Advances in Alzheimer's Disease
Vol.2 No.3(2013), Article ID:37292,5 pages DOI:10.4236/aad.2013.23015
BLMH and APOE genes in Alzheimer Disease: A possible relation
![]()
1Graduate Program in Oral Biology, University of the Sacred Heart, Sao Paulo, Brazil; *Corresponding Author: slmpayao@famema.br
2Blood Center, Facultyof Medicine of Marília, Sao Paulo, Brazil
3Radiotherapy/Oncology, Facultyof Medicine of Marilia, Sao Paulo, Brazil
4Neurology / Neurosurgery, Federal University of Sao Paulo, Sao Paulo, Brazil
5Morphology, Federal UniversityofSao Paulo, Sao Paulo, Brazil
Copyright © 2013 J. P. B. Ximenez et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 12 June 2013; revised 22 July 2013; accepted 31 July 2013
Keywords: Alzheimer Disease; Bleomycin Hydrolase; Apolipoprotein E; Expression Gene; Polymorphism
ABSTRACT
Alzheimer disease (AD) is a progressive and irreversible neurodegenerative disorder that is characterized by cognitive decline, memory loss and confusion. The E4 allele of the apolipoprotein E gene (APOE) is associated with AD and it is the main genetic risk factor for disease. Although the exact physiological function is unknown, bleomycin hydrolase (BLMH) may also be associated with AD development, although previous immunohistochemical findings have been inconsistent. Therefore, the purpose of this study was to evaluate the genotypic and allele frequencies of the APOE gene and BLMH 1450 G > A polymorphism and assess BLMH expression using PCR-RFLP and RT-qPCR analyses of blood samples from patients with Alzheimer disease (AD), healthy elderly adults (EC) and healthy young subjects (YC). BLMH expression was significantly different among groups (p = 0.015) and there was substantial reduction with age and with AD. The APOE and BLMH genotype frequency did not diverge from the Hardy-Weinberg equilibrium. There was a higher frequency of genotype 3/3 in all subjects (61.1%) and the AD group demonstrated a higher frequency of allele 4; however, differences in genotype and allele distributions were statistically different among groups.
1. INTRODUCTION
Alzheimer’s disease (AD) is the most common form of dementia and both environmental and genetic factorsare contributing to risk, which increases with age [1]. Prevalence of Alzheimer’s disease is 0.8% in individuals aged 65 - 69 years and upwards of 28.5% in persons aged 90 years and older [2]. Brain anatomy of patients with AD has two hallmark neuropathological characteristics, neuritic plaques and the neurofibrillary tangles. Amyloid-β (Aβ) peptide is the major plaque component, while hyperphosphorylated tau protein is the major tangle component [3-5].
The apolipoprotein E (APOE) gene is the most important genetic risk factor for sporadic AD and it is located on chromosome 19q13.2 and consists of 4 exons. The 3 APOE (ε2, ε3, and ε4) are defined by 2 SNP, which encode 3 protein isoforms (E2, E3, and E4). Only about 20% - 25% of the general population carries one or more ε4 alleles, whereas 50% - 65% of people with AD are ε4 carriers [6,7].
Bleomycin hydrolase (BLMH) enzyme is a cysteine protease of the papain superfamily encoded by the BLMH gene. This enzyme is expressed in most human tissues, including the hippocampus and amygdala. Although the exact physiological function is unknown, BLMH is associated with AD and the expression of this enzyme is substantially lower in AD brains [10,11]. BLMH gene is located on chromosome 17q BLMH 11.2 and its enzyme exposes 455 amino acids [8]. BLMH 1450 G > A is its main polymorphism, in which there is the substitution of nucleotide Adenine by Guanine at position 1450. This subsequently leads to either isoleucine or valine as the amino acid in position 443 (Ile443Val) [9]. In patients with the polymorphism, the biotransformation is inefficient and resulting in an accumulation and toxic action of the bleomycin hydrolase [9,19].
Although bleomycin hydrolase is widely distributed in most human tissues, previous studies have demonstrateedthat the BLMH gene is associated with AD. It is hypothesized that the BLMH gene alters the processing of amyloid precursor protein (APP) and significantly increases the release of its proteolytic fragment, Amyloid-β (Aβ) [10,11]. Despite this potential relationship, the specific role of this altered gene expression has not been fully elucidated and the relationship between BLMH and the development of AD remains a source of controversy [10-12].
Given the potential relationship between APOE, BLMH and the development of AD, the purpose of this paper was to determine whether differences exist in the APOE allele frequency, BLMH genotype distribution, and BLMH gene expression in healthy and pathological subject groups. PCR-RFLP and qRT-PCR techniques were to compare allele frequencies, genotypes distribution and gene expression in the peripheral blood of healthy young (YC), healthy elderly (EC) and AD subject groups. We hypothesized that the significant differences would exist between the AD and control groups and, in particular, subjects in the AD group would demonstrate less BLMH expression.
2. MATERIALS AND METHODS
2.1. Samples
Ninety-one subjects with AD (31 ♂ and 60 ♀, mean age of 74.54 ± 7.58 years), 93 EC subjects (38 ♂ and 55 ♀, mean age of 71.52 ± 8.02 years) and 86 YC subjects (31 ♂ and 55 ♀, mean age of 24.60 ± 1.92 years) participated in this study. BLMH expression was analyzed in 164 subjects: 56 subjects with AD (13 ♂ and 43 ♀, mean age of 74.54 ± 7.66 years), 59 EC (22 ♂ and 35 ♀, mean age of 71.52 ± 8.22 years) and 52 YC (23 ♂ and 28 ♀, mean age of 21.18 ± 1.83 years). The three subject groups had similar ethnic origins, 95% of the total sample were of European origin, 2.5%, were of Japanese origin and 2.5% were multi-ethnic. The ethnic origins were determined by self-report and by the geographical origin of family.
Subjects with AD were evaluated using Mini-Mental State Examination (MMSE) and Katz index [13,14]. AD patients were diagnosed according to National Institute of Neurological and Communicative Disorders and Stroke - Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRDA) criteria and The Diagnostic and Statistical Manual of Mental Disorders IV (DSM-IV) for probable AD [15]. All patients were recruited from the Department of Neurology at the Federal University of São Paulo (UNIFESP), São Paulo-SP, Brazil. The Institutional Research Ethics Committee approved this study and all subjects or their legal representatives signed an informed consent according to the Declaration of Helsinki.
2.2. DNA/RNA Extraction and cDNA Synthesis
Genomic DNA was extracted from blood samples using QIAamp DNA Blood Midi Kit (Qiagen, Germany), following the protocol provided by the manufacturer. All samples were stored at −20˚C until analyzed. Total RNA was extracted using the RiboPure™ Blood Kit (Ambion, USA) and RNeasy Lipid Tissue Mini Kit (Qiagen, Germany), respectively, according to the manufacturer’s protocol. Total RNA was quantified using Spectrophotometer NanoDrop-2000 (NanoDrop, USA). The concentrations were adjusted and the samples were stored at −80˚C until use. The cDNA synthesis was carried out using High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems™, USA), following the protocol provided by the manufacturer. All cDNA was quantified using Spectrophotometer NanoDrop - 2000 (NanoDrop, USA). The concentrations were adjusted and stored at −80˚C.
2.3. Genotyping
Genotypes for the APOE gene (rs429358 e rs7412) were determined by polymerase chain reaction (PCR) amplification using the primers 5’-TAA GCT TGG CAC GGC TGT CCA AGG A-3’ (sense) and 5’-ACA GAA TTC GCC CCG GCC TGG TAC AC-3’ (antisense) according to a previously described protocol (16). The amplified products were digested by RFLP with HhaI restriction enzyme (Fermentas, Canada) overnight at 37˚C, producing fragments of 91 bp, 81 bp, 72 bp, 42 bp and 33 bp [16]. Fragments were separated by electrophoresis in 3% agarose gels and visualized by ethidium bromide staining.
To determine the genotypes for the BLMH 1450 G > A polymorphism (rs1050565), PCR was carried out using the primers 5’-GTC GTG TTA GAG CAG GAA CCC AAT T-3’ (sense) and 5’-CCT GGA TCT GTC CTT TGC AGC TAC G-3’ (antisense) according to a previously described protocol [9,17]. The amplified products were digested with MunI restriction enzyme (New England Biolabs, Ipswich, MA) for 3 hours at 37˚C, producing fragments of 130 bp, 106 bp and 24 bp [9,17]. Fragments were separated by electrophoresis in 2.5% agarose gels and visualized by ethidium bromide staining.
2.4. Gene Expression Analysis
All gene expression was measured by RT-qPCR on the Applied Biosystems 7500 Fast Real-Time PCR system (Applied Biosystems™, USA), according to cycling conditions recommended by Applied Biosystems. We used BLMH (target; assay id: Hs00166071_m1), TM9 (endogenous control; assay id: Hs00207196_m1) and B2M (endogenous control; assay id: Hs99999907_m1) inventoried TaqMan® Gene Expression Assays.
The threshold values were uniformly set for all assays. All reactions were performed in duplicate. Replicates with standard deviations (SD) greater than 0.5 for the cycle threshold (CT) value were repeated or excluded from the analysis. The amplification curve of each group was created, and the CT values were obtained for all genes (BLMH, TM9 and B2M). Relative quantification was calculated using the comparative CT method (2−∆∆CT).
2.5. Statistical Analyses
Descriptive analyses were performed using means and standard deviations for continuous variables and proportions (%) for dichotomous variables. A one-way analysis of variance (ANOVA) was used to determine if significant differences existed between in BLMH expression among the three groups. Genotype distribution for each gene was assessed for deviation from the Hardy-Weinberg equilibrium. Differences in genotype and allele frequencies among groups were assessed using χ2-tests. Differences were considered significant with a p value less than 0.05. All statistical analyses were performed with SPSS version 20.0.
3. RESULTS
BLMH expression, as measured using relative quantification values, was significantly different between groups. (Table 1) Subjects in the AD group had the lowest mean expression, followed by the EC group. Expression was highest in the YC group (see Figure 1).
The distribution of genotypes for the two polymorphisms did not diverge from the Hardy-Weinberg equilibrium (Table 2). The frequencies of the BLMH genotypes in the all groups were similar to those previously reported in the literature [18]. There were no significant differences in APOE genotypes or allele distributions between groups (Table 2); however, the genotype 3/3 was most frequent for all subjects (61.1%). Although the distribution was not significantly different, allele 4 was much more common in the AD group (67.4%) compared to the EC (14.0%) and YC (18.6%) groups.
4. DISCUSSION
We hypothesized that subjects with AD would demonstrate reduced BLMH expression. This hypothesis was supported by our results and corroborated previous immunohistochemical findings. Immunohistochemical analyses have found a predominantly astrocytic expression of BH with a reduction in signal intensity [10,11]. However,
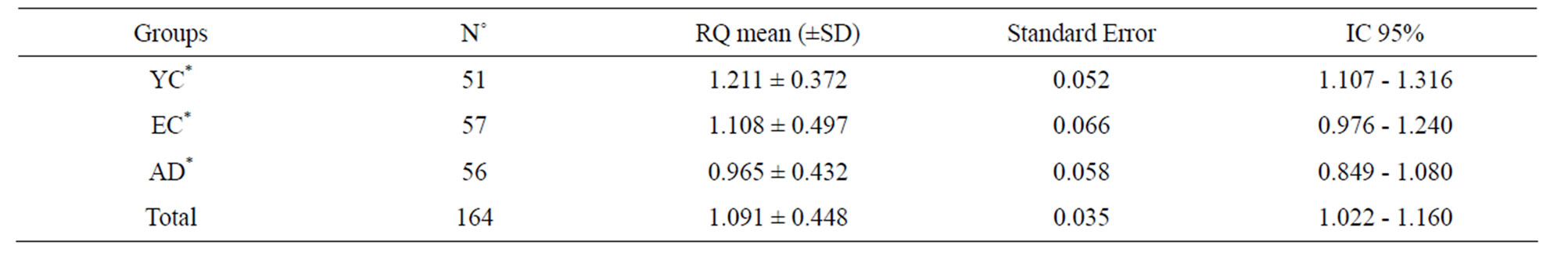
Table 1. BLMH expression in Alzheimer disease (AD), Early Control (EC) and Young Control (YC) groups.
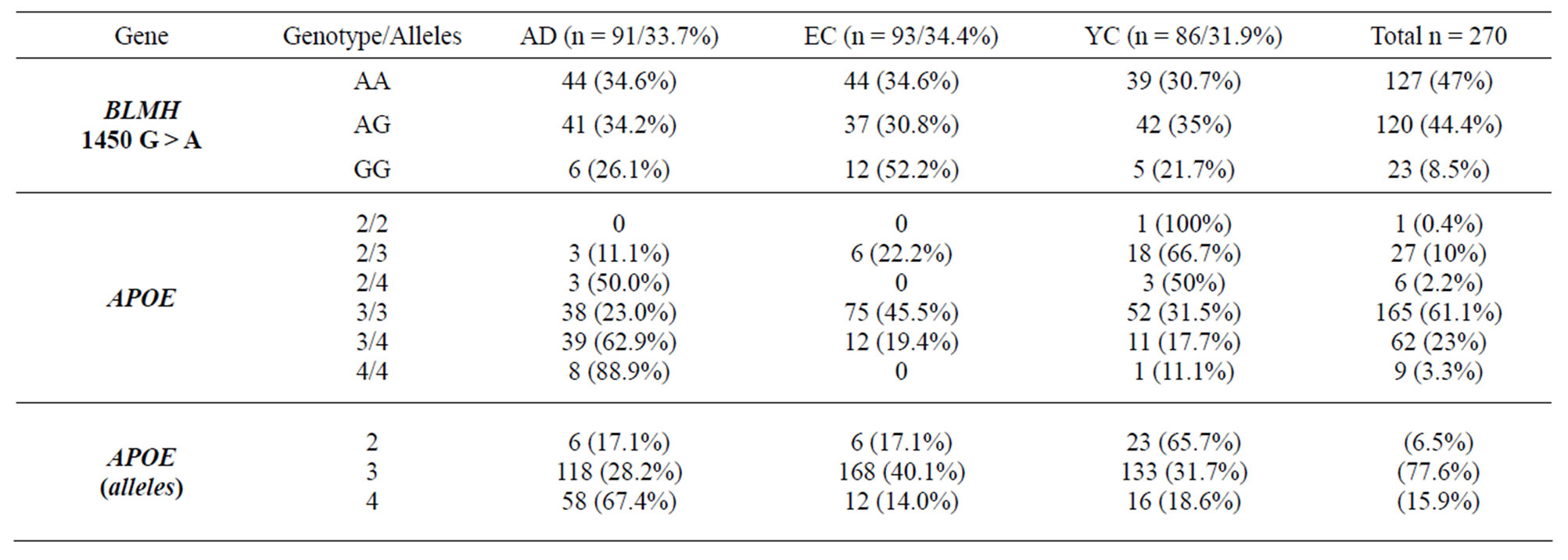
Table 2. Genotype and allele frequencies for BLMH and APOE in Alzheimer disease (AD), Early Control (EC) and Young Control (YC) groups.
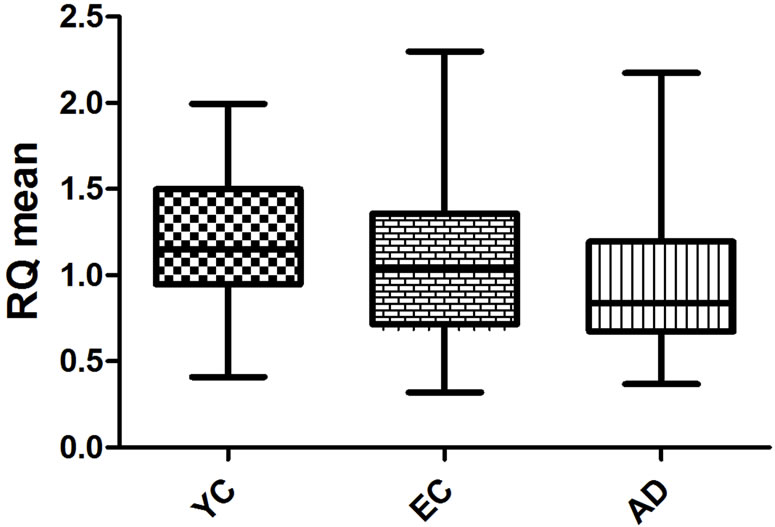
Figure 1. Box-plots representing mean, interquartile, and total range of BLMH expression for the three groups.
this reduction in signal intensity was not statistically significant, owing to the large variability among controls subjects [11]. While immunohistochemical analysis can be used to identify BH availability in specific tissues and determine regional and cellular distribution, our approach analyzed peripheral blood samples to identify systemic differences in BLMH expression. Our results support findings from the previous immunohistochemical studies because we also found a decrease in BLMH expression.
Reduced BLMH expression may affect the activity of the APP pathway in patients with AD. In particular, reduced expression of BLMH may alter the development of amyloid peptides Aβ1-40 and Aβ1-42, which are major components of plaques in AD brain [20,21]. However, this theory requires additional research as APP processing is complex and includes APP posttranslational modifications and the activity of at least three secretases [12,22]. Future work should evaluate the relationship between BLMH and APP expression in patients with AD to determine if lower BLMH expression occurs with a concomitant increase APP expression.
The BLMH 1450 G > A is localized in the C-terminal domain of the gene, which gives rise to Val443Ill isoforms and covers the active site of the enzyme [9,19]. In our analysis of the frequency of single nucleotide polymorphisms (SNP), allele 4 was most common in the AD groups, while for all subjects collectively, the 3/3 genotype was most frequent. Although different groups had different alleles that were most common, there were no statistically different distribution frequencies among the three groups in this study.
Previous studies investigating BLMH 1450 G > A in AD and control subjects have found that subjects with the G/G alleles and without APOE allele 4 had four times greater likelihood to have AD [23-25]. However, this association between BLMH genotypes and AD was not found by other researchers [18,26-28]. The relationship between BLMH expression and Val443Ill isoforms with amount of patients bigger than our study, it still is an alternative for another researchinasmuch as the 1450 G > A covers the BH’s active and it can have an influence about the disease.
In conclusion, our study was the first to identify a relationship between BLMH expression and Alzheimer’s disease using RT-qPCR. BLMH expression was significantly lower in subjects with the disease compared to healthy elderly adults and a healthy young sample. These findings corroborate with previous results and may offer a new direction of study to identify the underlying causes of pathological changes in individuals with AD.
REFERENCES
- Burns, A. and Iliffe, S. (2009) Alzheimer’s disease. BMJ, 338, b158. doi:1 0.1136/bmj.b158
- Mayeux, R. and Stern, Y. (2012) Epidemiology of Alzheimer disease. Nature Reviews Neurology, 7, 137-152.
- Gandy, S. (2005) The role of cerebral amyloid beta accumulation in common forms of Alzheimer disease. Journal of Clinical Investigation, 115, 1121-1129.
- Nelson, P.T., Head, E., Schmitt, F.A., Davis, P.R., Neltner, J.H., Jicha, G.A., et al. (2011) Alzheimer’s disease is not “brain aging”: Neuropathological, genetic, and epidemiological human studies. Acta Neuropathologica, 21, 571- 587. doi:10.1007/s00401-011-0826-y
- Kaden, D., Munter, L.M., Reif, B. and Multhaup, G. (2012) The amyloid precursor protein and its homologues: structural and functional aspects of native and pathogenic oligomerization. European Journal of Cell Biology, 91, 234-239. doi:10.1016/j.ejcb.2011.01.017
- Bu, G. (2009) Apolipoprotein E and its receptors in Alzheimer’s disease: Pathways, pathogenesis and therapy. Nature Reviews Neuroscience, 10, 333-344. doi:10.1038/nrn2620
- Abasolo, N., Torrell, H., Roig, B., Moyano, S., Vilella, E. and Martorell, L. (2011) RT-qPCR study on post-mortem brain samples from patients with major psychiatric disorders: Reference genes and specimen characteristics. Journal of Psychiatric Research, 45, 1411-1418. doi:10.1016/j.jpsychires.2011.06.001
- Hu, C.J., Sung, S.M., Liu, H.C., Hsu, W.C., Lee, L.S., Lee, C.C., Tsai, C.H. and Chang, J.G. (2000) Genetic risk factors of sporadic Alzheimer’s disease among Chinese in Taiwan. The Journal of Neuroscience, 181, 127-131. doi:10.1016/S0022-510X(00)00443-3
- Haas, E.C., Zwart, N., Meijer, C., Nuver, J., Boezen, H.M., Suurmeijer, A.J., Hoekstra, H.J., van der Steege, G., Sleijfer, D.T. and Gietema, J.A. (2008) Variation in bleomycin hydrolase gene is associated with reduced survival after chemotherapy for testicular germ cell cancer. Journal of Clinical Oncology, 26, 470-476
- Namba, Y., Ouchi, Y., Takeda, A., Ueki, A. and Ikeda K. (1999) Bleomycin hydrolase immunoreactivity in senile plaque in the brains of patients with Alzheimer’s disease. Brain Research, 830, 200-202. doi:10.1016/S0006-8993(99)01435-3
- Malherbe, P., Faull, R.L. and Richards, J.G. (2000) Regional and cellular distribution of bleomycin hydrolase mRNA in human brain: comparison between Alzheimer’s diseased and control brains. Neuroscience Letters, 3, 281, 37-40.
- Kajiya, A., Kaji, H., Isobe, T. and Takeda, A. (2006) Processing of amyloid beta-peptides by neutral cysteine protease bleomycin hydrolase. Protein & Peptide Letters, 13, 119-123. doi:10.2174/092986606775101562
- Katz, S., Ford, A.B., Moskowitz, R.W., Jackson, B.A. and Jaffe, M.W. (1963) Studies of Illness in the Aged. The Index of Adl: A Standardized Measure of Biological and Psychosocial Function. Jama, 185, 914-919. doi:10.1001/jama.1963.03060120024016
- Bertolucci, P.H., Okamoto, I.H., Brucki, S.M., Siviero, M.O., Toniolo, J. and Ramos, L.R. (2001) Applicability of the CERAD neuropsychological battery to Brazilian elderly. Arquivos de Neuro-Psiquiatria, 59, 532-536. doi:10.1590/S0004-282X2001000400009
- Mckhann, G., Drachman, D., Folstein, M., Katzman, R., Price, D. and Stadlan, E.M. (1984) Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology, 34, 939-944. doi:10.1212/WNL.34.7.939
- Hixson, J.E. and Vernier, D.T. (1990) Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with Hhai. The Journal of Lipid Research, 31, 545-548.
- Tuimala, J., Szekely, G., Gundy, S., Hirvonen, A. and Norppa, H. (2002) Genetic polymorphisms of DNA repair and xenobiotic-metabolizing enzymes: Role in mutagen sensitivity. Carcinogenesis, 23, 1003-1008. doi:10.1093/carcin/23.6.1003
- Llorca, J., Rodríguez-Rodríguez, E., Dierssen-Sotos, T., Delgado-Rodríguez, M., Berciano, J. and Combarros, O. (2008) Meta-analysis of genetic variability in the bamyloid production, aggregation and degradation metabolic pathways and the risk of Alzheimer’s disease. Acta Neurologica Scandinavica, 117, 1-14.
- Altés, A., Paré, L., Esquirol, A., Xicoy, B., Rámila, E., Vicente, L., López, R., Orriols, J., Vall-Llovera, F., Sánchez-González, B., Del Río, E., Sureda, A., Páez, D. and Baiget, M. (2013) Pharmacogenetic analysis in the treatment of Hodgkin lymphoma. Leukemia & Lymphoma, 54, 1706-1712. doi:10.3109/10428194.2012.752080
- Lefterov, I.M., Koldamova, R.P., Lefterova, M.I., Schwartz, D.R. and Lazo, J.S. (2001) Cysteine 73 in bleomycin hydrolase is critical for amyloid precursor protein processing. Biochemical and Biophysical Research Communications, 283, 994-999.
- Chen, J. and Stubbe, J. (2005) Bleomycins: towards better therapeutics. Nature Reviews Cancer, 5, 102-112. doi:10.1038/nrc1547
- Lefterov, I.M., Koldamova, R.P. and Lazo, J.S. (2000) Human bleomycin hydrolase regulates the secretion of amyloid precursor protein. The FASEB Journal, 14, 1837- 1847. doi:10.1096/fj.99-0938com
- Montoya, S.E., Aston, C.E., DeKosky, S.T., Kamboh, M.I., Lazo, J.S. and Ferrell, R.E. (1998) Bleomycin hydrolase is associated with risk of sporadic Alzheimers disease. Nature Genetics, 18, 211-212. doi:10.1038/ng0398-211
- Farrer, L.A., Abraham, C.R., Haines, J.L., et al. (1998) Association between bleomycin hydrolase and Alzheimer’s disease in Caucasians. Annals of Neurology, 44, 808-811. doi:10.1002/ana.410440515
- Papassotiropoulos, A., Bagli, M., Jessen, F., Frahnert, C., Rao, M.L., Maier, W. and Heun R. (2000) Confirmation of the association between bleomycin hydrolase genotype and Alzheimer’s disease. Molecular Psychiatry, 5, 213- 215. doi:10.1038/sj.mp.4000656
- Thome, J., Gewirtz, J.C., Sakai, N., Zachariou, V., RetzJunginger, P., Retz, W., Duman, R.S. and Rösler, M. (1999) Polymorphisms of the human apolipoprotein E promoter and bleomycin hydrolase gene: Risk factors for Alzheimer’s dementia? Neuroscience Letters, 274, 37-40. doi:10.1016/S0304-3940(99)00662-X
- Prince, J.A., Feuk, L., Sawyer, S.L., Gottfries, J., Ricksten, A., Nägga, K., Bogdanovic, N., Blennow, K. and Brookes, A.J. (2001) Lack of replication of association findings in complex disease: An analysis of 15 polymorphims in prior candidate genes for sporadic Alzheimer disease. European Journal of Human Genetics, 9, 437- 444. doi:10.1038/sj.ejhg.5200651
- Smach, M.A., Charfeddine, B., Lammouchi, T., Othman, L.B., Letaief, A., Nafati, S., Dridi, H., Bennamou, S. and Limem, K. (2010) Analysis of association between bleomycin hydrolase and apolipoprotein E polymorphism in Alzheimer’s disease. Neurological Sciences, 31, 687-691. doi:10.1007/s10072-010-0234-4

