Open Journal of Medical Microbiology
Vol.05 No.01(2015), Article ID:55073,14 pages
10.4236/ojmm.2015.51006
Molecular and Susceptibility Analysis of Toxigenic Clostridium difficile Obtained from Adult Patients Suspected of CDI in Trinidad
Lemar Wayne Blake1*, Patrick Eberechi Akpaka1, Adesh Ramsubhag2, Reneasha Kevi-Ann Steffi Hitlal-Blake3
1Microbiology & Pathology Unit, Department of Para-Clinical Sciences, The University of the West Indies, St. Augustine, Trinidad & Tobago
2Department of Life Sciences, The University of The West Indies, St. Augustine, Trinidad and Tobago
3Department of Medical Sciences, The University of the West Indies, Mona, Jamaica
Email: *lemar_blake@hotmail.com
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 8 March 2015; accepted 22 March 2015; published 26 March 2015
ABSTRACT
Clostridium difficile is a Gram positive rod-shaped bacterium that produces two major toxins, A and B. The detection of the organism and its toxins has been widely carried out using specialized Enzyme-Linked Immunosorbent Assay (ELISA) kits; however these generally have been unsuccessful in identifying all Clostridium difficile positive samples. In this study, fifteen clinically sym- ptomatic patients from three of the five major regional hospitals in Trinidad were investigated for Clostridium difficile infections. Stool samples were assessed by ELISA and cultured isolates were characterized using agar dilution antibiotic sensitivity assays, conventional Polymerase Chain Reaction (PCR), DNA sequencing and phylogenetic analysis of toxin A and B genes. All 15 patient stool samples and isolates were positive for toxigenic Clostridium difficile via ELISA and PCR respectively. All isolates were positive for the housekeeping tpi and Toxin B genes by PCR but only three of these were positive for the Toxin A gene. The Toxin B gene sequences showed 100% similarity levels among isolates while the Toxin A gene sequences showed 99% similarity among isolates. Phylogenetic analysis showed that the strains isolated in Trinidad most likely belonged to the same strain/group. Agar dilution sensitivity tests showed highest susceptibility to Pipercillin/ Tazobactam and Meropenem (87%) and the highest resistance was seen with Cefotaxime in 93%. These results indicate that similar virulent strains of C. difficile are present in the Trinidad population and that pathogenic strains are more likely to be susceptible to Pipercillin/Tazobactam and Meropenem.
Keywords:
Clostridium difficile Infection (CDI), Toxigenic Clostridium difficile, Toxin A Gene, Toxin B Gene, Housekeeping Gene (tpi), Antibiotic Susceptibility Testing, Phylogenetic, Sequencing

1. Introduction
Clostridium difficile is a spore―forming, anaerobic, Gram―positive rod or bacillus that causes gastrointestinal infections in humans, which range in severity from asymptomatic colonization to chronic diarrhea, toxic megacolon, pseudomembraneous colitis, colonic perforation and death [1] . Clostridium difficile is a part of the normal gut flora; however it can multiply to infectious amounts when competing bacteria are wiped out by antibiotics through collateral damage by such agents [2] . Its major virulence is the large clostridial toxins A and B; these share up to 60% similarities in their amino acid sequence [3] . Affected persons may experience bloating and diarrhea with abdominal pain which may become severe. Significant to clinicians are combined symptoms of intense diarrhea, recent antibiotic exposure, colitis, fever and a foul stool odor [3] . Clostridium difficile is a major cause of nosocomial diarrhea, especially in institutionalized patients over 65 years of age [4] . Episodes of outpatient diarrhea with concomitant antibiotic use (of less than 8 weeks duration) are most often due to Clostridium difficile infection [5] .
The diagnosis of CDAD is dependent on molecular analysis, antigen detection and cytotoxicity tests [6] . Most of the laboratories in Trinidad and Tobago do not use any of these methods; therefore, the number of misdiagnosed cases is likely to be high. Clostridium difficile-associated diarrhea (CDAD) is most strongly linked to fluoroquinolone administration [7] - [11] . Fluoroquinolones are antimicrobial agents that may be used as first-line treat- ment for serious cases of Acute Pyelonephritis or Bacterial Prostatitis [7] . Cancer patients and others treated with fluoroquinolones (e.g. Ciprofloxacin) are indicated for increased risk of C. difficile-associated diarrhea.
Clostridium difficile infections are also proved to be difficult to treat with antimicrobials and many times a less efficacious drug is given; hence its antibiotic susceptibility profile is of massive importance in therapeutics [8] [10] - [12] . Thus far the most common control measure for reduction of associated disease symptoms is withdrawal of antibiotics; however routine treatment may soon include the administration of competing bacteria by fecal transplant [11] - [15] .
Molecular analysis of Clostridium difficile can be carried out in many ways; the most popular is via Polymerase Chain Reaction (PCR) [12] [13] . Using this method, regions of interest on the organism’s DNA are amplified by the use of specific DNA primers. The differences in these amplified products are studied [13] . This method is used for both research and clinical diagnosis. Molecular sequencing and phylogenetic analysis are relatively new techniques that are used in research; these techniques allow for more beneficial information such as: determining genetic similarities and, evolutionary traits among organisms [14] . In this study, we will assess the similarities and antimicrobial susceptibility of the Clostridium difficile isolates in Trinidad.
2. Methods
Ethical approval was obtained from the various health authorities, and the University of the West Indies to collect and process the patient’s samples. Patient’s consent was also obtained; samples were collected between October 2012 and September 2013. A total of 15 patients who were suspected of having CDI were surveyed and their stool samples collected in sterile universal containers (no preservatives added) and analyzed. All subjects were examined once and no follow up was done. Both disease and study factors of interest were assessed in the shortest possible time. The medical histories were taken from the patients by use of a questionnaire.
2.1. Collection, Processing and Culturing of Specimens
The methods used for collection, culturing and processing stool samples and isolates were based on guidelines of the Clinical and Laboratory Standard Institute (CLSI) recommendations for carrying out antimicrobial testing for Gram positive anaerobes [15] -[21] . The detection and confirmation of bacteria were based on morphology, cultural characteristics, Gram stain and molecular analysis.
Diarrheal stool samples were collected from suspected CDI cases after patients gave their consents. Clinical specimens were transported to the Microbiology Laboratory (at the study sites or at the Teaching Laboratory, Pathology/Microbiology Units, Department of Para-Clinical Sciences, the UWI, St. Augustine) for analysis as soon as they were obtained. Specimens were transported in universal containers, enclosed in zip-lock bags; with the appropriate labels and forms. Samples that were not processed immediately were stored at 4˚C or at −70˚C for longer periods. Samples were visually analyzed for their consistency; hard, soft or watery. All samples were plated on Cycloserine Cefoxitin Fructose Agar (CCFA) supplemented with 7% horse blood and incubated for 72 hours under anaerobic conditions. Isolates were stored in Brain Heart Infusion Agar with 25% glycerol at −70˚C.
Clostridium difficile colonies were glossy, grey and circular with a ‘horse stable’ scent. Gram stains were positive for C. difficile characteristic shape.
2.2. Clostridium difficile Glutamate Dehydrogenase Testing
This is an EIA screening method that detects, common C. difficile producing enzyme. This was carried out using the C. diff Quick Check Complete (Techlabs). This test kit was able to detect both the presence of the organism as well as the toxin. A volume of 25 µl of liquid stool was added to a clean 2 ml cryovial; to which 750 µl of diluents and 1 drop of conjugate was added. Formed or semi-solid stool samples are first crushed with a sterile applicator to the appropriate amount. The mixture from the sterile cryovial was thoroughly mixed and 500 µl was transferred to the toxin region on the upper right hand corner of the kit. This was allowed to stand for 15 minutes. At the reaction window 300 µl of wash buffer was added; this was allowed to be absorbed after which 2 drops of substrate was added. The result was read after ten minutes.
2.3. Investigation of Clostridium difficile Toxin via EIA Kit
The commercially available toxin kit (C. diff Quick Check Complete Techlabs, Orlando Florida) was used for toxin detection. The method described in the test kit insert was strictly adhered to. This method is similar to what was seen above in the glutamate dehydrogenase test.
2.4. Quality Control Tests
The toxigenic C. difficile ATCC 9689 strain was subjected to similar analysis as the isolates being tested. This was done to verify the various methods used and to create a standard for analysis. Quality control procedures were carried out for each testing method used. The three major techniques employed for testing samples (EIA, culture and molecular), were selected based on international standards sets by Centre for Disease Control (CDC).
2.5. Toxin and tpi Gene Analysis
DNA Extraction was carried out using the DNeast Blood &Tissue-Qiagen Kit; the instructions were followed according to the user manual.
For DNA Amplification of the tpi housekeeping gene, ten micro liters (10 µl) of the resultant DNA extractions were added to separate PCR tubes containing 15 µl reaction buffer, which contained: 12.5 µl of GoTaq Green 2x master mix (Promega), 0.5 µl of each primer that was used for that particular reaction (Tpi-F and Tpi-R) and 1.5 µl of PCR water. This gave a total of 25 µl per reaction.
For the detection of toxin A and B gene, the same protocol that is used for tpi detection was used. However, the primer sets: NAK 9, NAK 11 and NAK 104 and NAK 105) were respectively used as previously described by others [12] [13] .
2.6. PCR Reaction Mixture Preparation
PCR assay was performed as described previously by Ludovic Leemee et al. [12] , with slight modification; as no glycerol was used in the PCR mixture. GoTaq Green Master Mix contains reaction buffer (pH 8.5), bacterially derived Taq DNA polymerase 0.63 units, 200 µM dATP, 200 µM dGTP, 200 µM dCTP, 200 µM dTTP and 1.5 mM MgCl2. PCR analysis for the toxin A and B genes were carried out by using the following primers NK11 (5_TGATGCTAATAATGAATCTAAAATGGTAAC-3_) and NK9 (5_CCACCAGCTGCAGCCATA_3); for tcdB detection were: NK104 (sequence-5_-GTGTAGCAATGAAAGTCCAAGTTTACGC-3_) and NK105 (se- quence-5_-CACTTACTTCCTACATTATCTGAAGG-3_), as previously reported by [14] . PCR analysis was carried out for the house keeping gene (tpi), which is specific for Clostridium difficile. The following primer set was used tpi-F 5-AAAGAAGCTACTAAGGGTACAAA-3 and tpi-R 5-CATAATATTGGGTCTATTCCTAC-3.
2.7. DNA Amplification Settings
PCR conditions were as follows: denaturation for 3 minutes at 95˚C, 30 seconds at 95˚C, annealing for 1 minute at 55˚C, final extension step of 72˚C for 30 seconds. A total of 35 cycles were performed as previously recommended.
The strain ATCC 9689 was used as a control for the two major toxin genes (A & B) and the house keeping gene.
2.8. DNA Sequencing and Phylogenetic Analysis
Amplified DNA products from the positive samples were sent to Macrogen, South Korea for sequencing of the toxin A and B gene fragments. After trimming either ends of the sequences, DNA sequence alignment and phylogenetic analysis were carried out using BioEdit software and DNA MEGA 6 according to the programs instructions.
2.9. Statistical Analysis
The results were entered in to the SPSS version 17 software (IBM Armonk, NY, USA) and statistical analysis was carried out by using the paired and one sample t-test; with a statistical significant P-value of ≤0.05. Information compared and analyzed were antibiotic history and antibiotic susceptibility. The specificity, sensitivity, turn-around time and cost of the three assays used for sample analysis were recorded. Specificity and sensitivity were measured by the following formulae:
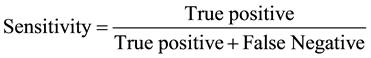
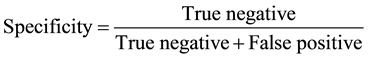
The time it takes to use an assay to process a sample and receiving the result is referred to as the turn-around time. Cost is the money required for one analysis.
3. Antibiotic Susceptibility Testing
Antimicrobial susceptibility of C. difficile was determined by minimum inhibitory concentration obtained by agar dilution method; as recommended by the Clinical and Laboratory Standard Institute (CLSI). A 105 bacteria inocula was prepared by the direct suspension of the bacterial colonies, into 10 ml peptone water, equivalent to a 0.5 McFarlane standard (Biomerieux Vitek Inc., Hazel Hood Missouri). Fresh bacterial colony was taken from that which was grown for a 48 - 72 hour period on the CCFA agar. The suspended colonies were then inoculated at 105 bacteria per spot on the Mueller Hinton supplemented with 7% horse blood and a 2 fold increase in varying antibiotic concentration (µg/mL). Antibiotic concentrations used were: Metronidazole (4, 8, 16, 32, and 64) Ampicillin (2, 4, 8, 16 and 32), Piperacillin-Tazobactam (16/2, 32/4, 64/8, 128/16, and 156/32), Meropenem (2, 4, 8, 16 and 32), Penicillin G (0.25, 0.5, 1, 2, and 4), Ceftriaxone (8, 16, 32, 64 and 32), Cefotaxime (8, 16, 32, 64 and 32) and Ciprofloxacin (1, 2, 4, 8 and 16). A total of 5 different concentrations were used for each antibiotic according to the CLSI recommendations for anaerobes. Inoculated plates were incubated at 37˚C for 48 hours.
4. Results
Table 1 illustrates the amount of toxin positive samples that were obtained from the three hospitals investigated. The highest number of cases came from SFGH, all of which were confirmed the three test methods used: culture EIA and PCR.
Table 2 is showing just how effective the various methods are for detecting and analyzing Clostridium difficile.
N = Total number tested; Pos = Total number positive; Sens = Sensitivity in percentage; Spec = Specificity in percentage; PPV = Positive predictive value; NPV = Negative predictive value; TT Time taken to complete procedure in hrs; Cost = Amount of money for the use of consumables in US dollars; Performance parameter was done at 95% Confidence interval (CI). PCR proved to be the cheapest, effective and less time consuming.
Figure 1 illustrates isolates tested for the tpi gene. Fifteen (15) isolates were tested for the presence of the housekeeping gene (tpi), which is specific for recognizing Clostridium difficile; from this group, 100% (15/15) showed positive tpi genes. PCR analysis was repeated several times to confirm these findings.
Wells 1 - 15 are positive samples, lambdas (λ) is where 100 bp DNA ladders was placed. Wells 16 - 18 are negative controls. Wells 19 and 20 are positive controls. The positive results for the tpi gene in all isolates suggest the absence of DNA degradation or PCR inhibitors.
Figure 2 shows the difference in the toxin A gene fragment obtained from the control strain ATCC 9689 and that obtained from diarrheal patient; the patient sample showed a truncated toxin A gene. Toxin A gene is responsible for toxin A production.
Lambda (λ) refers to the lane in which the 1 kb DNA ladder was loaded. Sample 1 was positive for the Toxin
Table 1. Test methods used for samples that were obtained from the three investigation sites EWMSC, SFGH and POSGH.
EWMSC―Eric Williams Medical Sciences Complex; SFGH―San Fernando General Hospital; POSGH―Port of Spain General Hospital.
Table 2. Comparison of the various test assays used for analyzing the samples received.
Figure 1. The presence of the tpi gene in patient C. difficile samples and control strain ATCC 9689.
Figure 2. PCR of Clostridium difficile toxin A gene from control strain and that isolated from a patient.
A gene. Whereas, samples 2 (negative control) 3 and 4 were negative for the toxin a gene. Lane 5 was that from the control strain ATCC 9689.
Figure 3 shows the similarities between toxin A gene fragments from patient samples.
Lambda (λ) refers to the lane in which the 1 kb DNA ladder was loaded. Lanes 1 - 16 refers to the different columns in which DNA samples were loaded. Samples 1 - 3 and 8 (repeat of sample 1) are positive for the Toxin A gene. Whereas, samples 4 - 7 and 9 - 16 are negative for the toxin A gene. Samples 5 and 6 are negative controls.
Figure 4 compares the toxin A positive sequences obtained from this research, to known toxin A sequences on National Center for Biotechnology Information (NCBI) database. Toxin A gene was positive on DNA molecular sequencing (Macrogen Inc., South Korea) for 3 of the isolates. ClustalW alignment analysis using DNA Bioedit software showed 99% similarities among samples 2, 8 and 13. However slight variations were visible between the samples and a few of the known (NCBI) strains.
Figure 5 uses a phylogenetic tree to group the isolates (3, 8 and 13) and compare them with known Clostridium difficile strains. This shows the strain/group the isolates are most closely related to. Phylogenetic analysis was done using DNA Mega6 software. All the toxin A genes from the isolates were most closely related to the Clostridium difficile 630 strain according to the National Center for Biotechnology Information (NCBI) database; with over 99% similarity.
Figure 6 illustrates isolates tested for the toxin B gene. Fifteen (15) isolates were tested; from this group, 100% (15/15) showed positive toxin B gene fragments. PCR analysis was repeated several times to confirm the findings. Toxin B gene is responsible for toxin B production. Lambda (λ) refers to the lane in which the 100 bp DNA ladder was loaded. Lanes 1 - 17 refers to the different columns in which DNA samples were loaded. Lanes 1 (control). Lanes 1 - 9, 12 - 17 are positive, lanes 10 - 11 are negative controls; (not all the positives are shown in figure). All submitted samples were positive and corresponded to the to the DNA band pattern obtained from the control strain.
Figure 7 compares the toxin B positive sequences obtained from this research are to known toxin B sequences on NCBI database. Toxin B gene was positive on molecular sequencing (Macrogen Inc., South Korea) for all the samples. ClustalW alignment analysis using DNA Bioedit software showed 100% similarities between the toxin B gene fragments from the isolates. Slight differences were however visible between the samples and a few of the control strains.
Figure 3. The presence of toxin A gene from patient samples.
Figure 4. Alignment between Clostridium difficile toxin A gene sequences from samples 3, 8 and 13 vs control strains from NCBI.
Figure 5. Phylogenetic tree comparing isolates to known C. difficile strains.
Figure 6. The presence of toxin B gene from patient isolates.
Figure 7. Alignment between Clostridium difficile toxin B gene sequences from samples and that of Known control strain from Gene Bank (NCBI).
Figure 8 uses a phylogenetic tree to group the isolated isolates and compare them to known Clostridium difficile strains. This shows the strain/group the isolates are most closely related to.
Phylogenetic analysis was done using DNA Mega6 software; all the isolates belonged to the same species and are most closely related to the Clostridium difficile IS25 strain (over 99% similar).
From Figure 9, the analyzed data revealed that 100% of the positive isolates were from patients with a history
Figure 8. Phylogenetic tree comparing isolates to known C. difficile strains.
of antibiotic use. However, the highest number of positive CDI cases came from patients who had used Ceftriaxone and Metronidazole antibiotics. Both antibiotics showed a 35% association in CDI cases, while Ciprofloxacin was 30%.
The antibiotics used are shown in Table 3. Two susceptibility groups were established; sensitive and resistant. Each group consisted of the same 15 isolates. The highest resistance was seen with the antibiotic Cefotaxime and was associated with 93% of the isolates. The highest susceptibility was seen with the antibiotics Piperacillin/Tazobactam and Meropenen; these were associated with 87% of the isolates. The antibiotic sensitive group was compared with the antibiotic resistant group and there was no significant difference between the two groups (p-value = 0.9).
Figure 10 represents a graphical display of the amount (%) of the samples that showed susceptibility and resistant results. The highest resistance frequency was seen with the antibiotic Cefotaxime (93%). Highest susceptibility was seen with the antibiotic Piperacillin/Tazobactam and Meropenem (87% each).
Figure 9. The major antibiotics that were associated with C. difficile infection in patients from the three major regional hospitals in Trinidad 2012-2013.
Table 3. Antibiotic susceptibility pattern of 15 Clostridium difficle isolates (%).
Figure 10. The percentage of isolates that showed susceptibility and resistance to different antibiotics.
5. Discussion
Due to the lack of previously documented research on Clostridium difficile at the three hospitals investigated, accurate comparison of the trend in Clostridium difficile infection (CDI) in Trinidad & Tobago cannot be made. For the purpose of this study Clostridium difficile, most commonly referred to as C. difficile was isolated from patients suspected of having CDI. Toxigenic Clostridium difficile was present in 100% of the antibiotic-asso- ciated diarrheal cases found in this study.
EIA and culture methods are less specific and sensitive; therefore they do lack the consistency in providing adequate and accurate data for patients [22] . There have also being reports of cross-reactivity between Clostridium sordellii lethal toxin and Clostridium difficile toxin B [22] -[26] . However, the possibility of cross-reactivity occurring in this investigation is low. This is due to the following reasons:
1) EIA kits used tested for both toxins and glutamate dehydrogenase, which is specific to Clostridium difficile.
2) All culture positive isolates were positive by PCR and DNA sequencing.
The trend and demographics of C. difficile could be used as an important epidemiological tool to characterize and differentiate strains from the different hospitals based on agar dilution and PCR technique. Both EIA and PCR are important, fast and effective tools in detecting toxigenic C. difficile (see Table 2). However, EIA has its advantage of being able to detect C. difficile and its toxins directly from stool in a short period of time. PCR on the other hand is more sensitive, specific, cheap, and is able to detect the presence of C. difficile from stool as well as the toxin genes of the organism [23] .
PCR is important for testing the presence of toxin genes as well as housekeeping genes that are specific for Clostridium difficile. PCR testing is very precise and sensitive (100% respectively) for the detection of Clostridium difficile [18] -[21] . According to the Centre of Disease Control and Prevention (CDC), 15% - 25% of toxigenic C. difficile retrieved from nosocomial and antibiotic associated diarrheal patients are responsible for the diarrheal disease being experienced [22] -[25] [27] . The PCR method was used on all culture positive isolates in this study.
The results obtained showed that all the culture positive isolates were positive for the C. difficile housekeeping gene (see Figure 1). This new finding, for the isolates obtained in our small geographical location, can be used to promote the use of molecular techniques for properly identifying Clostridium difficile. The tpi gene can also be considered the major identifying marker in this case. However more analysis will have to be carried out, using a larger sample size. Three of the isolates gave 289 bp DNA bands for the toxin A gene, which differed significantly from the expected 1266 bp fragment as seen for the ATCC 9689 C. difficile control strain. This finding indicates possible gene deletions/truncation. All three toxin A+B+ isolates showed slight variations in their 289 bp toxin A genes fragment, when compared among themselves. However greater variations were seen when the toxin A gene fragments obtained were compared with gene sequences from known C. difficile strains. The variability in the C. difficile DNA fragments amplified and sequenced, may have occurred due to horizontal gene transfer; similar truncated fragments have been reported by researchers who have conducted molecular studies on Clostridium difficile [13] . Lateral gene transfer via transduction, is a likely cause for bacterial gene deletions and insertions; Phages are virus materials that are known to insert their DNA into bacteria and produce changed in bacterial DNA [28] . Phages also produce enzymes such as nucleases which cuts DNA and may for truncated form of a gene [28] .
The toxin B gene fragments from the 15 toxin B+ isolates (including the 3 A+B+), were 100% similar when compared among themselves. However slight variations were seen when compared with some of the known strains from the NCBI database. The geographical location of the Clostridium difficile samples obtained may have played a vital role in the gene fragment variations seen. It is important to note that all the toxin B fragments isolated gave the correct 204 bp amplified base pair products. Therefore, we can conclude that truncation and variation in the toxin A gene is more likely to occur than that of the toxin B gene.
DNA sequencing procedure was important to the research as it without a doubt proves that the method used for CDI analysis was accurate, the primers used were correct for identifying specific toxin genes. Therefore, if this PCR method is implemented in investigated institutions it will be effective in confirming and analyzing CDI.
Phylogenetic analysis was used to analyze all the strains isolated; this was done to show whether or not they all belong to the same species/group of Clostridium difficile. Performing a clustal W alignment of the Toxin gene sequences showed that there were little variations in the strains present in Trinidad & Tobago. The isolates from this study showed great similarities to the Clostridium difficile 630 and IS25 strains; these have been isolated in Zurich Switzerland and China respectively. Further investigation is however needed in this field. The ease of travelling and the movement of people over extensive geographical regions are possible explanation for the similarity seen in local Clostridium difficile strains to that of the Clostridium difficile 630 and IS25 strains.
Other studies have shown that the predominance of toxigenic Clostridium difficile in stool cultures has been accredited to various factors. These factors include extensive hospital stays, being older than age 65, overuse or misuse of antibiotics and having an underlying disease such as chronic renal failure or HIV [29] . Other factors include the presence of an environment which contains C. difficile spores that can survive extreme conditions, as well as the use of medical devices on different patients without proper sterilization between usage [9] [10] [20] -[23] .
Surgical patients are at higher risk for CDI because of surgical interventions and high antibiotic use. This finding was in conformity with other studies conducted at various hospitals [24] -[27] .
Surgical patients need to be properly evaluated and followed up; therefore patients from the surgical wards may have experienced a longer hospital stay than patients from the other wards. These patients are therefore more predisposed to developing nosocomial CDI [5] [28] .
High resistance frequencies were seen with antibiotics such as Ceftriaxone, Ciprofloxacin and Cefotaxime in Clostridium difficile isolates obtained encountered in this study. Cefotaxime resistant isolates that were associated with diarrhea was an important finding (Table 2). Similarly, Cefotaxime resistant isolates were found in other studies [29] . These antibiotics are among the experient first line treatment for Clostridium difficile infection at the hospitals investigated. Other studies showed similar results with a high frequency of resistance to these antibiotics [29] [30] . The wide spread use of these antimicrobial agents now pose a great risk for patients, due to the fact that as these agents disrupts the bacterial flora of the intestine; Clostridium difficile is not only left to thrive but has the added capability to reproduce at a faster rate due to resistance. There has also been reports of increased CDI in hospitals where Pipercillin/Tazobactam was replaced by cephalosporins such as Cefotaxime [31] .
An important observation in this study was the moderate susceptibility rate to Piperacillin/Tazobactam (87%) and Meropenem (87%). This may be due to the fact that these drugs are not readily used as first line treatment for CDI at the hospitals investigated and there is a precautionary use of these as dictated by institutional drug policies. These drugs are not readily used for treating CDI as reported in the literature [25] .
6. Conclusion
The existence of Clostridium difficile toxin genes in hospitalized patients in Trinidad & Tobago is clinically significant. All the culture positive isolates obtained are toxigenic; this is determined by the presence of one or more toxin genes. All the toxigenic isolates are from patients with prior antibiotic exposure. Routine DNA testing is important and should be a part of the detection algorithm. There are great similarities between local strains based on the PCR and DNA sequencing patterns. Therefore, the extent of the implications of these strains in causing CDAD needs to be further assessed.
Acknowledgements
We thank the staff of the Microbiology Laboratories at Eric Williams Medical Sciences Complex, San Fernando General Hospital and the Port of Spain General Hospital. Many thanks to staff at Pathology and Microbiology University of the West Indies (UWI), Eric Williams Medical Sciences Complex and to Prof. Francis Dziva of the School of Veterinary Medicine UWI for his valuable suggestions during the study.
References
- Danese, S. and Fiocchi, C. (2011) Ulcerative Colitis. The New England Journal of Medicine, 365, 1713-1725. http://dx.doi.org/10.1056/NEJMra1102942
- Baxter, R.G., Ray, G.T. and Fireman, B.H. (2008) Case-Control Study of Antibiotic Use and Subsequent Clostridium difficile-Associated Diarrhea in Hospitalized Patients. Infection Control and Hospital Epidemiology, 29, 44-50. http://dx.doi.org/10.1086/524320
- Bartlett, J.G. and Gerding, D.N. (2008) Clinical Recognition and Diagnosis of Clostridium difficile Infection. Clinical Infectious Diseases, 46, S12-S18. http://dx.doi.org/10.1086/524320
- Simor, A.E., Yake, S.L. and Tsimidis, K. (1993) Infection Due to Clostridium difficile among Elderly Residents of a Long-Term-Care Facility. Clinical Infectious Diseases, 17, 672-678. http://dx.doi.org/10.1093/clinids/17.4.672
- Fekety, R., McFarland, L.V., Surawicz, C.M., Greenberg, R.N., Elmer, G.W. and Mulligan, M.E. (1997) Recurrent Clostridium difficile Diarrhea: Characteristics of and Risk Factors for Patients Enrolled in a Prospective, Randomized, Double-Blinded Trial. Clinical Infectious Diseases, 24, 324-333. http://dx.doi.org/10.1093/clinids/24.3.324
- Pépin, J., Saheb, N., Coulombe, M.-A., Alary, M.-E., Corriveau, M.-P., Authier, S., et al. (2005) Emergence of Fluo- roquinolones as the Predominant Risk Factor for Clostridium difficile-Associated Diarrhea: A Cohort Study during an Epidemic in Quebec. Clinical Infectious Diseases, 41, 1254-1260. http://dx.doi.org/10.1086/496986
- Pépin, J., Valiquette, L., Alary, M.-E., Villemure, P., Pelletier, A., Forget, K., et al. (2004) Clostridium difficile-Asso- ciated Diarrhea in a Region of Quebec from 1991 to 2003: A Changing Pattern of Disease Severity. Canadian Medical Association Journal, 171, 466-472. http://dx.doi.org/10.1503/cmaj.1041104
- Dzink, J. and Bartlett, J.G. (1980) In Vitro Susceptibility of Clostridium difficile Isolates from Patients with Antibiotic- Associated Diarrhea or Colitis. Antimicrobial Agents and Chemotherapy, 17, 695-698. http://dx.doi.org/10.1128/AAC.17.4.695
- McDonald, L.C., Killgore, G.E., Thompson, A., Owens Jr., R.C., Kazakova, S.V., Sambol, S.P, et al. (2005) An Epi- demic, Toxin Gene-Variant Strain of Clostridium difficile. The New England Journal of Medicine, 353, 2433-2441. http://dx.doi.org/10.1056/NEJMoa051590
- Hubert, B., Loo, V.G., Bourgault, A.-M., Poirier, L., Dascal, A., Fortin, É., et al. (2007) A Portrait of the Geographic Dissemination of the Clostridium difficile North American Pulsed-Field Type 1 Strain and the Epidemiology of C. dif- ficile-Associated Disease in Quebec. Clinical Infectious Diseases, 44, 238-244. http://dx.doi.org/10.1086/510391
- Gerding, D.N., Johnson, S., Peterson, L.R., Mulligan, M.E. and Silva Jr., J. (1995) Clostridium difficile-Associated Diarrhea and Colitis. Infection Control and Hospital Epidemiology, 16, 459-477. http://dx.doi.org/10.2307/30141083
- Lemee, L., Dhalluin, A., Testelin, S., Mattrat, M-A., Maillard, K., Lemeland, J-F., et al. (2004) Multiplex PCR Targeting tpi (Triose Phosphate Isomerase), tcdA (Toxin A), and tcdB (Toxin B) Genes for Toxigenic Culture of Clostridium difficile. Journal of Clinical Microbiology, 42, 5710-5714. http://dx.doi.org/10.1128/JCM.42.12.5710-5714.2004
- Kato, H., Kato, N., Watanabe, K., Iwai, N., Nakamura, H., Yamamoto, T., et al. (1998) Identification of Toxin A- Negative, Toxin B-Positive Clostridium difficile by PCR. Journal of Clinical Microbiology, 36, 2178-2182.
- Drmanac, R. (2011) The Advent of Personal Genome Sequencing. Genetics in Medicine, 13, 188-190. http://dx.doi.org/10.1097/GIM.0b013e31820f16e6
- Poilane, I., Humeniuk-Ainouz, C., Durand, I., Janoir, C., Cruaud, P., Delmée, M., et al. (2007) Molecular Characterization of Clostridium Difficile Clinical Isolates in a Geriatric Hospital. Journal of Medical Microbiology, 56, 386-90. http://dx.doi.org/10.1099/jmm.0.46608-0
- Gursoy, S., Guven, K., Arikan, T., Yurci, A., Torun, E., Baskol, M., et al. (2007) Clostridium difficile Infection Frequency in Patients with Nosocomial Infections or Using Antibiotics. Hepatogastroenterology, 54, 1720-1724.
- Bartlett, J.G. (1994) Clostridium difficile: History of Its Role as an Enteric Pathogen and the Current State of Knowledge about the Organism. Clinical Infectious Diseases, 18, S265-S272. http://dx.doi.org/10.1093/clinids/18.Supplement_4.S265
- Bartlett, J.G. (2002) Antibiotic-Associated Diarrhea. New England Journal of Medicine, 346, 334-339. http://dx.doi.org/10.1056/NEJMcp011603
- Miller, M.A., Hyland, M., Ofner-Agostini, M., Gourdeau, M. and Ishak, M. (2002) Morbidity, Mortality, and Healthcare Burden of Nosocomial Clostridium difficile―Associated Diarrhea in Canadian Hospitals. Infection Control and Hospital Epidemiology, 2, 137-140. http://dx.doi.org/10.1086/502023
- Weber, D.J., Rutala, W.A., Miller, M.B., Huslage, K. and Sickbert-Bennett, E. (2010) Role of Hospital Surfaces in the Transmission of Emerging Health Care-Associated Pathogens: Norovirus, Clostridium difficile, and Acinetobacter Species. American Journal of Infection Control, 38, S25-S33. http://dx.doi.org/10.1016/j.ajic.2010.04.196
- Higazi, T.B., Mohannad, A-S., Burkett, M. and Pusok, R. (2011) PCR Detection of Clostridium difficile and Its Toxigenic Strains in Public Places in Southeast Ohio. International Journal of Microbiological Research, 2, 105-111.
- Kuntz, J.L., Johnson, E.S., Raebel, M.A., Petrik, A.F., Yang, X., Thorp, M.L., et al. (2012) Epidemiology and Healthcare Costs of Incident Clostridium Difficile Infections Identified in the Outpatient Healthcare Setting. Infection Control and Hospital Epidemiology, 33, 1031-1038. http://dx.doi.org/10.1086/667733
- Tabaqchali, S. and Wilks, M. (1992) Epidemiological Aspects of Infections Caused by Bacteroides fragilis and Clostridium difficile. European Journal of Clinical Microbiology and Infectious Diseases, 11, 1049-1057. http://dx.doi.org/10.1007/BF01967798
- Samore, M.H., DeGirolami, P.C., Tlucko, A., Lichtenberg, D.A., Melvin, Z.A. and Karchmer, A.W. (1994) Clostridium difficile Colonization and Diarrhea at a Tertiary Care Hospital. Clinical Infectious Diseases, 18, 181-187. http://dx.doi.org/10.1093/clinids/18.2.181
- Clabots, C.R., Johnson, S., Olson, M.M., Peterson, L.R. and Gerding, D.N. (1992) Acquisition of Clostridium difficile by Hospitalized Patients: Evidence for Colonized New Admissions as a Source of Infection. Journal of Infectious Diseases, 166, 561-567. http://dx.doi.org/10.1093/infdis/166.3.561
- Sloan, L.M., Duresko, B.J., Gustafson, D.R. and Rosenblatt, J.E. (2008) Comparison of Real-Time PCR for Detection of the tcdC Gene with Four Toxin Immunoassays and Culture in Diagnosis of Clostridium difficile Infection. Journal of Clinical Microbiology, 46, 1996-2001. http://dx.doi.org/10.1128/JCM.00032-08
- Songer, J.G., Trinh, H.T., Killgore, G.E., Thompson, A.D., McDonald, L.C. and Limbago, B.M. (2009) Clostridium difficile in Retail Meat Products, USA, 2007. Emerging Infectious Diseases, 15, 819. http://dx.doi.org/10.3201/eid1505.081071
- Kato, H., Kato, N., Katow, S., Maegawa, T., Nakamura, S. and Lyerly, D.M. (1999) Deletions in the Repeating Sequences of the Toxin A Gene of Toxin A-Negative, Toxin B-Positive Clostridium difficile Strains. FEMS Microbiology Letters, 175, 197-203. http://dx.doi.org/10.1111/j.1574-6968.1999.tb13620.x
- Alston, W.K. and Ahern, J.W. (2004) Increase in the Rate of Nosocomial Clostridium difficile-Associated Diarrhoea during Shortages of Piperacillin-Tazobactam and Piperacillin. Journal of Antimicrobial Chemotherapy, 53, 549-550. http://dx.doi.org/10.1093/jac/dkh127
- Varughese, C.A., Vakil, N.H. and Phillips, K.M. (2013) Antibiotic-Associated Diarrhea a Refresher on Causes and Possible Prevention with Probiotics―Continuing Education Article. Journal of Pharmacy Practice, 26, 476-482. http://dx.doi.org/10.1177/0897190013499523
- Nelson, J., Reynolds-Kohler, C., Margaretten, W., Wiley, C., Reese, C. and Levy, J. (1988) Human Immunodeficiency Virus Detected in Bowel Epithelium from Patients with Gastrointestinal Symptoms. The Lancet, 331, 259-262. http://dx.doi.org/10.1016/S0140-6736(88)90348-0
NOTES

*Corresponding author.












