Advances in Microbiology
Vol.08 No.01(2018), Article ID:82160,12 pages
10.4236/aim.2018.81006
A Review of Staphylococcus aureus and the Emergence of Drug-Resistant Problem
Zhu Li
The Hong Kong Polytechnic University, Hong Kong, China
Copyright © 2018 by author and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: December 7, 2017; Accepted: January 28, 2018; Published: January 31, 2018
ABSTRACT
There are various bacteria living in this world. The most common one is Staphylococcus aureus. Almost everyone has heard of it. It is easy to find their habitats, such as hospitals, homes, parks, schools etc. Some of them are difficult to be eliminated because of drug-resistant mutations. Hence, lots of researchers devoted their efforts to eliminate them. This review illustrates the characteristics of the Staphylococcus aureus and the main threat of their drug-resistant strains, especially methicillin-resistant S. aureus. What’s more, the article also highlights the plight in the drug development.
Keywords:
Features of Staphylococcus aureus (S. aureus), Drug-Resistant S. aureus, Pharmacotherapy to Drug-Resistant S. aureus

1. Introduction
Staphylococcus has caused diseases in human beings for centuries [1] . These bacteria were first defined as Staphylococcus (from the Greek staphylos “grape” and kokkos “berry or seed”) in 1882 by the Scottish surgeon Sir Alexander Ogston [2] . A German physician, Friedrich J. Rosenbach, described 2 pigmented colonies of staphylococci and classified them as Staphylococcus albus (Latin for “white”) and Staphylococcus aureus (from the Latin “gold”) [3] . From then on, Staphylococcus aureus (S. aureus) has surprised scientists and physicians continuously, because they killed millions of patients [4] .
S. aureus is a gram-positive coccal bacterium and is the most common opportunistic pathogens of humans. They colonize on approximately 30% of the human population [5] . Furthermore, it refers to a large range of diseases from mild skin infections to life-threatening diseases [6] . Taking the human skin as an example, the bacteria contribute to folliculitis, furuncles, and carbuncles, impetigo, mastitis, wound infections, and staphylococcal scalded skin syndrome. There are also many other existing infections, such as bacteremia, pneumonia, endocarditis, osteomyelitis, meningitis, urinary tract infection, septic thrombophlebitis, cellulitis, abscesses, and sepsis, necrotizing fasciitis, and toxic shock syndrome [6] [7] [8] .
According to these diseases, it is easy to conclude that the infection site determines the type of disease. The capacity of S. aureus causing diseases depends on a lot of virulence factors which colonize, disseminate and evade the immune system of the hosts [9] .
The distinguishing feature of S. aureus: biofilm formation
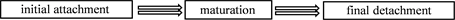
For keeping themselves away from hostile environmental effects, the agglomerate bacteria have been described as “biofilms”. And it is common to see the biofilms of S. aureus. They usually come into being surface-attached communities which embedded in an extracellular matrix [10] . There are 2 main advantages for S. aureus. First, it protects S. aureus from being washed or scraped away to enhance the survival rate. Second, it helps S. aureus to flee the host defense [11] .
On the basis of Figure 1, the biofilm formation contains 3 stages, initial attachment, maturation and final detachment. The first attachment occurs to a surface (like polymeric surface) or to a conditioning film (such as host matrix proteins). The procedure of maturation relies on those adhesive factors, while the process of detachment depends on disruptive factors. Furthermore, abbreviations in Figure 1 are shown here: Aap, accumulation-associated protein; eDNA, extracellular DNA; SCRAMMs, microbial surface components recognizing adhesive matrix molecules; PIA, polysaccharide intercellular adhesin; PSMs, phenol-soluble modulins [11] .
Figure 1. The process of biofilm formation (S. aureus) [11] .
2. The Immune Response to S. aureus: Neutrophils
Neutrophils play an important role in phagocytizing cells so that they can defend the host against acute bacterial infection. If patients have congenital neutrophil deficiencies, bacterial infections will be fatal to them [12] . Thus a healthy neutrophil-mediated killing system is the critical defense to eliminate gram-positive S. aureus in the host [13] .
As is shown in Figure 2, it illustrates how neutrophils can kill S. aureus. All things begin with the activation of the endothelium. There are some neutrophils which roll on the activated endothelium. Then they stop rolling and adhere to the activated endothelium firmly. After that, neutrophils transmigrate through the endothelium into the tissue, named as extravasation. Neutrophils keep migrating to an infected tissue in terms of a chemotactic gradient. When they arrive at an infected area, there are 2 directions for them to eliminate S. aureus. One of them is phagocytosis process to phagocytize S. aureus directly. Neutrophils with complement receptors (CRs) and Fcγ receptors (FcγRs) are able to recognize and phagocytize S. aureus owing to attached antibodies and complements on the bacteria. Inside the neutrophils, the bacteria are sequestered by phagosomes. Subsequently, S. aureus is killed by granule fusion and NADPH oxidase, which release antimicrobial proteins (AMPs) and reactive oxygen species (ROS) respectively. The other one is called NETosis process for killing S. aureus extracellularly. Neutrophils expel their DNA which is decorated with histones and AMPs. Therefore, these special neutrophils have the ability to capture and kill the bacteria, namely, entrapment to S. aureus [13] . Besides, in the
Figure 2. The process of neutrophils eliminating S. aureus [13] .
process of phagocytosis, the apoptotic cell death (the neutrophils) follows eradication of ingested bacteria [14] .
3. The Emergence of MRSA
As is known to all, S. aureus is famous for the pathogen causing numerous diseases to human. More seriously, its impact prominently enhanced by the antibiotic-resistant ability. Besides, one of the most common types is methicillin-resistant S. aureus (MRSA). The first appearance is in 1960. It has the capacity of causing serious healthcare-associated and community-associated infections, such as skin and soft-tissue infection (SSTIs), pneumonia, endocarditis and osteomyelitis [15] . Actually, one of the most common causes associated with bone and joint infection is also MRSA [16] [17] . MRSA is a great challenge for high mortality, limited therapeutic options, and the heavy cost burden [15] [18] . That is to say, compared with methicillin-susceptible S. aureus infections, patients with MRSA infections are easier to be seriously ill, and face a higher risk of death and more expensive costs [19] [20] [21] . For example, recent years, researchers in the USA indicate that MRSA causes approximately 95 000 invasive infections and 19,000 deaths per year [22] .
By definition, MRSA is resistant to all β-lactam antibiotics, including oxacillin, nafcillin, dicloxacillin, and cefazolin [23] . Normally, we use oxacillin and/or cefoxitin for susceptibility testing. For methicillin-susceptible S. aureus, ß-lactams are able to bind to the penicillin-binding proteins (PBP) which are essential for cell wall biosynthesis. Furthermore, the peptidoglycan crosslink formation is also inhibited. Therefore, this results in lysis of bacterial cells. However, MRSA has a mobile genetic element called staphylococcal cassette chromosome (SCCmec). The SCCmec carries the mecA gene to encode altered PBP (PBP2a) so that the affinity to β-lactam antibiotics is decreased prominently. Consequently, the inhibition of cell wall biosynthesis fails, and MRSA strains are capable of surviving even in the presence of β-lactam antibiotics [24] .
MRSA strains are the most common bacteria among those healthcare-associ- ated patients, and they are named as HA-MRSA [25] [26] [27] . Then, the first report about community-associated MRSA (CA-MRSA) infections was published at the beginning of the 20th century, the infections happened on healthy individuals (with no risk factors of HA-MRSA infections), injection drug users, incarcerated people, and athletes [28] [29] . HA-MRSA strains are genetically different from CA-MRSA, and one significant difference is the SCCmec. So far, 11 SCCmec types have been identified [30] [31] . CA-MRSA strains are mainly related to SCCmec types IV and V while researchers characterize SCCmec types I, II and III from HA-MRSA [32] .
The attention of scientific literature and the press are paid to MRSA has reduced the burden of hospital-acquired MRSA or healthcare-associated MRSA (HA-MRSA) infections, such as the infection of USA100 in hospitals [33] [34] [35] [36] . Nevertheless, CA-MRSA is spreading rapidly throughout many industrialized regions of the world [37] [38] [39] . On the basis of some reports, the increasing trend of CA-MRSA indicates that these clones may ultimately replace HA-MRSA clones in hospitals [40] . Actually, USA300 is now commonly found in both the healthcare setting and the community setting [41] .
Dating back to the beginning of CA-MRSA, there were rare reports related to it [42] . However, after the discovery of a unique MRSA clone in the community in Western Australia, the situation has changed [43] . Several years later, the CA-MRSA clones were recognized in Europe, the United States, Latin America, and Asia [28] [44] [45] [46] . Thus, it is crucial to understand the epidemiology of CA-MRSA for controlling MRSA. According to most reports, the CA-MRSA infections are implicated in USA 300. CA-MRSA emerged, and the number of cases escalated, rapidly in the USA in the early 2000s. For example, a survey of 11 US hospitals confirmed that 97 percent of CA-MRSA isolates were the USA300 [47] [48] . Not only are they popular in the US, but also they contribute to a global epidemic threat [49] [50] .
It is well accepted that the earliest cases of USA 300 occurred in a collegiate football team in Pennsylvania, then several outbreaks happened among prisoners in Mississippi and Los Angeles. The USA 300 strains originally have a close relationship with military personnel, prisoners, athletes, intravenous drug users, the homeless, urban populations, and men who have sex with men [38] . But it soon has expanded quickly to the general population [51] . It is easy to find the reports of USA300 from other countries besides the United States. For instance, USA300 was discovered in neighboring countries of Colombia since 2006 [52] . The predominant CA-MRSA clone, USA300, has become the primary cause of community-associated skin infection [48] . What is more, with strong virulence [53] , little improvement was made for controlling them effectively.
4. The Epidemiology of MRSA
There are a variety of reports related to MRSA. In 2011, approximately 80461 cases of MRSA infections occurred and caused over 11000 deaths in the USA [54] . MRSA also exists in Iran, and the prevalence is in the middle of Australia (lower) and the United States (higher) [55] [56] . Over 9000 cases of S. aureus bacteraemia (SAB) happened per year in England, Wales and Northern Ireland, and 12.7% of these owed to MRSA in 2012 [57] . In Taiwan, the MRSA infection rate increased dramatically from 9.8% in 1999-2000 to 56% in 2004-2005 [58] . In addition, the MRSA infection rate in Europe is not high but rising day by day [59] . There are few reports about MRSA in developing countries, but it is certain that MRSA will lead to devastating consequences if it becomes prevalent there since resource-poor settings.
5. The History and the Mechanism of the Pharmacotherapy to Drug-Resistant S. aureus
The penicillin G was introduced for improving prognosis prominently in the early 1940s, however, the resistant S. aureus appeared 2 years later [60] . Penicillinase (β-lactamase enzyme) is the crucial reason which is responsible for the drug-resistant problem. These enzymes hydrolyze the β-lactam ring so that the drug is inactivated [61] [62] . Besides, they are encoded by blaZ, which exists on a large transposon on a plasmid. Unfortunately, the rate of drug-resistant in human S. aureus isolates is over 90%. That is to say, penicillin almost lost therapeutical effect [7] . Worse to come, S. aureus was capable of resisting other antibiotics such as erythromycin, streptomycin, and the tetracyclines [63] [64] [65] .
In 1959, a semisynthetic antibiotic called methicillin was used to resolve the spread of penicillin-resistant dilemma. Unfortunately, the methicillin-resistant S. aureus (MRSA) was isolated in 1960 [66] . The mechanism of MRSA resistance has already shown above (the part of the emergence of MRSA). Then MRSA keeps spreading in many countries, and it evolves to be a worldwide problem. In spite of useless methicillin, the term of MRSA is remembered by lots of people because of the remaining problem. Subsequently, MRSA shows drug-resistance to an entire class of penicillin-like antibiotics including penicillin, amoxicillin, oxacillin, meticillin, and others [67] .
So far, vancomycin is considered as the perfect treatment choice for severe MRSA infections. With development, the susceptibility to vancomycin is decreasing in terms of some studies. In fact, full vancomycin-resistant S. aureus (VRSA) emerged clinically in 2002 [68] . And another kind of alternative antibiotic (daptomycin) also has the drug-resistant problem [4] .
Table 1 as shown below, it summarizes the mechanisms between drug-resistant S. aureus and antibiotics.
6. Conclusions
The most serious problem we are facing is the drug-resistant problem. The fundamental reason is that using antibiotics caused the drug-resistant problem. Furthermore, the chronic misuse and overuse of antibiotics have deteriorated the drug-resistant problem. The development of new antibiotics falls behind the urgent need for treatment. It is possible to speed up the development of new antibiotics. However, it may make the drug-resistant problem worse in future.
Therefore, the way of developing novel drugs to resist the MRSA needs to be diversified. Some of the drugs should be used to eliminate the MRSA before the infection; some of the drugs ought to be utilized for treatment. Also, the policy should be made for avoiding abuse of antibiotics. To sum up, we have to create various ways to prevent infection and treat the patients at the same time so that we may get the chance to diminish the negative effect from MRSA and other drug-resistant S. aureus.
This review retrospects the background of S. aureus and their drug-resistant problem. The aim of this article is appealing more people to focus on S. aureus. With the bacterial evolution, the drug-resistant S. aureus is spreading wider and wider. For the public, we ought to learn how to prevent the infection from
Table 1. Representative mechanisms of Staphylococcus aureus resistance to antimicrobials [69] [70] .
S. aureus. With regard to researchers, it is urgent to find an appropriate way to control the drug-resistant S. aureus.
Cite this paper
Li, Z. (2018) A Review of Staphylococcus aureus and the Emergence of Drug-Resistant Problem. Advances in Microbiology, 8, 65-76. https://doi.org/10.4236/aim.2018.81006
References
- 1. Kloos, W.E. (1997) Taxonomy and Systematic of Staphylococci Indigenous to Humans. In: Crossley, K.B. and Archer, G.L., Eds., The Staphylococci in Human Disease, Churchill Livingstone, New York, 113-137.
- 2. Wilson, LG. (1987) The Early Recognition of Streptococci as Causes of Disease. Medical History, 31, 403-414. https://doi.org/10.1017/S0025727300047268
- 3. Rosenbach, F.J. (1884) Microorganisms in the Wound Infections Diseases of Man. J.F. Bergmann, Wiesbaden, 18.
- 4. Stryjewski, M.E. and Corey, G.R. (2014) Methicillin-Resistant Staphylococcus aureus: An Evolving Pathogen. Clinical Infectious Diseases, 58, S10-S19. https://doi.org/10.1093/cid/cit613
- 5. Wertheim, H.F., Melles, D.C., Vos, M.C., et al. (2005) The Role of Nasal Carriage in Staphylococcus aureus Infections. The Lancet Infectious Diseases, 5, 751-762. https://doi.org/10.1016/S1473-3099(05)70295-4
- 6. Lowy, F.D. (1998) Staphylococcus aureus Infections. The New England Journal of Medicine, 339, 520-532. https://doi.org/10.1056/NEJM199808203390806
- 7. Peacock, S.J. and Paterson, G.K. (2015) Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annual Review of Biochemistry, 84, 577–601. https://doi.org/10.1146/annurev-biochem-060614-034516
- 8. Tong, S.Y.C., Davis, J.S., Eichenberger, E., et al. (2015) Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clinical Microbiology Reviews, 28, 603-661. https://doi.org/10.1128/CMR.00134-14
- 9. Vandenesch, F., Lina, G. and Henry, T. (2012) Staphylococcus aureus Hemolysins, bi-Component Leukocidins, and Cytolytic Peptides: A Redundant Arsenal of Membrane-Damaging Virulence Factors? Frontiers in Cellular and Infection Microbiology, 2, 2-13. https://doi.org/10.3389/fcimb.2012.00012
- 10. Costerton, J.W., Lewandowski, Z., Caldwell, D.E., et al. (1995) Microbial Biofilms. Annual Review of Microbiology, 49, 711–745. https://doi.org/10.1146/annurev.mi.49.100195.003431
- 11. Otto, M. (2013) Staphylococcal Infections: Mechanisms of Biofilm Maturation and Detachment as Critical Determinants of Pathogenicity. Annual Review of Medicine, 64, 175-188. https://doi.org/10.1146/annurev-med-042711-140023
- 12. Amulic, B., Cazalet, C., Hayes, G.L., Metzler, K.D. and Zychlinsky, A. (2012) Neutrophil Function: From Mechanisms to Disease. Annual Review of Immunology, 30, 459-489. https://doi.org/10.1146/annurev-immunol-020711-074942
- 13. Spaan, A.N., Surewaard, B.G.J., Nijland, R., et al. (2013) Neutrophils versus Staphylococcus aureus: A Biological Tug of War. Annual Review of Microbiology, 67, 629-650. https://doi.org/10.1146/annurev-micro-092412-155746
- 14. Greenlee-Wackera, M., DeLeod, F.R. and Nauseef, W.M. (2015) How Methicillin-Resistant Staphylococcus aureus Evade Neutrophil Killing. Wolters Kluwer Health, 22, 31-35.
- 15. Boucher, H.W. and Corey, G.R. (2008) Epidemiology of Methicillin-Resistant Staphylococcus aureus. Clinical Infectious Diseases, 46, S344-S349. https://doi.org/10.1086/533590
- 16. Archer, G.L. (1998) Staphylococcus aureus: A Well-Armed Pathogen. Clinical Infectious Diseases, 26, 1179-1181. https://doi.org/10.1086/520289
- 17. Baker, A.D.L. and Macnicol, M.F. (2008) Haematogenous Osteomyelitis in Children: Epidemiology, Classification, Aetiology and Treatment. Journal of Paediatrics and Child Health, 18, 75-84. https://doi.org/10.1016/j.paed.2007.11.002
- 18. Shorr, A.F., Tabak, Y.P., Gupta, V., et al. (2006) Morbidity and Cost Burden of Methicillin-Resistant Staphylococcus aureus in Early Onset Ventilator Associated Pneumonia. Critical Care, 10, R97. https://doi.org/10.1186/cc4934
- 19. Lambert, M.L., Suetens, C., Savey, A., et al. (2011) Clinical Outcomes of Health-Care-Associated Infections and Antimicrobial Resistance in Patients Admitted to European Intensive-Care Units: A Cohort Study. The Lancet Infectious Diseases, 11, 30-38. https://doi.org/10.1016/S1473-3099(10)70258-9
- 20. Cosgrove, S.E., Qi, Y., Kaye, K.S., et al. (2005) The Impact of Methicillin Resistance in Staphylococcus aureus Bacteremia on Patient Outcomes: Mortality, Length of Stay, and Hospital Charges. Infection Control and Hospital Epidemiology, 26, 166-174. https://doi.org/10.1086/502522
- 21. McHugh, C.G. and Riley, L.W. (2004) Risk Factors and Costs Associated with Methicillin-Resistant Staphylococcus aureus bloodstream infections. Infection Control and Hospital Epidemiology, 25, 425-430. https://doi.org/10.1086/502417
- 22. Klevens, R.M., Morrison, M.A., Nadle, J., et al. (2007) Invasive Methicillin Resistant Staphylococcus aureus Infections in the United States. JAMA, 298, 1763-1771. https://doi.org/10.1001/jama.298.15.1763
- 23. Chang, V.S., Dhaliwal, D.K., et al. (2015) Antibiotic Resistance in the Treatment of Staphylococcus aureus Keratitis: A 20-Year Review. Wolters Kluwer Health, 34, 698-703. https://doi.org/10.1097/ICO.0000000000000431
- 24. Paterson, G.K., Harrison, E.M. and Holmes, M.A. (2014) The Emergence of mecC Methicillin-Resistant Staphylococcus aureus. Trends in Microbiology, 22, 42-47. https://doi.org/10.1016/j.tim.2013.11.003
- 25. National Nosocomial Infections Surveillance (NNIS) (2004) System Report, Data Summary from January 1992 through June 2004, Issued October 2004. American Journal of Infection Control, 32, 470-485. https://doi.org/10.1016/j.ajic.2004.10.001
- 26. Chambers, H.F. (2001) The Changing Epidemiology of Staphylococcus aureus? Emerging Infectious Diseases, 7, 178-182. https://doi.org/10.3201/eid0702.010204
- 27. Grundmann, H., Aires-de-Sousa, M., Boyce, J., et al. (2006) Emergence and Resurgence of Meticillin-Resistant Staphylococcus aureus as a Public-Health Threat. The Lancet, 368, 874-885. https://doi.org/10.1016/S0140-6736(06)68853-3
- 28. Centers for Disease Control and Prevention (1999) Four Pediatric Deaths from Community-Acquired Methicillin-Resistant Staphylococcus aureus-Minnesota and North Dakota, 1997-1999. JAMA, 282, 1123-1125.
- 29. Herold, B.C., Immergluck, L.C., Maranan, M.C., et al. (1998) Community-Acquired Methicillin-Resistant Staphylococcus aureus in Children with No Identified Predisposing Risk. JAMA, 279, 593-598. https://doi.org/10.1001/jama.279.8.593
- 30. Boye, K., Bartels, M.D., Andersen, I.S., et al. (2007) A New Multiplex PCR for Easy Screening of Methicillin-Resistant Staphylococcus aureus SCCmec Types I-V. Clinical Microbiology and Infection, 13, 725-727. https://doi.org/10.1111/j.1469-0691.2007.01720.x
- 31. Gostev, V.V. and Sidorenko, S.V. (2012) Staphylococcal Cassette Chromosome Mec, Evolution and Genetic Lines of Methicillin-Resistant Staphylococcus aureus. Antibiotiki i khimioterapiia, 57, 38-46.
- 32. Al-Zubeidi, D., Hogan, P.G., Boyle, M., et al. (2014) Molecular Epidemiology of Methicillin-Resistant Staphylococcus aureus Isolated in Serial Cultures from the Respiratory Tract of Children with Cystic Fibrosis. Pediatric Infectious Disease Journal, 33, 549-553. https://doi.org/10.1097/INF.0000000000000204
- 33. Anonymity (2010) MRSA Infections Decline, But New Bacterial Strains Threaten. You Can Take Action to Protect Yourself against Drug-Resistant Bacteria. Duke Medicine Health News, 16, 7.
- 34. Perencevich, E.N. and Diekema, D.J. (2010) Decline in Invasive MRSA Infection: Where to Go from Here? JAMA, 304, 687-689. https://doi.org/10.1001/jama.2010.1125
- 35. Usui, J., Kobayashi, M., Ebihara, I., et al. (2011) Methicillin-Resistant Staphylococcus-aureus-Associated Glomerulonephritis on the Decline: Decreased Incidence since the 1990s. Clinical and Experimental Nephrology, 15, 184-186. https://doi.org/10.1007/s10157-010-0369-x
- 36. Eells, S.J., Mckinnell, J.A., Wang, A.A., et al. (2013) A Comparison of Clinical Outcomes between Healthcare-Associated Infections Due to Community-Associated Methicillin-Resistant Staphylococcus aureus Strains and Healthcare-Associated Methicillin-Resistant S. aureus Strains. Epidemiology and Infection, 141, 1-14. https://doi.org/10.1017/S0950268812002634
- 37. Gottlieb, T., Su, W.Y., Merlino, J., et al. (2008) Recognition of USA300 Isolates of Community-Acquired Methicillin-Resistant Staphylococcus aureus in Australia. The Medical Journal of Australia, 189, 179-180.
- 38. Tenover, F.C. and Goering, R.V. (2009) Methicillin-Resistant Staphylococcus aureus Strain USA300: Origin and Epidemiology. Journal of Antimicrobial Chemotherapy, 64, 441-446. https://doi.org/10.1093/jac/dkp241
- 39. Larsen, A.R., Bocher, S., Stegger, M., et al. (2008) Epidemiology of European Community-Associated Methicillin-Resistant Staphylococcus aureus Clonal Complex 80 Type IV Strains Isolated in Denmark from 1993 to 2004. Journal of Clinical Microbiology, 46, 62-68. https://doi.org/10.1128/JCM.01381-07
- 40. Moellering, R.C. (2010) Why Has Methicillin-Resistant Staphylococcus aureus Become Such a Successful Pathogen in Adults? Infectious Diseases in Clinical Practice, 18, 286-291. https://doi.org/10.1097/IPC.0b013e3181efebca
- 41. Uhlemann, A.-C., Otto, M., Lowy, F.D., et al. (2014) Evolution of Community- and Healthcare-Associated Methicillin-Resistant Staphylococcus aureus. Infection, Genetics and Evolution, 21, 563-574. https://doi.org/10.1016/j.meegid.2013.04.030
- 42. Levine, D.P., Cushing, R.D., Jui, J., et al. (1982) Community-Acquired Methicillin-Resistant Staphylococcus aureus Endocarditis in the Detroit Medical Center. Annals of Internal Medicine, 97, 330-338. https://doi.org/10.7326/0003-4819-97-3-330
- 43. Udo, E.E., Pearman, J.W. and Grubb, W.B. (1993) Genetic Analysis of Community Isolates of Methicillin-Resistant Staphylococcus aureus in Western Australia. Journal of Hospital Infection, 25, 97-108. https://doi.org/10.1016/0195-6701(93)90100-E
- 44. Dufour, P., Gillet, Y., Bes, M., et al. (2002) Community-Acquired Methicillin-Resistant Staphylococcus aureus Infections in France: Emergence of a Single Clone That Produces Panton-Valentine Leukocidin. Clinical Infectious Diseases, 35, 819-824. https://doi.org/10.1086/342576
- 45. Ma, X.X., Galiana, A., Pedreira, W., et al. (2005) Community-Acquired Methicillin-Resistant Staphylococcus aureus, Uruguay. Emerging Infectious Diseases, 11, 973-976. https://doi.org/10.3201/eid1106.041059
- 46. Wang, C.C., Lo, W.T., Chu, M.L., et al. (2004) Epidemiological Typing of Community-Acquired Methicillin-Resistant Staphylococcus aureus Isolates from Children in Taiwan. Clinical Infectious Diseases, 39, 481-487. https://doi.org/10.1086/422642
- 47. Pallin, D.J., Egan, D.J., Pelletier, A.J., et al. (2008) Increased US Emergency Department Visits for Skin and Soft Tissue Infections, and Changes in Antibiotic Choices, during the Emergence of Community-Associated Methicillin-Resistant Staphylococcus aureus. Annals of Emergency Medicine, 51, 291-298. https://doi.org/10.1016/j.annemergmed.2007.12.004
- 48. Moran, G.J., Krishnadasan, A., Gorwitz, R.J., et al. (2006) Methicillin-Resistant S. aureus Infections among Patients in the Emergency Department. New England Journal of Medicine, 355, 666-674. https://doi.org/10.1056/NEJMoa055356
- 49. Nimmo, G.R. (2012) USA300 Abroad: Global Spread of a Virulent Strain of Community-Associated Methicillin-Resistant Staphylococcus aureus. Clinical Microbiology and Infection, 18, 725-734. https://doi.org/10.1111/j.1469-0691.2012.03822.x
- 50. Thurlow, L.R., Joshi, G.S. and Richardson, A.R. (2012) Virulence Strategies of the Dominant USA300 Lineage of Community Associated Methicillin Resistant Staphylococcus aureus (CA-MRSA). FEMS Immunology & Medical Microbiology, 65, 5-22. https://doi.org/10.1111/j.1574-695X.2012.00937.x
- 51. King, M.D., Humphrey, B.J., Wang, Y.F., et al. (2006) Emergence of Community-Acquired Methicillin-Resistant Staphylococcus aureus USA 300 Clone as the Predominant Cause of Skin and Soft-Tissue Infections. Annals of Internal Medicine, 144, 309-317. https://doi.org/10.7326/0003-4819-144-5-200603070-00005
- 52. Reyes, J., Rincon, S., Diaz, L., et al. (2009) Dissemination of Methicillin-Resistant Staphylococcus aureus USA300 Sequence Type 8 Lineage in Latin America. Clinical Infectious Diseases, 49, 1861-1867. https://doi.org/10.1086/648426
- 53. Hota, B., Lyles, R., Rim, J., et al. (2011) Predictors of Clinical Virulence in Community-Onset Methicillin-Resistant Staphylococcus aureus Infections. Clinical Infectious Diseases, 53, 757-765. https://doi.org/10.1093/cid/cir472
- 54. Dantes, R., Mu, Y., Belflower, R., et al. (2013) National Burden of Invasive Methicillin-Resistant Staphylococcus aureus Infections, United States, 2011. JAMA Internal Medicine, 173, 1970-1978.
- 55. Nimmo, G.R., Pearson, J.C., Collignon, P.J., et al. (2011) Antimicrobial Susceptibility of Staphylococcus aureus Isolated from Hospital Inpatients, 2009: Report from the Australian Group on Antimicrobial Resistance. Communicable Diseases Intelligence, 35, 237-243.
- 56. Styers, D., Sheehan, D.J., Hogan, P., et al. (2006) Laboratory-Based Surveillance of Current Antimicrobial Resistance Patterns and Trends among Staphylococcus aureus: 2005 Status in the United States. Annals of Clinical Microbiology and Antimicrobials, 5, 1-9. https://doi.org/10.1186/1476-0711-5-1
- 57. Public Health England (2013) Voluntary Reporting of Staphylococcus aureus Bacteraemia in England, Wales and Northern Ireland, 2012. Health Protection Report.
- 58. Huang, Y.-C. and Chen, C.-J. (2011) Community-Associated Meticillin-Resistant Staphylococcus aureus in Children in Taiwan, 2000s. International Journal of Antimicrobial Agents, 38, 2-8. https://doi.org/10.1016/j.ijantimicag.2011.01.011
- 59. Otter, J.A. and French, G.L. (2010) Molecular Epidemiology of Community-Associated Meticillin-Resistant Staphylococcus aureus in Europe. Lancet Infectious Diseases, 10, 227-239. https://doi.org/10.1016/S1473-3099(10)70053-0
- 60. Rammelkamp, C.H. and Maxon, T. (1942) Resistance of Staphylococcus aureus to the Action of Penicillin. Experimental Biology and Medicine, 51, 386-389. https://doi.org/10.3181/00379727-51-13986
- 61. Bondi, A. and Dietz, C.C. (1945) Penicillin Resistant Staphylococci. Experimental Biology and Medicine, 60, 55-58. https://doi.org/10.3181/00379727-60-15089
- 62. Kirby, W.M. (1944) Extraction of a Highly Potent Penicillin Inactivator from Penicillin Resistant Staphylococci. Science, 99, 452-453. https://doi.org/10.1126/science.99.2579.452
- 63. Finland, M. (1955) Emergence of Antibiotic-Resistant Bacteria. New England Journal of Medicine, 253, 909-922. https://doi.org/10.1056/NEJM195511242532105
- 64. Brumfitt, W. and Hamilton-Miller, J. (1989) Methicillin-Resistant Staphylococcus aureus. New England Journal of Medicine, 320, 1188-1196. https://doi.org/10.1056/NEJM198905043201806
- 65. Jessen, O., Rosendal, K., Bulow, P., Faber, V., Eriksen, K.R. (1969) Changing Staphylococci and Staphylococcal Infections. A Ten-Year Study of Bacteria and Cases of Bacteremia. New England Journal of Medicine, 281, 627-635. https://doi.org/10.1056/NEJM196909182811201
- 66. Jevons, M.P. (1961) “Celbenin”-Resistant Staphylococci. British Medical Journal, 1, 124-125. https://doi.org/10.1136/bmj.1.5219.124-a
- 67. Stefani, S., Campanile, F., Santagati, M., et al. (2015) Insights and Clinical Perspectives of Daptomycin Resistance in Staphylococcus aureus: A Review of the Available Evidence. International Journal of Antimicrobial Agents, 8, 1-12.
- 68. Centers for Disease Control and Prevention (CDC) (2002) Staphylococcus aureus Resistant to Vancomycin—United States, 2002. MMWR. Morbidity and Mortality Weekly Report, 51, 565-567.
- 69. Lowy, F.D. (2003) Antimicrobial Resistance: The Example of Staphylococcus aureus. Journal of Clinical Investigation, 111, 1265-1273. https://doi.org/10.1172/JCI18535
- 70. Malachowa, N. and DeLeo, F.R. (2010) Mobile Genetic Elements of Staphylococcus aureus. Cellular and Molecular Life Sciences, 67, 3057-3071. https://doi.org/10.1007/s00018-010-0389-4




