomyelitis,” European Journal of Pediatrics, Vol. 167, No. 11, 2008, pp. 1335-1338. http://dx.doi.org/10.1007/s00431-008-0674-5
- M. Giese, “DNA-Antiviral Vaccines: New Developments and Approaches—A Review,” Virus Genes, Vol. 17, No. 3, 1998, pp. 219-232. http://dx.doi.org/10.1023/A:1008013720032
- C. N. Wu, Y. C. Lin, C. Fann, N. S. Liao, S. R. Shih and M. S. Ho, “Protection Against Lethal Enterovirus 71 Infection in Newborn Mice By Passive Immunization With Subunit VP1 Vaccines and Inactivated Virus,” Vaccine, Vol. 20, No. 5-6, 2001, pp. 895-904. http://dx.doi.org/10.1016/S0264-410X(01)00385-1
- S. T. Wong, S. AbuBakar, Z. Sekawi and R. Rosli, “DNA Vaccine Constructs against Enterovirus 71 Elicit Immune Response in Mice,” Genetic Vaccines and Therapy, 2007, Vol. 5, p. 6. http://dx.doi.org/10.1186/1479-0556-5-6
- W. W. Leitner, H. Ying and N. P. Restifo, “DNA and RNA-Based Vaccines: Principles, Progress and Prospects,” Vaccine, Vol. 18, No. 9-10, 1999, pp. 765-777. http://dx.doi.org/10.1016/S0264-410X(99)00271-6
- S. Manoj, L. A. Babiuk and S. D. Littel-van den Hurk, “Approaches to Enhance the Efficacy of DNA Vaccines,” Critical Reviews in Clinical Laboratory Sciences, Vol. 41, No. 1, 2004, pp.1-39. http://dx.doi.org/10.1080/10408360490269251
- J. Běláková, M. Horynová, M. Křupka, E. Weigl and M. Raška, “DNA Vaccines: Are They Still Just a Powerful Tool For The Future?” Archivum Immunologiae et Therapia Experimentalis, Vol. 55, No. 6, 2007, pp. 387-398. http://dx.doi.org/10.1007/s00005-007-0044-4
- N. Y. Sardesai and D. B. Weiner, “Electroporation Delivery of DNA Vaccines: Prospects for Success,” Current Opinion in Immunology, Vol. 23, No. 3, 2011, pp. 421- 429. http://dx.doi.org/10.1016/j.coi.2011.03.008
- K. Oosterhuis, J. H. van den Berg, T. N. Schumacher and J. B. Haanen, “DNA Vaccines and Intradermal Vaccination By DNA Tattooing,” Current Topic in Microbiology and Immunology, Vol 351, 2012, pp. 221-250. http://dx.doi.org/10.1007/82_2010_117
- S. A. Kalams, S. D. Parker, M. Elizaga, B. Metch, S. Edupuganti, J. Hural, S. De Rosa, D. K. Carer, K. Rybczyk, I. Frank, J. Fuchs, B. Koblin, D. H. Kim, P. Joseph, M. C. Keefer, L. R. Baden, J. Eldridge, J. Boyer, A. Sherwat, M. Cardinali, M. Allen, M. Pensiero, C. Butler, A. S. Khan, J. Yan, N. Y. Sardesai, J. G. Kublin, D. B. Weiner and the NIAID HIV Vaccine Trials Network, “Safety and Comparative Immunogenicity of an HIV-1 DNA Vaccine in Combination With Plasmid Interleukin 12 and Impact of Intramuscular Electroporation For Delivery,” The Journal of Infectious Diseases, Vol. 208, No. 5, 2013, pp. 818-829. http://dx.doi.org/10.1093/infdis/jit236
- E. J. Yager, C. Stagnar, R. Gopalakrishnan, J. T. Fuller and D. H. Full, “Optimizing Particle-Mediated Epidermal Delivery of an Influenza DNA Vaccine in Ferrets,” In: S. Sudowe and A. B. Reske-Kunz, Eds., Biolistic DNA Delivery: Methods and Protocols, Methods in Molecular Biology, Springer Science+Business Media, Vol. 940, 2013, pp. 223-237. http://dx.doi.org/10.1007/978-1-62703-110-3_19
- V. Douin, S. Bornes, L. Creancier, P. Rochaix, G. Favre, A. C. Prats and B. Couderc, “Use and Comparison of Different Internal Ribosomal Entry Sites (IRES) in Tricistronic Retroviral Vectors,” BMC Biotechnology, 2004, Vol. 4, p. 16. http://dx.doi.org/10.1186/1472-6750-4-16
- J. C. Lee, T. Y. Wu, C. F. Huang, F. M. Yang, S. R. Shih and J. T. A. Hsu, “High-Efficiency Protein Expression Mediated By Enterovirus 71 Internal Ribosome Entry Site,” Biotechnology and Bioengineering, Vol. 90, No. 5, 2005, pp. 656-662. http://dx.doi.org/10.1002/bit.20440
- C. Jünemann, Y. Song, G. Bassili, D. Goergen, J. Henke and M. Niepmann, “Picornavirus Internal Ribosome Entry Site Elements Can Stimulate Translation of Upstream Genes,” The Journal of Biological Chemistry, Vol. 282, No. 1, 2007, pp. 132-141. http://dx.doi.org/10.1074/jbc.M608750200
- J. Attal, M. C. Theron and L. M. Houdebine, “The Optimal Use of IRES (Internal Ribosome Entry Site) in Expression Vectors,” Genetic Analysis: biomolecular engineering, Vol. 15, No. 3-5, 1999, pp. 161-165.
- L. K. Johansen and C. D. Morrow, “Inherent Instability of Poliovirus Genomes Containing Two Internal Ribosome Entry Site (IRES) Elements Supports a Role For the IRES in Encapsidation,” Journal of Virology, Vol.74, No. 18, 2000, pp. 8335-8342. http://dx.doi.org/10.1128/JVI.74.18.8335-8342.2000
- E. Martínez-Salas, R. Ramos, E. Lafuente and S. López de Quinto, “Functional Interactions in Internal Translation Initiation Directed by Viral and Cellular IRES Element,” The Journal of General Virology, Vol. 82, No. 5, 2001, pp. 973-984.
- E. Martínez-Salas, S. López de Quinto, R. Ramos and O. Fernández-Miragall, “IRES Elements: Features of The RNA Structure Contributing to Their Activity,” Biochimie, Vol. 84, No. 8, 2002, pp. 755-763. http://dx.doi.org/10.1016/S0300-9084(02)01408-6
- E. Y. Dobrikova, P. Flores and M. Gromeier, “Structural Determinants of Insert Retention of Poliovirus Expression Vectors with Recombinant IRES Elements,” Virology, Vol. 311, No. 2, 2003, pp. 241-253.
- K. Choi, J. H. Kim, X. Li, K. Y. Peak, S. H. Ha, S. H. Ryu, E. Wimmer and S. K. Jang, “Identification of Cellular Proteins Enhancing Activities of Internal Ribosomal Enty Sites by Competition With Oligodeoxynucleotides,” Nucleic Acids Research, Vol. 32, No. 4, 2004, pp. 1308-1317. http://dx.doi.org/10.1093/nar/gkh300
- N. Shafee and S. AbuBakar, “Immunization With DNA Vectors Consisting of Selected Dengue 2 Virus Genes Stimulated Antibody Responses in Mice,” International Journal of Virology, Vol. 2, No. 1, 2006, pp. 14-20. http://dx.doi.org/10.3923/ijv.2006.14.20
- V. Vorndam and M. Beltran, “Enzyme-Linked Immunosorbent Assay-Format Microneutralization Test for Dengue Viruses,” The American Journal of Tropical Medicine Hygiene, Vol. 66, No. 2, 2002, pp. 208-212.
- A. Jaegly, F. Mouthon, J. M. Peyrin, B. Camugli, J. P. Deslys and D. Dormont, “Search For a Nuclear Localization Signal in the Prion Protein,” Molecular and Cellular Neuroscience, Vol. 11, No. 3, 1998, pp. 127-133. http://dx.doi.org/10.1006/mcne.1998.0675
- A. B. Sachs, “Cell Cycle-Dependent Translation Initiation: IRES Elements Prevail,” Cell, Vol. 101, No. 3, 2000, pp. 243-245. http://dx.doi.org/10.1016/S0092-8674(00)80834-X
- M. J. Gale, S. L. Tan and M. G. Katze, “Translational Control of Viral Gene Expression in Eukaryotes,” Microbiology and Molecular Biology Review, Vol. 64, No. 2, 2000, pp. 239-280. http://dx.doi.org/10.1128/MMBR.64.2.239-280.2000
- K. M. Kean, “The Role of mRNA 5’-Noncoding and 3’- End Sequences on 40S Ribosomal Subunit Recruitment, and How RNA Viruses Successfully Compete With Cellular mRNAs To Ensure Their Own Protein Synthesis,” Biology of the Cell, Vol. 95, No. 3-4, 2003, pp. 129-139. http://dx.doi.org/10.1016/S0248-4900(03)00030-3
- A. M. Borman, P. L. Mercier, M. Girard and M. Kean, “Comparison of Picornaviral IRES-Driven Internal Initiation of Translation in Cultured Cells of Different Origins,” Nucleic Acids Research, Vol. 25, No. 5, 1997, pp. 925-932. http://dx.doi.org/10.1093/nar/25.5.925
- A. Tanghe, O. Denis, B. Lamvrecht, V. Motte, T. van de Berg and K. Huygen, “Tuberculosis DNA Vaccine Encoding Ag85A Is Immunogenic and Protective When Administered By Intramuscular Needle Injection But Not by Epidermal Gene Gun Bombardment,” Infection and Immunity, Vol. 68, No. 7, 2000, pp. 3854-3860. http://dx.doi.org/10.1128/IAI.68.7.3854-3860.2000
- C. K. Yu, C. C. Chen, C. L. Chen, J. R. Wang, C. C. Liu, J. J. Yan and I. J. Su, “Neutralizing Antibody Provided Protection Against Enterovirus Type 71 Lethal Challenge in Neonatal Mice,” Journal of Biomedical Science, Vol. 7, No. 6, 2000, pp. 523-528. http://dx.doi.org/10.1007/BF02253368
- R. Buglione-Corbett, K. Pouliot, R. Marty-Roix, K. West, S. Wang, E. Lien and S. Lu, “Serum Cytokine Profiles Associated With Specific Adjuvants Used in a DNA Prime-Protein Boost Vaccination Strategy,” PLoS ONE, Vol. 8, No. 9, 2013, Article ID: e74820. http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0074820 http://dx.doi.org/10.1371/journal.pone.0074820
- P. Muir, U. Kammerer, K. Korn, M. N. Mulders, T. Poyry, B. Weissbrich, R. Kandolf, G. M. Cleator and A. M. van Loon, “Molecular Typing of Enteroviruses: Current Status and Future Requirements,” Clinical Microbiology Reviews, Vol. 11, No. 1, 1998, pp. 202-227.
- Y. C. Chung, M. S. Ho, J. C. Wu, W. J. Chen, J. H. Huang, S. T. Chou and Y. H. Hu, “Immunization with Virus-Like Particles of Enterovirus 71 Elicits Potent Immune Responses and Protects Mice Against Lethal Challenge,” Vaccine, Vol. 26, No. 15, 2008, pp. 1855-1862. http://dx.doi.org/10.1016/j.vaccine.2008.01.058
- K. Fujii, N. Nagata, Y. Sato, K. C. Ong, K. T. Wong, S. Yamayoshi, M. Shimanuki, H. Shitara, C. Taya and S. Koike, “Transgenic Mouse Model for The Study of Enterovirus 71 Neuropathogenesis,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 110, No. 36, 2013, pp. 14753-14758. http://dx.doi.org/10.1073/pnas.1217563110

