Advances in Microbiology
Vol.3 No.4(2013), Article ID:35524,16 pages DOI:10.4236/aim.2013.34049
Genomic Analysis of Anabaena variabilis Mutants PK17 and PK84 That Are Characterised by High Production of Molecular Hydrogen
1Faculty of Biology, Moscow State University, Moscow, Russia
2Centre “Bioengineering”, Russian Academy of Sciences, Moscow, Russia
Email: *shestakovgen@mail.ru
Copyright © 2013 Sergey V. Shestakov et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 7 June, 2013; revised 7 July, 2013; accepted 15 July, 2013
Keywords: Cyanobacteria; Hydrogen Metabolism; Hydrogenases; Pyrosequencing; Mutants; Anabaena
ABSTRACT
The use of cyanobacteria for producing molecular hydrogen is one of the desirable tasks of photobiotechnology. Some years ago, we isolated several chemically induced mutants of the cyanobacterium Anabaena variabilis ATCC 29413 that exhibited a high level of H2-production; but the genetic nature of these mutants remained unresolved. To reveal mutations that could be responsible for enhancement of H2-production in two independent mutants, PK17 and PK84, the pyrosequencing of their entire genomes was performed. The results were analyzed on the basis of comparison with the complete genome sequence of the reference strain Anabaena variabilis ATCC 29413. The genomes of mutants PK17 and RK84 contain 107 and 104 point deviations from the reference genome, respectively. The most probable reason for the increase of H2-production in mutant PK17 is the mutation identified in the gene hupL encoding the large subunit of uptake hydrogenase. A high level of H2-production in mutant PK84 could be the result of a mutation in a conserved part of the gene hypF, which participates in the post-translation maturation of hydrogenase complexes.
1. Introduction
The development of method of exploiting cyanobacteria for H2-production on the basis of bioconversion of solar energy is a desirable task of photobiotechnology [1-3]. In heterocystous cyanobacteria, molecular hydrogen is produced by nitrogenase and reutilised by uptake (Hup) hydrogenase in heterocysts. Bidirectional (Hox) hydrogenase, which functions in heterocysts and vegetable cells, catalyses a reversible evolution/uptake reaction [3,4]. Hydrogen metabolism is a part of a complex gene network that coordinates processes of photosynthetic and respiration electron transport, carbon assimilation, redox cycle, sulfur and metal metabolism [5]. Therefore, investigation of the influence of mutations in genes involved in this network on the level of H2-production is important for the genetic creation of efficient producers of molecular hydrogen.
Some years ago, we isolated two novel mutants of Anabaena variabilis ATCC 29413 that exhibited high H2-production in the air atmosphere. The mutants were obtained by chemical mutagenesis of the wild-type strain using N-methyl-N-nitro-N-nitrosoguanidine (NTG) and selection of slower growing colonies on agar medium with limited concentrations of ammonium or nitrate [6,7]. One of them, PK84 with reduced activity of both (Hup and Hox) hydrogenases, was successively used for cultivation in photobioreactors [8,9]. The genetic nature of the mutations has remained unresolved. Proceeding from a key function of uptake hydrogenase in the recycling of hydrogen released by nitrogenase, a number of H2-producing mutants of heterocystous cyanobacteria were created by insertional mutagenesis of hupSL-genes [10-14]. However, most of these mutants were characterised by a lower degree of H2-production in comparison with the mutant PK84 [4,9].
By using a current high-throughput technique of pyrosequencing, we have attempted to reveal mutations that might be responsible for increased levels of hydrogen production in the PK17 and PK84 mutants. The results of re-sequencing of the genomes have been analysed on the basis of comparison with the complete genome sequence of the reference strain Anabaena variabilis ATCC 29413 (US DOE Joint Genome Institute, http://genome.jgi.doe.gov/anava/anava.info.html). The information obtained suggests that the most probable reason for the enhancement of H2-production in PK17 is a mutation in the hupL gene encoding the large subunit of uptake hydrogenase, while a high level of H2-production in mutant PK84 is associated with mutation of the hydrogenase accessory gene hypF, which is involved in the maturation of active complexes of both hydrogenases.
2. Materials and Methods
2.1. Strains and Growth Conditions
The wild-type strain of the filamentous heterocystous cyanobacterium Anabaena variabilis ATCC 29413 was kindly provided by Dr. C. P. Wolk in 1977. Since that time, this strain was cultivated in our laboratory with periodical passages on agar slices and plates. This strain will be hereafter named as A. variabilis strain PW to distinguish it from the reference wild-type strain Anabaena variabilis ATCC 29413 (A. variabilis strain TT), which was derived from the laboratory of Dr. T. Thiel for sequencing experiments in the US DOE Joint Genome Institute in 2005. (http://genome.jgi.doe.gov/anava/anava.info.html)
Two previously obtained H2-producing mutants of A. variabilis ATCC 29413, PK17 and PK84 [6,7], were chosen for whole-genomic analysis. Levels of their nitrogenase activity and H2-production are shown in Table S1.
For isolating of genomic DNA for pyrosequencing cultures were grown in BG11 medium [15] under fluorescent light (40 µE·m−2·s−1) at 28˚C - 30˚C in 1-L flasks containing 400 ml of magnetically stirred culture. Under these conditions the liquid cultures grew logarithmically with a generation time of approximately 20 - 24 h and after 7 - 10 days of growth contain 80 - 100 µg·ml−1 of protein. The total protein concentration in culture suspension was determined after the alkaline lysis of cells by a modification of the Lowry method.
2.2. Isolation of genomic DNA
Cultures of the wild-type A. variabilis strain PW and mutants PK17 and PK84 grown in BG11 medium for 7 - 10 days were harvested by centrifugation at 2800 g for 20 min, washed with 10 mM Tris∙HCl buffer at pH 8.0 and frozen at −20˚C overnight. The samples of genomic DNA were isolated according to the manufacturer’s specifications in the QIAamp DNA Stool Mini Kit (QIAGEN, Germany). The concentration of DNA was determined on a NanoDrop (ND 1000) micro volume spectrophotometer (PEQLAB, Germany). All DNA preparations were characterised by an average fragment length not less than 20 kb and had a OD260/OD280 ratio of about 2.05 ÷ 2.1.
2.3. Genome Sequencing and Analysis
Genomes of Anabaena variabilis ATCC 29413 strain PW, and mutants PK17 and PK84 were sequenced on a Roche GS FLX genome sequencer using the Titanium protocol for a shotgun genome library. The GS FLX runs resulted in the generation about 106 - 120 MB per genome, which corresponds to about 15 - 20 times genome coverage. For each analysed genome, the reads were mapped on the reference genome of Anabaena variabilis ATCC 29413 strain TT, available in GenBank (NC_007413, NC_007410, NC_007411 and NC_007412 for the chromosome, plasmid A, plasmid B and plasmid C, respectively) using the GS Reference mapper program (Roche, Switzerland). Only point differences supported by more then 80% of reads were recorded. Genome rearrangements were not found.
3. Results and Discussion
3.1. Mutations Revealed in Genomes of Mutants PK17 and PK84
Whole-genomic pyrosequencing has shown that A. variabilis ATCC 29413 strain PW studied in our laboratory differs from the reference A. variabilis ATCC 29413 strain TT in 14 positions in the chromosome (Tables 1, S2). One mutation was revealed in plasmid pAvaA, one of three plasmids reported in the A. variabilis ATCC 29413 strain TT (EMBL/GeneBank/DDBJ database). In strain PW and mutants PK17 and PK84, we found an additional plasmid named pAvaD (27501 bp), which structure that is described separately [16].
In comparison with the genome of the reference strain TT, the genome of mutant PK17 (Tables 1, S3) contains 107 differences including 97 in chromosome. The genome of mutant PK84 has 104 differences (including 96 in chromosome) from the reference strain TT (Tables 1, S4). A number of mutations, 26 in PK17 and 41 in PK84, were found in noncoding regions or in sites of synonymic substitutions of the chromosomes. Several mutations were revealed in plasmids A and C. Only two deviations from the reference sequence were present simultaneously in the genomes of A. variabilis ATCC 29413 strain PW and both mutants (Tables S2-S4), suggesting that most of the sequence variations between A. variabilis ATCC 29413 strains PW and TT had accumulated in the course of independent laboratory cultivation of these strains apparently after obtaining the PK17 and PK84 mutants. The number of variations between the genomes of strains PW and TT is similar to that found between the labora-
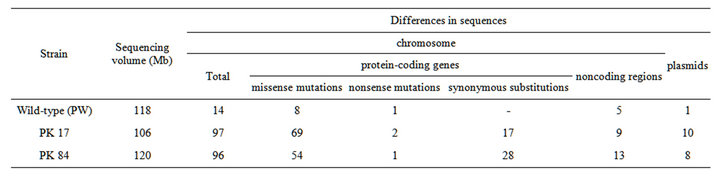
Table 1. Comparison of the genomes of the wild-type A. variabilis ATCC 29413 strain PW and NTG-induced mutants with the reference genome of A. variabilis ATCC 29413 strain TT.
tory strains of Synechocystis sp. PCC 6803, derived from a common ancestor and cultivated independently [17].
The genomes of strains PK17 and PK84 contain missense and nonsense mutations in 74 and 58 putative genes, respectively (Tables S3, S4). In the PK17 mutant, nonsynonymous substitutions were localised in 23 genes encoding hypothetical proteins or products with unknown functions. The genome of mutant PK84 contains mutations in 17 genes of this type (Table 2). Nonsynonymous substitutions were revealed mostly in genes coding for histidineand serine/threonine protein kinases, other proteins of signal transduction systems, transporters, various types of transferases and peptidases, enzymes of secondary metabolism and biosynthesis of cell walls. A number of mutations occurred in genes controlling processes of DNA repair, recombination, restriction-modification, translation etc. (Tables 2, S3, S4).
It should be noted that PK17 and PK84 are independently isolated mutants induced by NTG, causing multiple mutations. Therefore, it is not surprising that almost all mutations occurred in different genes (except only two of them) in the genomes of these two mutants. The absence of visible phenotypic alterations in both mutants indicates that mutations in most of the genes are located in functionally insignificant sites or in genes expressed under specific cultural conditions, for example, related to stress-induced adaptive responses. It is also possible that mutations are compensated for by other functionally similar genes. The mutant PK17 showed a decreased rate of growth (nearly 50% of wild-type strain) in nitrogenfree liquid medium and in medium supplemented with NH4, presumably because of its possible deficiency in glutamate synthase activity (Table S3). Preliminary data shows the approximately 50% decreasing of GOGAT enzyme activity in PK17 cells comparing the wild type level. The growth rate of the mutant PK84 in nitrogenfree liquid medium did not differ from the wild type strain [6,7].
3.2. Mutations in Genes Encoding Hup Hydrogenase and HypF Accessory Protein
Some missense mutations that might be related to an increase of H2-production in mutants PK17 and PK84 are represented in Table 3. The most probable gene-candidate for the mutation responsible for the enhancement of H2-production in PK17 is the gene hupL; it codes for the large subunit of uptake hydrogenase (Table 3). A mutation in this gene leads to the replacement of the polar amino acid cysteine with aromatic tyrosine at position 446, which is located in a highly conserved part of the enzyme, as determined by alignment analysis (Figure S1, Table S5). This site is situated near the C-terminal part of the protein and may be functionally important for its conformational state. It cannot be excluded that a mutation in this site prevents the cleavage of 16 terminal amino acids by a specific hupW-endonuclease essential for the maturation of active uptake hydrogenase [18].
The obviously predictive reason leading to the high level of H2-production in PK84 is a mutation in the gene hypF, the product of which plays an essential role in the post-translational modification of uptake hydrogenase as well as bidirectional hydrogenase. Inactivation of hydrogen uptake activity in the hypF-deficient mutant resulted in a dramatic increase in the hydrogen evolution capacity of purple sulfur phototrophic bacterium Thiocapsa roseopersicina under nitrogen-fixing conditions [19]. Deletion and insertion mutants of hypF, hypA1, hypB1, hypC, and other hyp genes of the cyanobacterium Synechocystis sp. PCC 6803 showed no hydrogenase activity [20]. The disruption of the hypF gene in the cyanobacterium Anabaena sp. PCC 7120 also resulted in low Hup and Hox activities without negative effects on growth rate and nitrogenase activity [21].
The substitution of a negatively charged aspartic acid to polar asparagine at position 374 was identified in the middle part of the Hyp F protein of mutant PK84 (Table 3). The mutation is localised in a highly conserved region according to alignment analysis (Figure S2, Table S6). Evident information about the function of this site is absent, but its location in the YrdC-like domain of the HypF protein (region of 213-391 amino acid sequence) indicates its possible participation in RNA binding [22, 23] and/or conversion of a carbamoyl moiety to a carbamoyl adenylate intermediate and allowing the invol-
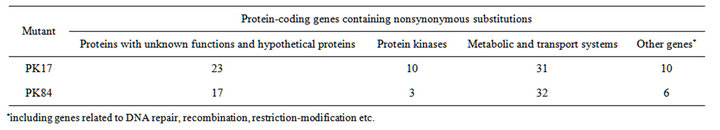
Table 2. Substitutions in the genomes of H2-producing mutants in comparison with the wild-type A. variabilis ATCC 29413 strain TT.
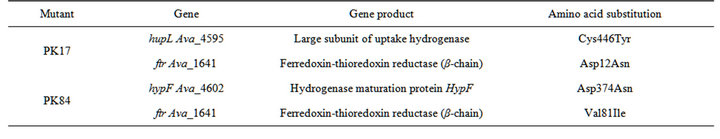
Table 3. Mutations in the PK17 and PK84 genomes that might be related to elevated levels of H2-production.
vement of HypF in the conversion of carbamoyl phosphate to the CO and CN ligands of [NiFe] cluster of hydrogenases. The blockage of such a function must cause repression of the activity of both hydrogenases. Actually, the level of phenazine-methosulfate dependent H2-uptake and activity of bidirectional hydrogenase measured by methylviologen-dependent H2 production were significantly decreased in mutant PK84 [7].
3.3. Mutations that Might be Related to the Network of Hydrogen Metabolism
As mentioned above, the genomes of the PK17 and PK84 mutants contain a large number of mutations in various genes (Tables S3, S4). It is possible that some of these mutations contribute to alterations in H2 photoproduction. A high level of hydrogen release might be caused by combination of hupL and hypF deficiency with other mutations that affect ferredoxin-driven electron transfer to nitrogenase and/or activity of enzymes controlling reductant formation. Hypothetically, this proposal concerns missense mutations in two genes, ftr, coding for the β- chain of ferredoxin-thioredoxine reductase (Table 3), and Ava_3964, coding for hypothetical protein without close homologs in the genomes of cyanobacteria as well as in other prokaryotes. They are possible candidates because mutations in only these genes were found in both mutants. The presence of mutations in ftr and Ava_3946 in these mutants may be just a matter of chance. However, it cannot be excluded that fixation specifically of these mutations in the genomes of both mutants resulted from a particular selection procedure for a decreased ability to reutilize molecular hydrogen produced by nitrogenase. In this case, the consideration of a possible involvement of ftr and Ava_3964 in the control of H2production deserves attention.
Ferredoxin-thioredoxin reductase (FTR) reduces thioredoxins through a disulfide-dithiol interexchange system. Thioredoxins participate in the regulation of several key enzymes of carbon metabolism (including the Calvin-Benson cycle) and some proteins that control redox status and, thus, are involved in the network of hydrogen metabolism [24,25]. Reduced thioredoxin activates uptake hydrogenase at the post-translational level [26]. It was also known that the disulfide redox cascade participates in the assembly of the hydrogenase maturation complex [27]. The active site of the FTR β-chain (13 kDa) comprises a [FeS] cluster and an adjacent redox-active disulfide responsible for interaction with thioredoxins. A mutation in the FTR protein of mutant PK84 was found at position 81 (substitution of valine to isoleucine) within the highly conserved part of the β-chain (Figure S3, Table S7). Mutation in PK17 occurred at position 12 (substitution of negative charged aspartic acid to polar asparagine), which is also localised in the conserved part. It is possible that both mutations in different sites of FTR can potentially lead to a lack/decrease in the activity of FTR, thus, they affect a regulatory function of thioredoxins and correspondingly a gene-metabolic network of hydrogen production.
In order to examine a hypothesis about a connection between the functions of the ftr and H2-photoproduction in A. variabilis, experiments for the construction and studies of mutants impaired in this gene and double mutants containing also hupL− or hypF− mutations have been started.
4. Acknowledgements
This work was supported by the Ministry of Education and Science of Russian Federation (contract 8272) and a Grant for Leading Scientific Schools “NSh-376.2012.4”.
REFERENCES
- R. C. Prince and H. S. Kheshgi, “The Photobiological Production of Hydrogen: Potential Efficiency and Effectiveness as a Renewable Fuel,” Critical Reviews in Microbiology, Vol. 31, No. 1, 2005, pp. 19-31. doi:10.1080/10408410590912961
- D. Dutta, D. De, S. Chaudhuris and S. K. Bhattacharya, “Hydrogen Production by Cyanobacteria,” Microbial Cell Factories, Vol. 4, 2005, pp. 36-46. doi:10.1186/1475-2859-4-36
- P. Tamagnini, E. Leitao, P. Oliveira, D. Ferreira, F. A. L. Pinto, D. J. Harris, T. Heidorn and P. Lindblad, “Cyanobacterial Hydrogenases: Diversity, Regulation and Application,” FEMS Microbiology Reviews, Vol. 31, No. 6, 2007, pp. 692-720. doi:10.1111/j.1574-6976.2007.00085.x
- H. Bothe, O. Schmitz, M. G. Yates and W. E. Newton, “Nitrogen Fixation and Hydrogen Metabolism in Cyanobacteria,” Microbiology Molecular Biology Reviews, Vol. 74, No. 4, 2010, pp. 529-551. doi:10.1128/MMBR.00033-10
- S. V. Shestakov and L. E. Mikheeva, “Genetic Control of Hydrogen Metabolism in Cyanobacteria,” Russian Journal of Genetics, Vol. 42, No. 11, 2006, pp. 1272-1284. doi:10.1134/S1022795406110093
- L. E. Mikheeva, O. A. Koksharova and S. V. Shestakov, “Mutant of the Cyanobacterium Anabaena variabilis ATCC 29413 that Produces Hydrogen,” Vestnik of Moscow University, Section Biology, No. 2, 1994, pp. 54-57 (in Russian).
- L. E. Mikheeva, O. Schmitz, S. V. Shestakov and H. Bothe, “Mutants of the Cyanobacterium Anabaena variabilis Altered in Hydrogenase Activity,” Zeitschrift für Naturforschung, Vol. 50C, 1995, pp. 505-510.
- V. B. Borodin, A. Tsygankov, K. K. Rao and D. O. Hall, “Hydrogen Production by Anabaena variabilis PK84 under Simulated Outdoor Conditions,” Biotechnology and Bioengineering, Vol. 69, No. 5, 2000, pp. 479-485. doi:10.1002/1097-0290(20000905)69:5<478::AID-BIT2>3.0.CO;2-L
- J. Liu, V. E. Bukatin and A. A. Tsygankov, “Light Energy Conversion into H2 by Anabaena variabilis Mutant PK84 Dense Cultures Exposed to Nitrogen Limitation,” International Journal of Hydrogen Energy, Vol. 31, No. 11, 2006, pp. 1591-1596. doi:10.1016/j.ijhydene.2006.06.025
- T. Happe, K. Schutz and H. Bohme, “Transcriptional and Mutational Analysis of the Uptake Hydrogenase of the Filamentous Cyanobacterium Anabaena variabilis,” Journal of Bacterioogy, Vol. 182, No. 6, 2000, pp. 1624-1631. doi:10.1128/JB.182.6.1624-1631.2000
- H. Masukawa, M. Mochimaru and H. Sakurai, “Disruption of the Uptake Hydrogenase Gene, But Not of the Bidirectional Hydrogenase Gene, Leads to Enhanced Photobiological Hydrogen Production by Nitrogen-Fixing Cyanobacterium Anabaena sp. PCC 7120,” Applied Microbiology and Biotechnology, Vol. 58, No. 5, 2002, pp. 618-624. doi:10.1007/s00253-002-0934-7
- P. Lindberg, K. Schutz, T. Happe and P. Lindblad, “A Hydrogen-Producing Hydrogenase-Free Mutant Strain of Nostoc punctiforme ATCC 29133,” International Journal of Hydrogen Energy, Vol. 27, No. 11-12, 2002, pp. 1291- 1296. doi:10.1016/S0360-3199(02)00121-0
- P. Lindblad, K. Christensson, P. Lindberg, A. Fedorov, F. Pinto and A. Tsygankov, “Photoproduction of H2 by Wild Type Anabaena PCC 7120 and a Hydrogen Uptake Deficient Mutant: From Laboratory Experiments to Outdoor Culture,” International Journal of Hydrogen Energy, Vol. 27, No. 11-12, 2002, pp. 1271-1281. doi:10.1016/S0360-3199(02)00111-8
- F. Yoshino, H. Ikeda, H. Masukawa and H. Sakurai, “High Photobiological Hydrogen Production Activity at a Nostoc sp. PCC 7422 Uptake Hydrogenase-Deficient Mutant with High Nitrogenase Activity,” Marine Biotechnology, Vol. 9, No. 1, 2007, pp. 101-112. doi:10.1007/s10126-006-6035-3
- R. Rippka, J. Deruelles, B. Waterbury, M. Herdman and R. Y. Stanier, “Generic Assignment, Strain Histories and Properties of Pure Cultures of Cyanobacteria,” Journal of General Microbiology, Vol. 11, No. 1, 1979, pp. 1-61. doi:10.1099/00221287-111-1-1
- A. V. Mardanov, A. V. Beletsky, V. M. Gumerov, E. A. Karbysheva and L. E. Mikheeva, “The New Low-Copy Number Plasmid of Cyanobacterium Anabaena variabilis,” Genetika, Vol. 49, No. 8, 2013, pp. 921-929 (in Russian).
- Y. Kanesaki, Y. Shiwa, N. Tajima, M. Suzuki, S. Watanabe, N. Sato, M. Ikeuchi and H. Yoshikawa, “Identification of Substrain-Specific Mutations by Massively Parallel Whole-Genome Resequencing of Synechocystis sp. PCC 6803,” DNA Research, Vol. 19, No. 1, 2012, pp. 67- 79. doi:10.1093/dnares/dsr042
- R. Wunschiers, M. Batur and P. Lindblad, “Presence and Expression of Hydrogenase Specific C-Terminal Endopeptidases in Cyanobacteria,” BMC Microbiology, Vol. 3, 2003, pp. 8-20. doi:10.1186/1471-2180-3-8
- B. Fodor, G. Rakhely, A. Kovacs and K. L. Kovacs, “Transposon Mutagenesis in Purple Sulfur Photosynthetic Bacteria: Identification of hypF, Encoding a Protein Capable of Processing [NiFe] Hydrogenases in Alpha, Beta, and Gamma Subdivisions of the Proteobacteria,” Applied and Environmental Microbiology, Vol. 67, No. 6, 2001, pp. 2476-2483. doi:10.1128/AEM.67.6.2476-2483.2001
- D. Hoffmann, K. Gutekunst, M. Klissenbauer, R. SchulzFriedrich and J. Appel, “Mutagenesis of Hydrogenase Accessory Genes of Synechocystis sp. PCC 6803. Additional Homologues of hypA and hypB are not Active in Hydrogenase Maturation,” FEBS Journal, Vol. 273, No. 19, 2006, pp. 4516-4527. doi:10.1111/j.1742-4658.2006.05460.x
- M. Wakai, S. Davar, H. Masukawa and H. Sakurai, “Effects of Disruption of HypF Gene on Photobiological Hydrogen Production in Anabaena sp. PCC 7120,” Abstracts of 13th International Congress on Photosynthesis, Montréal, 28 August-3 september 2004, p. 769.
- S. Petkun, R. Shi, Y. Li, A. Asinas, C. Munger, L. Zhang, M. Waclawek, B. Soboh, G. Sawers and M. Cygler, “Structure of Hydrogenase Maturation Protein HypF Reaction Intermediates Shows Two Active Sites,” Structure, Vol. 19, No. 12, 2011, pp. 1773-1783. doi:10.1016/j.str.2011.09.023
- K. A. Harris, V. Jones, Y. Bilbille, M. A. Swairjo and P. F. Agris, “YrdC Exhibits Properties Expected of a Subunit for a tRNA Threonylcarbamoyl Transferase,” RNA, Vol. 17, No. 9, 2011, pp. 1678-1687. doi:10.1261/rna.2592411
- S. Dai, C. Schwendtmayer, K. Johansson, S. Ramaswamy, P. Schurmann and H. Eklund, “How Does Light Regulate Chloroplast Enzymes? Structure—Function Studies of the Ferredoxin/Thioredoxin System,” Quarterly Reviews of Biophysics, Vol. 33, No. 1, 2000, pp. 67-108. doi:10.1017/S0033583500003607
- M. Lindal and F. J. Florencio, “Thioredoxin-Linked Processes in Cyanobacteria Are as Numerous as in Chloroplasts, But Targets Are Different,” Proceedings of National Academy Sciences of USA, Vol. 100, No. 26, 2003, pp. 16107-16112. doi:10.1073/pnas.2534397100
- H. Papen, T. Kentemich, T. Schmulling and H. Bothe, “Hydrogenase Activities in Cyanobacteria,” Biochemie, Vol. 68, No. 1, 1986, pp. 121-132. doi:10.1016/S0300-9084(86)81077-X
- S. Watanabe, R. Matsumi, T. Arai, H. Atomi, T. Imanaka and K. Miki, “Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling,” Molecular Cell, Vol. 27, No. 1, 2007, pp. 29-40. doi:10.1016/j.molcel.2007.05.039
Supplementary Material
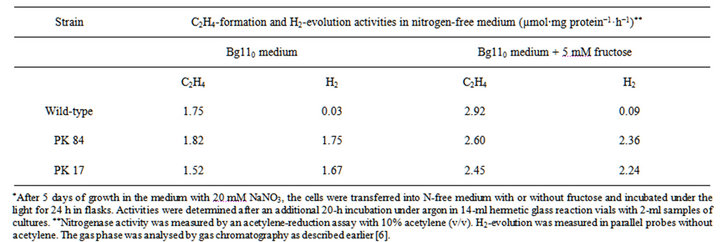
Table S1. . The nitrogenase activity and H2-evolution in the wild-type PW strain and mutants PK 17 and PK 84 in the course of nitrogenase derepression*.
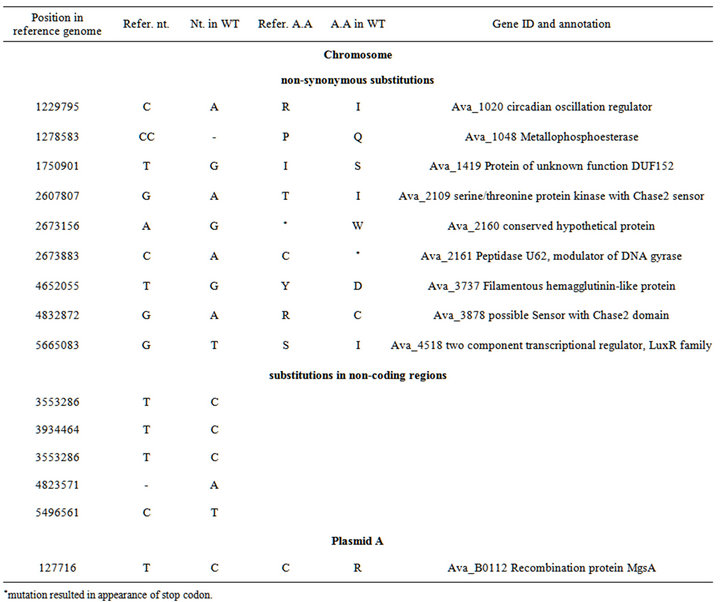
Table S2. . Identified differences between nucleotide sequences of wild-type Anabaena variabilis ATCC 29413 (strain PW) and the reference genome of Anabaena variabilis ATCC 29413 (strain TT).
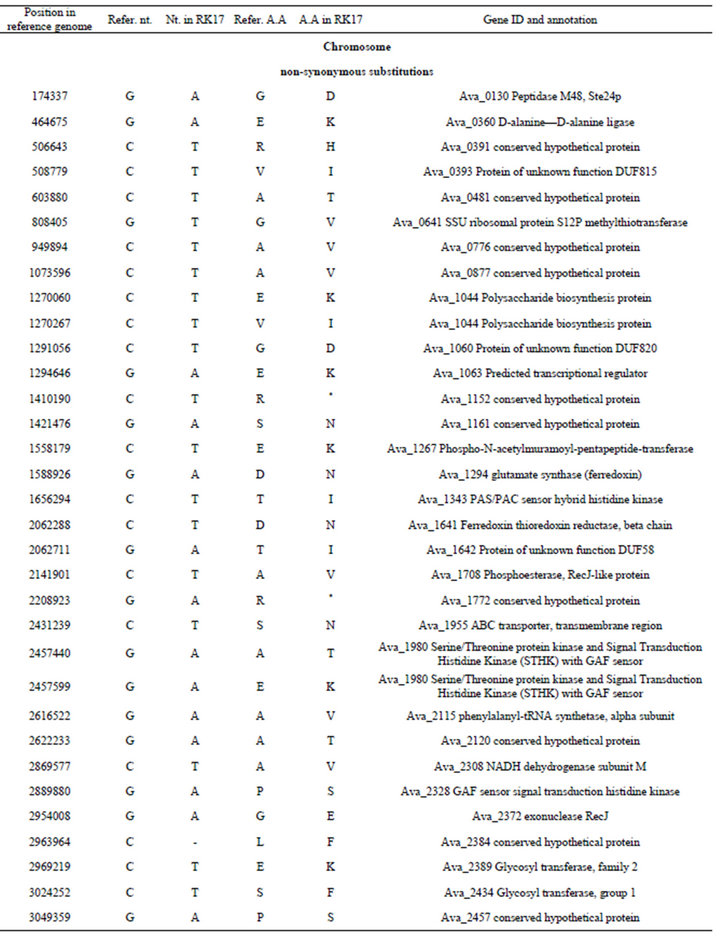
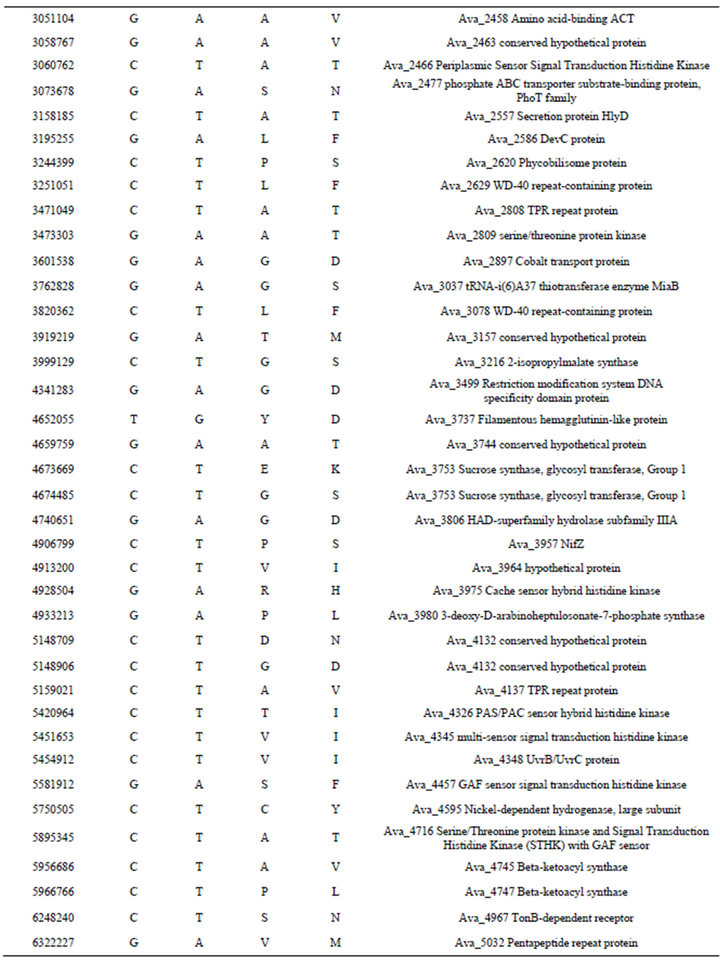
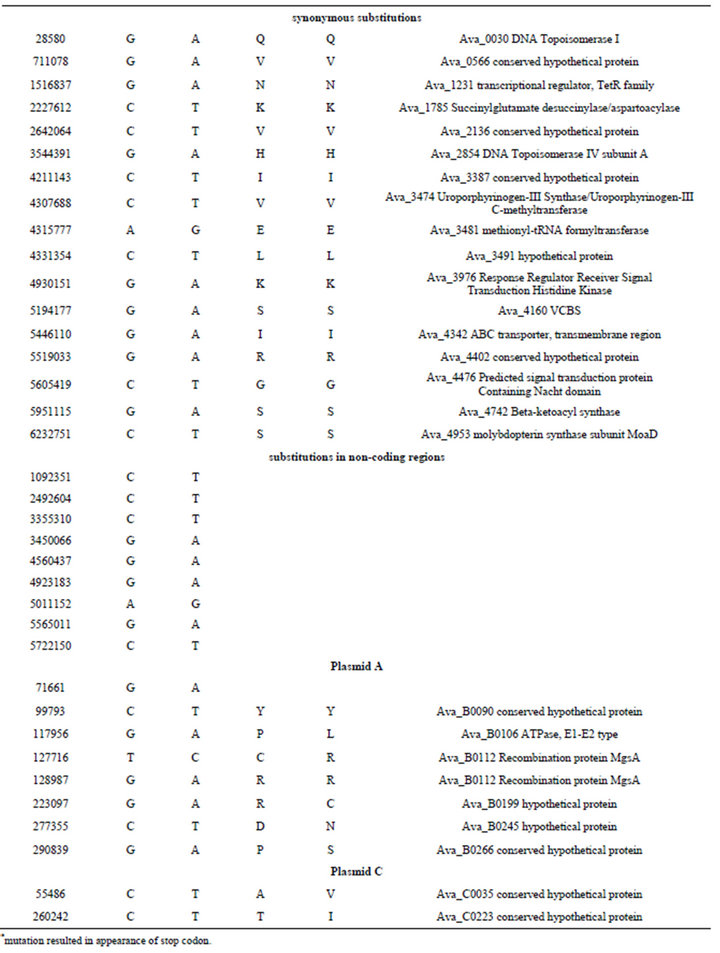
Table S3. . Identified differences between nucleotide sequences of mutant PK17 (strain PW) and the reference genome of Anabaena variabilis ATCC 29413 (strain TT).
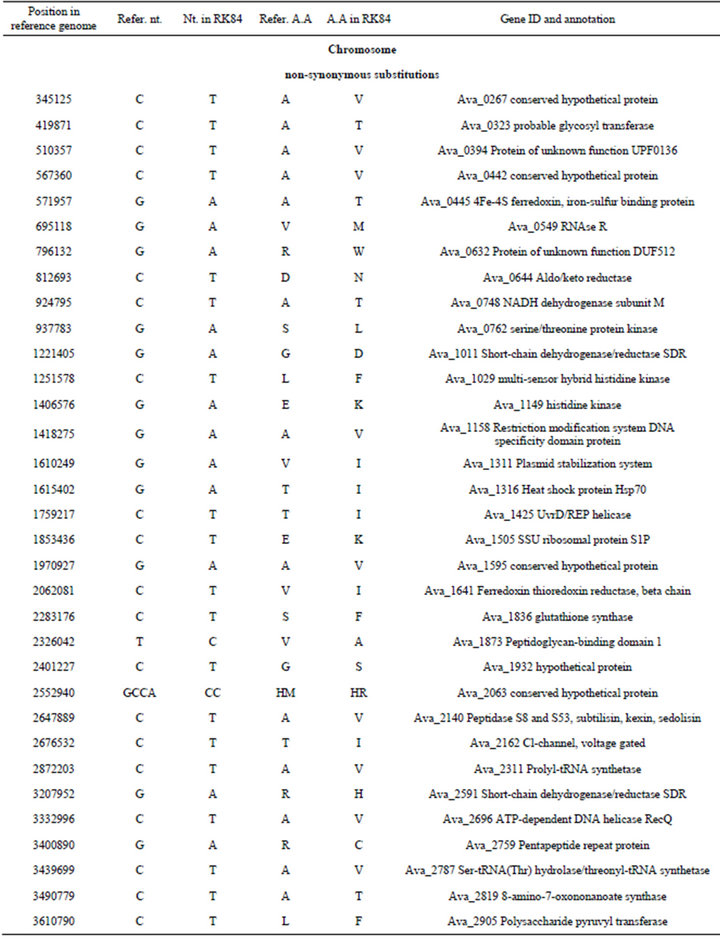
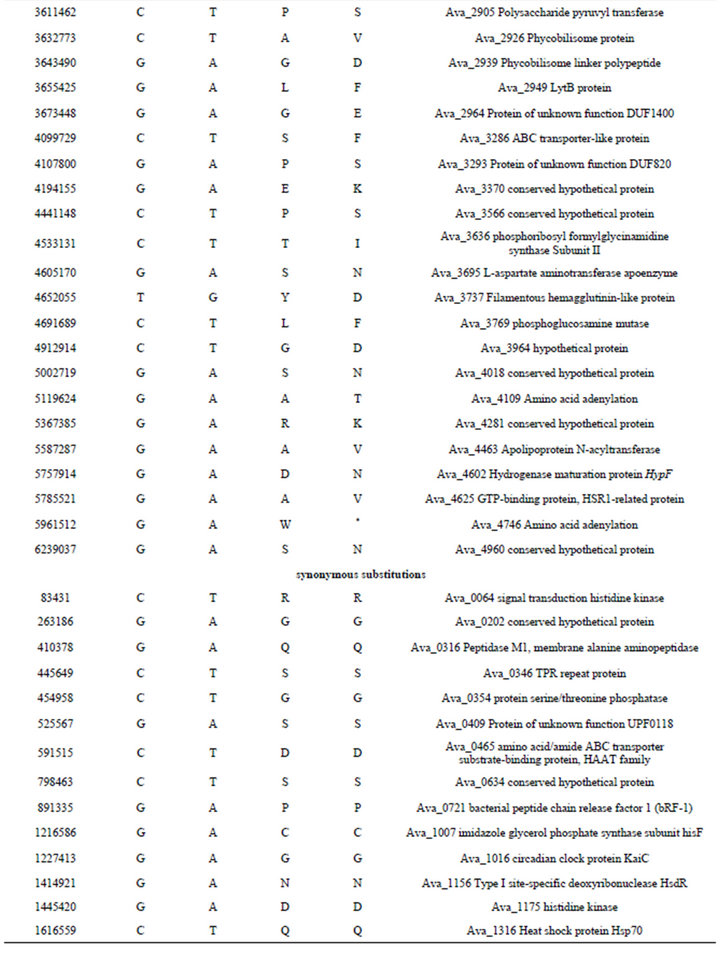
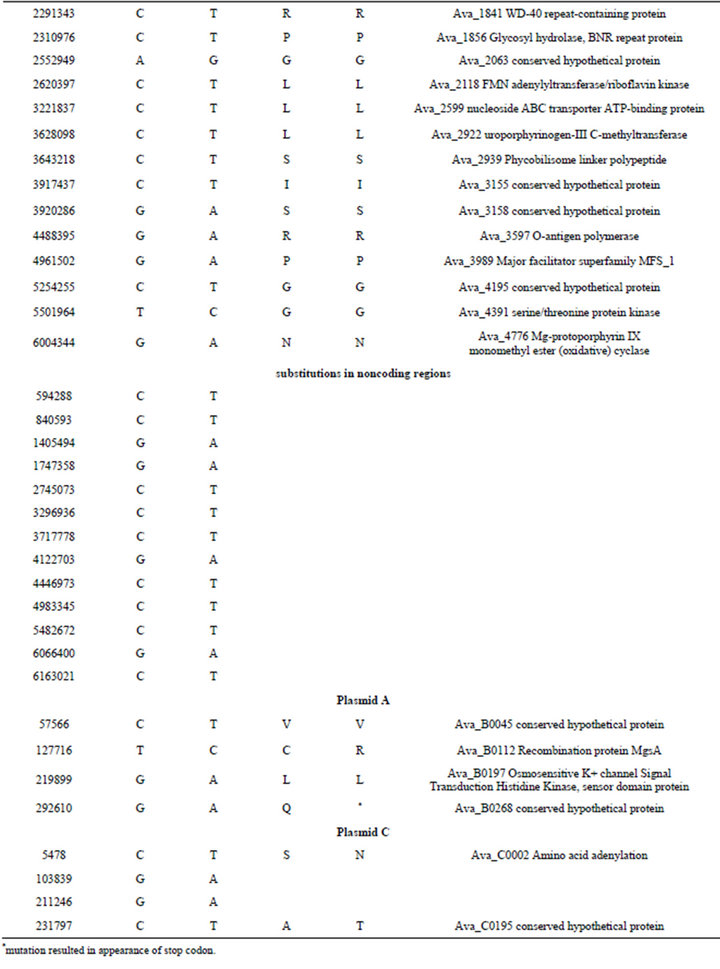
Table S4. .Identified differences between nucleotide sequences of mutant PK84 (strain PW) and the reference genome of Anabaena variabilis ATCC 29413 (strain TT).
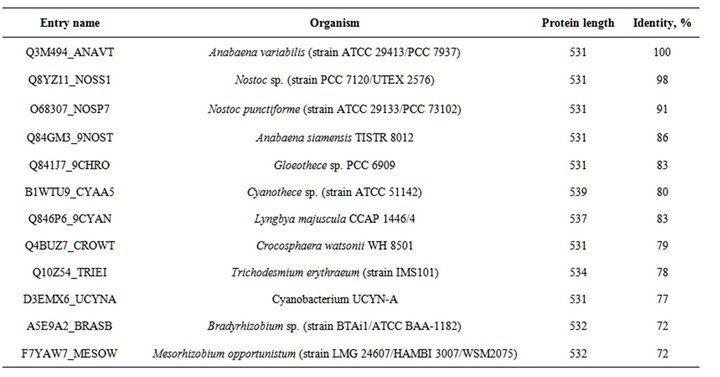
Table S5. . The organisms, proteins and % of identity of enzymes indicated in the Figure S1.
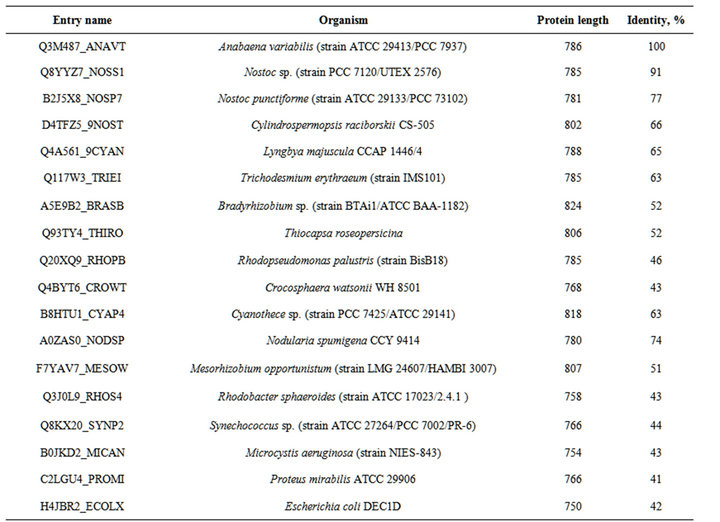
Table S6. . The organisms, proteins and % of identity of enzymes indicated in the Figure S2.
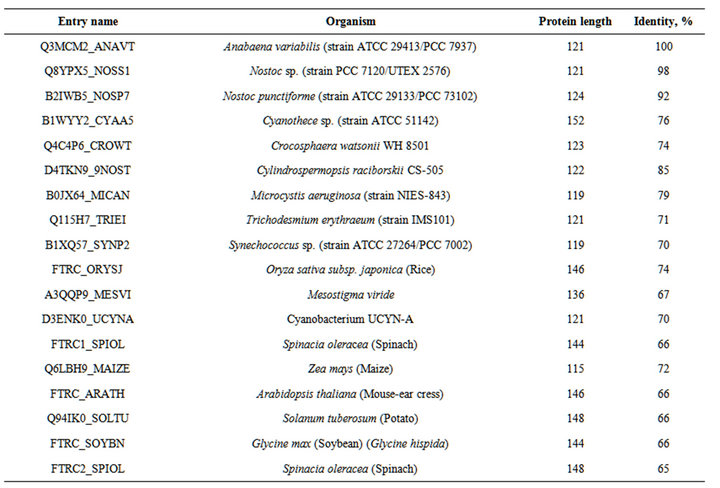
Table S7.. The organisms, proteins and % of identity of enzymes indicated in the Figure S3.

Figure S1. . Alignment of the amino acid sequences of the C-terminal regions of various HupL proteins. Amino acids highlighted in grey are analogues of Cys 446 of HupL of A. variabilis. Residues that are identical (*), conserved (:), or semiconserved (.) in the sequences are indicated below the sequences. Conserved aminoacids of the active sites are shown bold and underlined. The C-end segment that is cleaved by the HupW-endopeptidase is shown in bold italics. (Abbreviations after enzyme sequences are shown in the Table S5).

Figure S2. . Alignment of the amino acid sequences in the middle part of various HypF proteins. Amino acids highlighted in grey are analogues of Asp -374 of Hyp F of A.variabilis. Residues that are identical (*), conserved (:), or semiconserved (.) in the sequences are indicated below the sequences. (Abbreviations after enzyme sequences are shown in the Table S6).

Figure S3. . Alignment of the amino acid sequences of various ferredoxin-thioredoxin reductases, beta chain proteins. Amino acids highlighted in grey are analogues of Asp 12 and Val 81 of A. variabilis Ftr. Residues that are identical (*), conserved (:), or semiconserved (.) in the sequences are indicated below the sequences. (Abbreviations after enzyme sequences are shown in the Table S7).
NOTES
*Corresponding author.

