Advances in Breast Cancer Research
Vol. 2 No. 3 (2013) , Article ID: 33818 , 8 pages DOI:10.4236/abcr.2013.23014
Downregulation of Telomerase Activity in Breast Cancer Impairs Cells Proliferation, Invasive Ability and Sensitizes Cells to Ultraviolet-Radiation and Adriamycin-Chemotherapy in Vitro*
1Division of Plastic and Reconstructive Surgery, First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China 2Division of Breast Surgery, First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China
3Department of Anaesthesiology, First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China
4Division of Vasular, Thyroid and Hernia, Guangdong General Hospital, Guangzhou, China
Email: *shenmingwang@vip.sohu.com
Copyright © 2013 Xiangxia Liu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received April 5, 2013; revised May 7, 2013; accepted May 15, 2013
Keywords: breast cancer; telomerase; rna interference; radiation; chemotherapy
ABSTRACT
Backgroud: Telomerase activity, mainly regulated by the human telomerase reverse transcriptase (hTERT) gene, plays critical roles in tumor growth and progression through the maintenance of telomere length and structure. Telomerase is elevated in most malignant tumors as well as in breast cancer, the ubiquitous expression of telomerase makes it a promising target for cancer therapy. With the goal of down regulating telomerase activity, RNA interference technology has been applied to specifically knockdown the hTERT gene expression in breast cancer cell line MCF-7 and MDA-MB- 231 and determine whether h TERT-specific RNA interference technology serve as an effective method of telomerase-based cancer therapy. Methods: Interfering p Super-retro-puro-hTERT-RNA and the control were transfected into breast cancer cell line MCF-7 and MDA-MB-231. The telomerase activity, cell proliferation, invasive ability and cell response to ultraviolet-radiation or adriamycin-chemotherapy in vitro were recorded in transfected, untransfeced and empty-transfected cells respectively. Results: Telomerase activity was successfully suppressed in transfected cells (P < 0.005). Decreased expression of telomerase activity was associated with reduced cell proliferation (P < 0.001), migration and invasive ability (P < 0.001) and enhanced sensitivity to ultraviolet-radiation or adriamycin-chemotherapy (P < 0.001). Conclusions: Telomerase activity down regulation inhibits breast cancer cell growth, impairs cell migration, invasion and sensitizes cancer cells to radiotherapy and chemotherapy. The hTERT-specific RNA interference technology combined with radio-therapy and/or chemotherapy may serve as an effective method of telomerase-based therapy in breast cancer.
1. Introduction
Breast cancer is the most common diagnosed malignant tumor in women and by far the second most frequent cancer in the world [1,2]. In the past nearly fifty years, the treatment of breast cancer has advanced greatly in surgery as well as in chemotherapy, radiotherapy and endocrine therapy. But there are still a substantial number of breast cancer patients die of recurrence and metastasis despite adequate surgical intervention and adjuvant therapy [3]. The primary function of telomere, mainly regulated by telomerase, is to maintain the stabilization of chromosome [4,5]. Progressive shortening of telomere with cell division causes chromosomal instability and cell senescence [6]. Telomerase consists of two functional components, the telomerase reverse transcriptase (TERT, h TERT in human) and telomerase RNA template. Telomerase activity is activated in most malignant tumors including breast cancer [7]. Telomerase activity correlates with tumor aggressiveness and reflects therapy effect in breast cancer [8] and is believed as a prognostic marker in breast cancer [9]. The ubiquitous expression of telomerase makes it a promising target for cancer therapy [10].
With the goal of investigating the role of telomerase activity in breast cancer progression, RNA interference technology was applied to specifically inhibit the expression of telomerase in breast cancer cell line MCF-7 and MDA-MB-231 which express high level of telomerase activity.
2. Methods
2.1. Cell Lines and Culture
Breast cancer cell line MCF-7 and MDA-MB-231 were kindly provided by Professor Li Meng-feng (Sun Yat-sen University, China), and cultured with Dulbecco’s modified Eagle’s medium (DMEM, high glucose, Invitrogen, USA) supplemented with 10% fetal bovine serum (HyClone, USA) and 1% penicillin (100 IU/ml)/streptomycin (100 µg/ml) (complete media). Cells were cultured in incubator at 37˚C, 5% CO2, and 95% humidity.
2.2. Plasmids Construction and Transfection
To establish stable cell lines, we used the pSUPER-retropuro plasmids obtained from Professor Li Mengfeng’s laboratory and the hTERT-targeted shRNA sequence were as follows: no.1, sense: 5’-gatcccc TTT CATCAGCA AGTTTGGAttcaaggaTCCAAACTTGCTGATGAAAtttt ta-3’ and anti-sense:’-agctta aaaaTTTCATCAGCAAGT TTGGAtctcttgaa TCAAACTTGCTGATG-AAAggg; no. 2, sense: 5’-gatccccAACCTTCCTCAGGACCCTGttcaa gagaCAGGGTCCTGAGAAGGTT ttttta-3’ and antisense: 5’-agcttaaaaaAACCTTCCTCAGGACCCTGtctct tgaaCAGGGTCCTGAGGAAGGTTggg-3’, the sequence were designed as previously described [11,12] and synthesized through Invitrogen. The vectors were reconstructed according to the protocols of Psu-PER-retropuro RNAi system (Oligo Engine, Seattle, USA) and recombinant retroviral vectors were produced by transient cotransfection as described previously [13]. Viral infections were done serially, and stable cell lines expression hTERT RNAi (s) were selected for 14 days with 0.5 mg/ml of puromycin 72 hr after transfection to establish a stable cell line. The sequence pSuper-retropuro after cloning was verified by enzymatic digestion and DNA sequencing.
2.3. Semi-Quantitativereverse-Transcription Polymerase Chain Reaction (RT-PCR)
The expression of hTERT mRNA was semi-quantitatively evaluated by RT-PCR. Briefly, total RNA from cells was isolated using Trizol® reagent (Invitrogen, USA) according to the manufacturer’s instruction. Total RNA (2 µg) was used as the template in reverse-transcription using MMLV reverse transcriptase (Promega, USA) at 42˚C for 50 min and inactiviated at 95˚C for 10 min, 2 µl of RT reaction mixture and 10 µl Mix were mixed and amplified with PCR, The primer for hTERT were as follows: Forward 5’-ACCAAGCATTCCTGCTCAAGCT G-3’ and Reverse 5’-CGGCAGGTGTGCTGGACACT C-3’. The condition for PCR amplification of hTERT cNDA were denaturation at 94˚C for 10 min, followed by 35 cycles of denaturation at 94˚C for 30 sec, annealing at 68˚C for 45 sec and extension at 72˚C for 45 sec, and the last extension step was 72˚C for 10 min. 13 Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified as an internal control, the primers for GAPDH were as follows: Forward 5’-AATCCCATCACCATCTTCC A-3’ and Reverse 5’-CCTGCTTCACCACCTTCTTG-3’. Applying 10 min at 94˚C, 35 cycles of 30 sec at 94˚C, 30 sec at 56˚C and 30 sec at 72˚C for PCR amplication and the last extension step was 72˚C for 10 min [14]. The amplified products were electrophoresed on a 2% agarose gel and stained with ethidium bromide. The relative band intensities were quantified applying Quality ONE® and normalized to GAPDH in the same cDNA sample.
2.4. Analysis of Telomerase Activity
The telomeric repeat amplification protocol (TRAP) was performed with Telo TAGGG Telomerase PCR ELISA Kit (Roche, Germany, Cat No.11854666910). Samples were extracted following the standard protocol. Briefly, cell samples containing 2 × 105 cells were transferred into a fresh Eppendorf tube and centrifuge at 3000 × g for 5 min at 4˚C, resuspended the cells in PBS and after repeated the centrifugation step, the pelleted cells were re-suspended in 200 µl Lysis reagent and centrifuged at 16000 × g for 20 min at 4˚C, then the supernatant were transferred to a fresh tube to perform the following TRAP reaction. Quantification of telomerase activity was performed as described in the previously published protocol [15,16]. All measurements were performed in trip-licates applying Synergy Multi-Detection Microplate Reader (BioTek, USA). Telomerase activity was normalized against protein concentrations of each sample.
2.5. In Vitro Proliferation Assays with MTT Assay
To evaluate cell viability, we used the colorimetric [3-(4, 5-dimethylthiazol-2-yle) 2, 5-diphenyltetrazolium bromide] (MTT) assay. To perform MTT assay, the culture medium was removed and 20 µl of MTT solution (5 mg/ ml; Sigma Chemical Co. USA) were added to each well and incubated for 4 hours. Then the plates were centrifuged at 450 × g for 5 min and the medium containing MTT solution was removed, 150 µl of 100% d-i methyl sulfoxide (DMSO, Sigma Chemical Co, USA) were added and the plates were shaken at 100 rpm/min for 10 min on a plate shaker. The optical density (OD) was determined at a wavelength of 490 nm.
2.6. Soft Agar Colony Formation
Cells trypsined and re-suspended in 0.33% Noble agar (BD, France) containing DMEM supplemented with 20% FBS were overlaid on the top of 0.66% base agar in DMEM supplemented with 10% FBS, at cell concentration of 1 × 104 per well of a six-well plate, and incubated for 2 weeks. Colonies containing more than 50 cells were taken as positive colonies. Numbers of positive colonies were photographed and quantified.
2.7. Wound-Healing and Transwell Migration Assays
For wound-healing migration assay, confluent monolayer cells were serum-free starved for 24 hr and washed with phosphate-buffered saline, a scrape in the form of a cross was made with a P-200 pipette tip, then the medium was replaced with DMEM supplemented with 5% FBS, wounded areas were marked for observation and photographed at indicated time after the scratch. Invasion assay were performed using 24-well polycarbonate filter (12- μm pore size) Transwell (Corning-Costar, USA) coated with matrigel (BD Pharmingen, USA), the upper sides of the membranes were pre-coated with Matrigel matrix, the coated insert was placed in each well filled with 600 µl of complete medium, the upper chambers contained a 200 µl suspension of cells (1 × 105 cell/ml, in DMEM), the cells were incubated for 24 hr at 37˚C to allow invasion of the cells to the underside of the precoated filter. Non-invading cells on the upper surface were removed with a cotton swab. The filter were fixed, mounted and stained. The cells invaded through Matrigel were counted. Three invasion chambers were used for each experimental condition.
2.8. Radiosensitivity and Chemosensitivity of Cell Lines by MTT Hybrid Assay
Cells (1 × 104) were seeded in a 96-well micro-titer plate (Corning-Costar, USA) in a total volume of 200 µl per well and cultured for 18 hr to perform the following experiments of irradiation or chemotherapy. Irradiations of UV were performed at a dose of 40 J/cm2 (UVP UV crosslink, USA), Adriamycin were added at a concentration of 1.0 mmol/L. After irradiation or chemotherapy, the plates were subsequently cultured for various periods (24 hr, 48 hr and 72 hr). Then the MTT assay was per formed as previously described, the radiosensitivity or chemosensitivity curve was drawn according to the OD value of each well.
2.9. Statistical Analysis
The one-way ANOVA test was used to compare difference among hTERT-shRNA transfected groups, empty transfected group and the untransfected group (data from untransfected groups were designated as “control”). Differences were considered to be statistically significant if P-value < 0.05.
3. Results
3.1. Characterization of HTERT-ShRNA Stable Transfectant
Stable transfectant cell lines were obtained by transfecting MCF-7 and MDA-MB-231 cells with pSuper-retropuro-hTERT-RNAi#1 (RNAi#1), pSuper-retro-puro-hTERT-RNAi#2 (RNAi#2) (transfected group) and pSuperretro-puro (empty-transfected group, Vector). Positive clones were confirmed by EcoRII and HindIII bi-enzyme digestion, which showed a 280 bp fragment, and DNA sequencing showed correct insertion site and sequence. After 2 weeks of puromycin selection, the stable transfected cell lines were established and expanded individually.
3.2. Down Regulation of HTEET Gene Inhibits HTERT MRNA Expression and Telomerase Activity
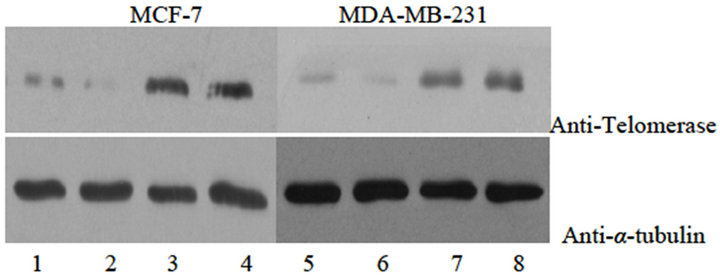
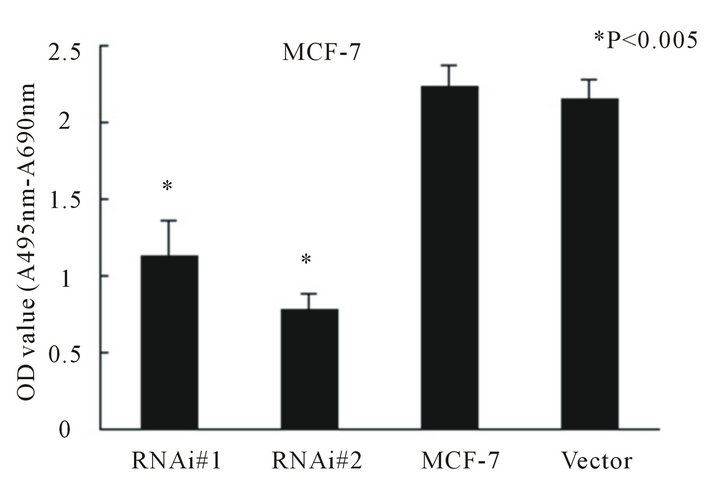
In the present study, the levels of hTERT mRNA expres sion and telomerase activity were quantitatively assessed by employing RT-PCR. As shown in Figure 1, there was a considerable reduction of hTERT mRNA in both transfected MCF-7 and MDA-MB-231 cell line compared with untransfected (MCF-7 or MDA-MB-231 cell line) or empty-transfected cells (P < 0.001). Reduction of hTERT mRNA resulted in a significant decrease in telomerase activity assessed by TRAP in both MCF-7 and MDA-MB-231 cell line (Figures 2(a) and (b), P < 0.005).
3.3. Telomerase Activity Suppression Inhibits Cell Proliferation and Clone Formation
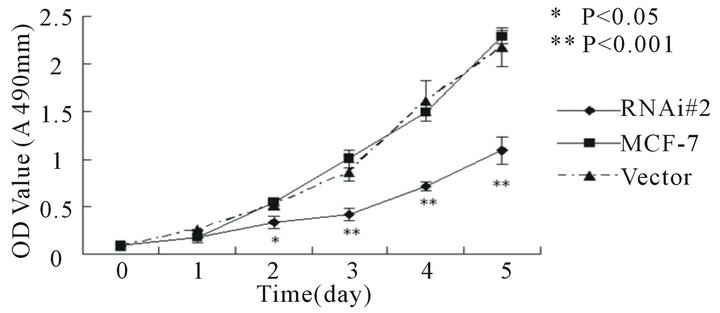
To determine the effect of telomerase activity on cellproliferation and clone formation we measured the growth rate of MCF-7 and MDA-MB-231 and their transfected stable cell lines by MTT assay and soft agar clone formation experiment respectively. In both cell lines, the hTERTRNAi-transfected stable cell lines proliferate slowly (Figures 3(a) and (b)) and form small and less clones (Figures 3(c) and (d)) compared with untransfected or empty-transfected cell lines, this indicated that inhibition of telomerase activity suppress cell proliferation and clone formation.
3.4. Telomerase Activity Suppression Inhibit Breast Cancer Cell Migration and Invasion
In this study, we use wound healing experiment and matrigel Transwell experiment to determine the ability of MCF-7 cells to migrate and MDA-MB-231 cells to in-
 (a)
(a)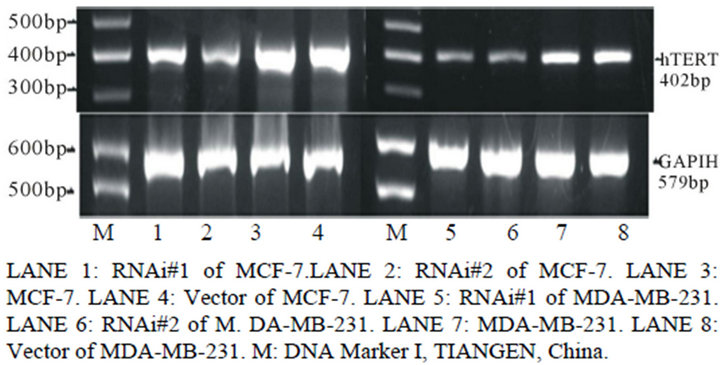 (b)
(b) (c)
(c)
Figure 1. Expression analysis of Telomerase protein and mRNA in different groups by Western blotting (a), reverse transcription-PCR (b) and relative band intensity (c). A, expression of telomerase protein in breast cancer cell line MCF-7 and MDA-MB-231 stably transducted with retrovirus vectors expressing hTERT RNAi(s) (hTERT RNAi#1 and hTERT RNAi#2, respectively) or with control vector virus (Vector). B, expression of hTERT mRNA in different groups of breast cancer cell line MCF-7 and MDA-MB-231. C, the relative band intensity read by Quantity One®, normalized for group of untransfected cell line MCF-7 and MDA-MB-231. Columns mean from three parallel experiments; bars, SD. *P < 0.001 (one-way ANOVA, Bonferroni).
 (a)
(a) (b)
(b)
Figure 2. Telomerase activity determined by Telomerase Repeat Amplification Protocol (TRAP) in breast cancer cell line MCF-7 (A) and MDA-MB-231 (B). Columns mean from three parallel experiments; bars, SD. *P < 0.005 (oneway ANOVA, Bonferroni).
vade through matrigel respectively. After 24 hr culture, the gap between the scrap in the group of htert-RNAitransfected is much larger than the group of untransfected (see Figure 4(a)), which indicates that suppression of telomerase activity does inhibit breast cancer cell migration. As shown in Figures 4(b) and (c), the effective suppression of telomerase in MDA-MB-231 cells resulted in a significant reduction in the ability of invasion (P < 0.001). This implies that suppression of telomerase can inhibit breast cancer cell invasion.
3.5. Telomerase Activity Suppression Sensitizes Cancer Cells to Radiotherapy and Chemotherapy
We supposed that telomerase-based therapy combined with other adjuvant therapies such as radiotherapy or chemotherapy might enhance the effects of tumor suppression. So we treated each cell line with increasing doses of
 (a)
(a)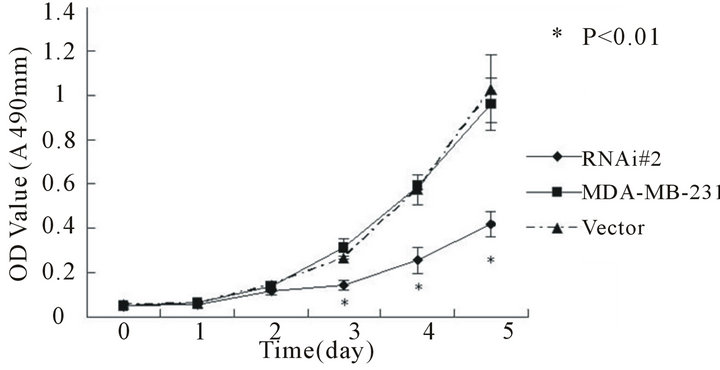 (b)
(b)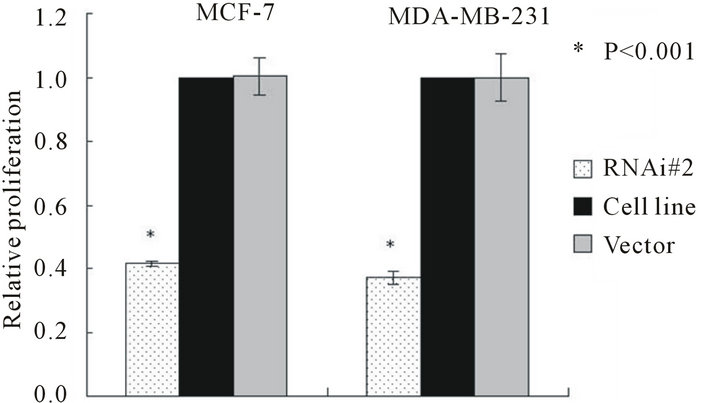 (c)
(c)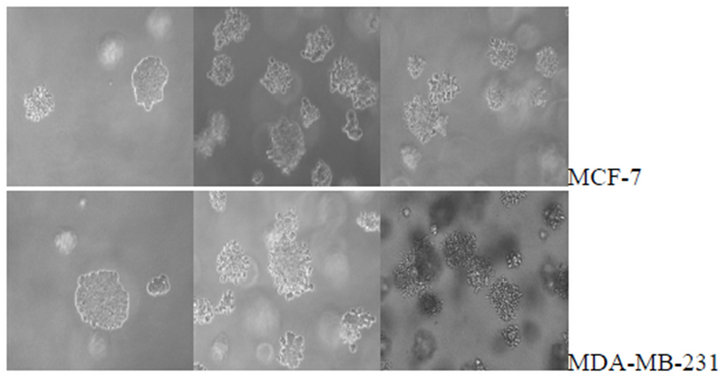 (d)
(d)
Figure 3. Cell proliferations of MCF-7(a) and MDA-MB- 231(b) determined by MTT method, soft agar colony formation experiment (c and d). A, Cell proliferation curve of MCF-7 among groups of hTERT-RNAi#2, untransfected and empty-transfected. B, Cell proliferation curve of MDAMB-231 among groups of hTERT-RNAi#2, untransfected and empty-transfected. C, relative number of positive colony formation among different groups controlled by untransfected group of each cell line. Columns mean from three parallel experiments; bars, SD. *P < 0.001 (one-way ANOVA, Bonferroni).
 (a)
(a)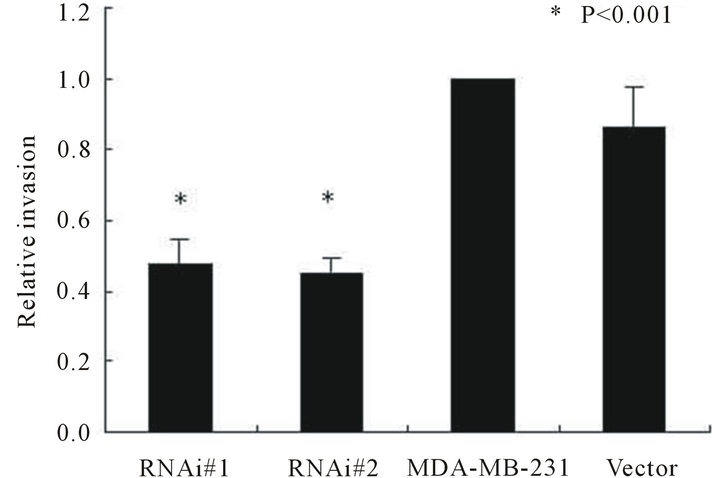 (b)
(b) (c)
(c)
Figure 4. Wound healing of MCF-7 cell line between groups of hTERT-RNAi-transfected and untransfected in different sessions (4A), and invasion ability of MDA-MB-231 cell line among groups of hTERT-RNAi-transfected, untransfected and empty-transfected (4(b) and 4(c)). Columns mean from three parallel experiments; bars, SD. *P < 0.001 (one-way ANOVA, Bonferroni).
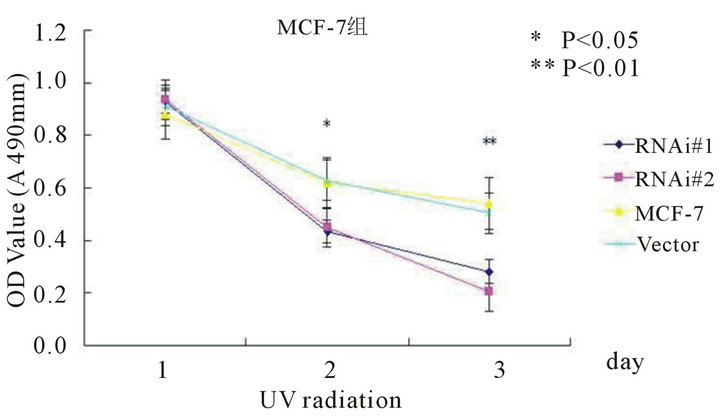
ionizing radiation and Adriamycin, and assessed the cell viability by MTT assays. As shown in Figure 5, after UV radiation or Adriamycin-chemotherapy, the htert-RNAi #2 transfected cells tend to be less viable compared with untransfected or empty-transfected cells (P < 0.001).
4. Discussion
Breast cancer is the most frequently diagnosed malignant tumor in women [1,2]. Telomerase activity has shown to be strongly increased in almost all human malignancies as well as in breast cancer [7,16]. Since the discovery of RNA interference technique, it has changed our under standing of how cells guard their genomes and led to the development of new strategies for blocking gene function [8,17-20]. Previous studies targeting telomerase by RNAi indicate that telomerase is a promising target for breast
 (a)
(a) (b)
(b)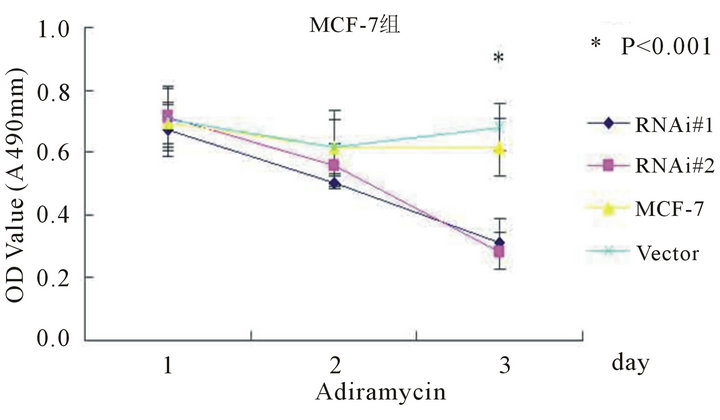 (c)
(c)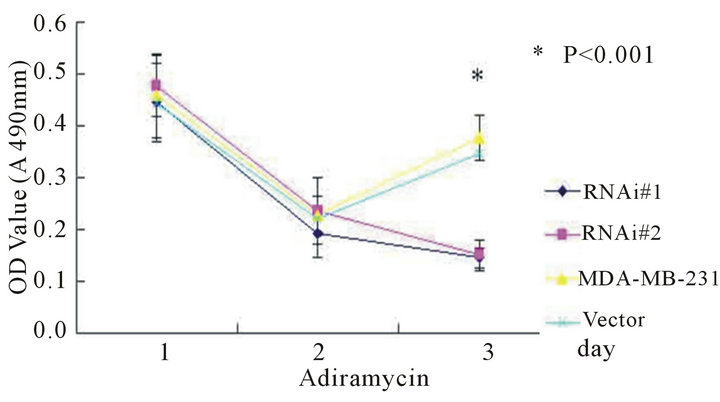 (d)
(d)
Figure 5. The cell viability determined by MTT method after UV radiation (5(a) and 5(b)) or adriamycin-chemothrapy (5(c) and 5(d)) in different groups of MCF-7 and MDA-MB-231 cell lines respectively. Columns mean from three parallel experiments; bars, SD one-way ANOVA, Bonferroni.
cancer therapy [10,21-23]. It is reported that more than 85% malignant tumors expressed hTERT positively [11,24]. Though the details of how telomerase is activated or regulated in cancer cells is not clear yet, it has generally been acknowledged that telomerase inhibition may hinder the growth of cancer cells and hTERT down-regulation will induce impairment of cell proliferation, apoptotic cell death in vitro and tumor growth in vivo [25]. The hTERT mRNA expression was found playing a critical role in the regulation of telomerase activity and appeared to be functionally equivalent to telomerase activity especially in cases when hTERT was overpressed [26,27].
In our present work, we successfully constructed pSuper-retro-puro-hTERT-shRNA vectors which were transfected into MCF-7 and MDA-MB-231 breast cancer cell lines to induce RNAi. hTERT mRNA was downregulated and telomerase activity was inhibited at various degrees, which were verified by Western-Blot, RT-PCR and TRAP. This manifest that the methods are feasible by constructing vectors targeting on hTERT gene, transfected into breast cancer cells and inducing RNAi effects in vitro, which are the same with research reported by different group [10,21]. Expression of hTERT gene may be affected by many factors in cells. Telomerase activity decreased corresponded with down-regulation of hTERT gene expression confirm that hTERT is the key factor in telomerase activity regulation [12,21,22,24]. The efficiency of RNAi by different templates targeting various regions of the same gene was different and was not thorough. As shown in our data, telomerase activity in transfected group declined to 45.3 percent of untransfected group. Breast cancer cells in transfected groups show retardation of cell proliferation and impaired ability to grow in soft agar, we conclude that suppression of hTERT expression can alter the proliferative potential of breast cancer cells and lead to decreased tumorigenic potential in vitro. In wound-healing and matrigel Transwell experiments applied to determine the migrate ability of MCF- 7 and invasive ability of MDA-MB-231 cells respectively, we observed that cells in transfected groups showed less ability of both cell migration and cell invasion, which means down-regulation of telomerase activity could inhibit breast cancer cells from migrating and invading in vivo. The mechanism is not clear yet, we suppose that expression of hTERT was coordinated with SMAD network (data not shown) which needs further investigation. Now most studies indicate suppression of hTERT gene expression does inhibit the proliferation of breast cancer cells, but these cells failed to exhibit immediate cell death, this phenotype may be attributed to the number of cell division required to shorten telomerase lengths to critical lengths, so the conception of suppression of hTERT expression combined with other adjuvant therapy such as irradiation and chemotherapy come into birth. Nakamura et al. [27] reported that inhibition of hTERT expression by RNA interference sensitizes cancer cells to ionizing radiation and chemotherapy. As shown in our study, after hTERT gene knock-down, the breast cancer cells are more fragile when exposed to certain dosage of UV-irradiation or chemo agents.
Taken together, our results revealed that transfected with specific shRNA targeting hTERT gene could inhibit breast cancer cell proliferation, impair cell migration, invasion and sensitize cancer cells to UV-radiotherapy and chemotherapy. Besides the surgical excision, downregulation of hTERT gene by RNAi conbimed with radiotherapy and/or chemotherapy could be a new effective therapy for breast cancer.
5. Acknowledgements
The authors appreciate for the help from Key Laboratory of Tropical Disease Control (Sun Yat-Sen University), Ministry of Education by providing the lab facilities and equipments. We give heartfelt thanks to Professor Jun Li for the advice of the article.
REFERENCES
- World Health Organization International Agency, “World Health Organization International Agency for Research on Cancer,” World Cancer Report, 2011.
- World Health Organization, “Fact Sheet No. 297: Cancer,” 2012.
- G. Curigliano, G. Spitaleri, E. Pietri, et al., “Breast Cancer Vaccines: A Clinical Reality or Fairy Tale?” Annals of Oncology, Vol. 17, No. 5, 2006, pp. 750-762.
- P. W. Nan and J. H. Richard, “The Role of Telomerase Expression and Telomere Length Maintenance in Human and Mouse,” Journal of Clinical Immunology, Vol. 20, No. 4, 2000, pp. 257-267. doi:10.1023/A:1017223602293
- W. Guenther, “The Viral Origins of Telomeres and Telomerases and Their Important Role in Eukaryogenesis and Genome Maintenance,” Biosemiotics, Vol. 1, No. 2, 2008, pp. 191-206. doi:10.1007/s12304-008-9018-0
- M. A. Blasco, “Telomeres and Human Disease: Ageing, Cancer and Beyond,” Nature Reviews. Genetics, Vol. 6, No. 8, 2005, pp. 611-622. doi:10.1038/nrg1656
- J. Liu, A. Baykal, K. M. Fung, et al., “Human Telomerase Reverse Transcriptase mRNA Is Highly Expressed in Normal Breast Tissues and Down-Regulated in Ductal Carcinoma in Situ,” International Journal of Oncology, Vol. 24, No. 4, 2004, pp. 879-884.
- J. Beesley, H. A. Pickett, S. E. Johnatty, et al., “Functional Polymorphisms in the TERT Promoter Are Associated with Risk of Serous Epithelial Ovarian and Breast Cancers,” PLoS One, Vol. 6, No. 9, 2001, Article ID: e24987. doi:10.1371/journal.pone.0024987
- C. Poremba, B. Heine, R. Diallo, et al., “Telomerase as a Prognostic Marker in Breast Cancer: High-Throughput Tissue Microarray Analysis of hTERT and hTR,” Journal of Pathology, Vol. 198, No. 2, 2002, pp. 181-189. doi:10.1002/path.1191
- K. Mokbel and N. J. Williams, “Telomerase and Breast Cancer: From Diagnosis to Therapy,” International Journal of Surgical Investigation, Vol. 2, No. 1, 2000, pp. 85- 88.
- K. Masutomi, E. Y. Yu, S. Khurts, et al., “Telomerase Maintains Telomere Structure in Normal Human Cells,” Cell, Vol. 114, No. 2, 2003, pp. 241-253. doi:10.1016/S0092-8674(03)00550-6
- P. de Souza Nascimento, G. Alves and W. Fiedler, “Telomerase Inhibition by an siRNA Directed against hTERT Leads to Telomere Attrition in HT29 Cells,” Oncology Reports, Vol. 16, No. 2, 2006, pp. 423-428.
- J. Li, N. Zhang, L. B. Song, et al., “Astrocyte Elevated Gene-1 Is a Novel Prognostic Marker for Breast Cancer Progression and Overall Patient Survival,” Clinical Cancer Research, Vol. 14, No. 11, 2008, pp. 3319-3326. doi:10.1158/1078-0432.CCR-07-4054
- R. Pallini, A. Sorrentino, F. Pierconti, et al., “Telomerase Inhibition by Stable RNA Interference Impairs Tumor Growth and Angiogenesis in Glioblastoma Xenografts,” International Journal of Cancer, Vol. 118, No. 9, 2006, pp. 2158-2167. doi:10.1002/ijc.21613
- N. W. Kim and F. Wu, “Advances in Quantification and Characterization of Telomerase Activity by the Telomeric Repeat Amplification Protocol,” Nucleic Acids Research, Vol. 25, No. 13, 1997, pp. 2595-2597. doi:10.1093/nar/25.13.2595
- H. Wege, M. S. Chui, H. T. Le, et al., “SYBR Green Real-Time Telomeric Repeat Amplification Protocol for the Rapid Quantification of Telomerase Activity,” Nucleic Acids Research, Vol. 31, No. 2, 2003, Article ID: e3-3. doi:10.1093/nar/gng003
- S. M. Hammond, E. Bernstein, D. Beach, et al., “An RNADirected Nuclease Mediates Post-Transcriptional Gene Silencing in Drosophila Cells,” Nature, Vol. 404, No. 6775, 2000, pp. 293-296. doi:10.1038/35005107
- P. D. Zamore, “RNA Interference: Listening to the Sound of Silence,” Nature Structural Biology, Vol. 8, No. 9, 2001, pp. 746-750. doi:10.1038/nsb0901-746
- P. Ahlquist, “RNA-Dependent RNA Polymerases, Viruses, and RNA Silencing,” Science, Vol. 296, No. 5571, 2002, pp. 1270-1273. doi:10.1126/science.1069132
- I. J. Macrae, K. Zhou, F. Li, et al., “Structural Basis for Double-Stranded RNA Processing by Dicer,” Science, Vol. 311, No. 5758, 2006, pp. 195-198. doi:10.1126/science.1121638
- L. Zou, P. Zhang, C. Luo, et al., “ShRNA-Targeted hTERT Suppress Cell Proliferation of Bladder Cancer by Inhibiting Telomerase Activity,” Cancer Chemotherapy and Pharmacology, Vol. 57, No. 3, 2006, pp. 328-334. doi:10.1007/s00280-005-0056-x
- K. Kurvinen, S. Syrjänen and B. Johansson, “Long-Term Suppression of Telomerase Expression in HeLa Cell Clones, Transfected with an Expression Vector Carrying siRNA Targeting hTERT mRNA,” International Journal of Oncology, Vol. 29, No. 1, 2006, pp. 279-288.
- A. P. Cunningham, L. G. Andrews and T. O. Tollefsbol, “Retrovirus-Mediated RNA Interference Targeting hTERT through Stable Expression of Short-Hairpin RNA,” Methods in Molecular Biology, Vol. 405, 2007, pp. 39-46. doi:10.1007/978-1-60327-070-0_5
- P. Phatak and A. M. Burger, “Telomerase and Its Potential for Therapeutic Intervention,” British Journal of Pharmacology, Vol. 152, No. 7, 2007, pp. 1003-1011. doi:10.1038/sj.bjp.0707374
- M. F. Li, M. Y. Fang and Y. Wang, “Plasmid-Mediated RNAi Targeting hTERT Inhibits Telomerase Activity in K562 Cell Line,” Zhongguo Shi Yan Xue Ye Xue Za Zhi, Vol. 16, No. 1, 2008, pp. 54-60.
- H. Chen, Y. Li and T. O. Tollefsbol, “Strategies Targeting Telomerase Inhibition,” Molecular Biotechnology, Vol. 41, No. 2, 2009, pp. 194-199. doi:10.1007/s12033-008-9117-9
- M. Nakamura, K. Masutomi, S. Kyo, et al., “Efficient Inhibition of Human Telomerase Reverse Transcriptase Expression by RNA Interference Sensitizes Cancer Cells to Ionizing Radiation and Chemotherapy,” Human Gene Therapy, Vol. 16, No. 7, 2005, pp. 859-68. doi:10.1089/hum.2005.16.859
NOTES
*Grant sponsor: Project of College Fundamental Research for Medical Young Teachers, Sun Yat-sen University, China; Grant number: 09 ykpy45.
#Corresponding author.

