Advances in Infectious Diseases
Vol.2 No.4(2012), Article ID:25308,9 pages DOI:10.4236/aid.2012.24019
Evaluation of Reverse Transcription Loop-Mediated Isothermal Amplification Assays for Rapid Detection of Human Enterovirus 71 and Coxsackievirus A16 in Clinical Samples*
![]()
1Department of Microbiology, Center for Disease Control and Prevention of HuNan, Changsha, China; 2State Key Laboratory for Molecular Virology and Genetic Engineering, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China.
Email: #gldlj@hotmail.com, #maxj2004@yahoo.com.cn
Received September 25th, 2012; revised October 27th, 2012; accepted November 29th, 2012
Keywords: Human Enterovirus 71; Coxsackievirus A16; Reverse Transcription Loop-Mediated Isothermal Amplification
ABSTRACT
A sensitive reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for human enterovirus 71 (EV71) and Coxsackievirus A16 (CVA16) infection was further evaluated. The one step reaction was performed in a single tube at 65˚C for 45 min for EV71 and 35 min for CVA16. The detection limits of RT-LAMP assays for both EV71 and CVA16 were 0.1 of a 50% tissue culture infective dose (TCID50) per reaction, based on 10—Fold dilutions of a titrated EV71 or CVA16 strain. The specific assay showed there were no cross-reactions with Coxsackievirus A (CVA) viruses (CVA 2, 4, 5, 7, 9, 10, 14, and 25), Coxsackievirus B (CVB) viruses (CVB 1, 2, 3, 4, and 5) or ECHO viruses (ECHO 3, 6, 11, and 19). In parallel with commercial quantitative real-time polymerase chain reaction (qRT-PCR) diagnostic kits for EV71 and CVA16, the RT-LAMP assay was evaluated with 515 clinical specimens, the results showed the RT-LAMP assay and the qRT-PCR assay were in complete agreement for 513/515 (99.6%) of the specimens. Two samples with discrepant results from two methods were further verified by nested reverse transcription polymerase chain reaction (nRT-PCR) assay and sequencing to be true positives for CVA16. In conclusion, RT-LAMP assay is demonstrated to be a sensitive and specific assay and have a great potential for the rapid and visual screening of EV71 and CVA16 in China, especially in those resource-limited hospitals and rural clinics of provincial and municipal regions.
1. Introduction
Human enteroviruses (HEVs) comprise more than 100 serotypes in four species (HEV-A to HEV-D) in the genus Enterovirus, family Picornaviridae. Hand-Foot-andMouth Disease (HFMD) is a common febrile illness in young children and is characterized by lesions on the skin and oral mucosa. HFMD cases caused by human enterovirus 71(EV71) infections have been found to be associated with severe neurological complications at much higher rates than those caused by coxsackievirus A16 (CVA16) infections in Asia Pacific region [1,2]. Outbreaks or sporadic cases may also be caused by some of the other HEV-A (CVA 2, 4, 5, 6, 8, and 10) or HEVB viruses (ECHO viruses 1, 4, 7, and 9, CVB3 and CVA9) [3-6].
The classical diagnostic method for enterovirus is propagation in cell culture followed by neutralization to confirm the serotype, which is time-consuming. A number of real-time RT-PCR assays for detection of EV71 and CVA16 with high specificities and sensitivities have been reported [1,7,8]. However, this method might not be suitable in primary clinical settings in developing countries or for field use because of the sophisticated instrumentation required and expensive reagents.
Loop-mediated isothermal amplification (LAMP) is a nucleic acid amplification method developed by Notomi et al. [9] and has been used increasingly for rapid detection of pathogens [10-19] due to its simplicity, speedhigh specificity and cost-effectiveness. A reverse transcription LAMP (RT-LAMP) is carried out in a single tube by mixing primers, reverse-transcriptase and Bst DNA polymerase together with RNA extracted from the sample and the mixture was incubated at 65˚C for 60 min. Compared to other molecular detection methods, the advantage of the RT-LAMP assay is due to continuous amplification under isothermal conditions using six primers that recognize eight distinct regions of the target gene and visual observation of the detection results without post-amplification operation. Visual LAMP assays using hydroxyl naphthol blue dye (HNB) to detect pandemic influenza A H1N1 virus, EV71, CVA16 and high-risk human papillomavirus have been successfully reported by our research group [20-22]. As limited number of predetermined EV71 and CVA16 clinical samples were tested using home-made reaction mixture in our previous report [22], the reliability of RT-LAMP assays in the clinical application and a comprehensive comparison with commercial qRT-PCR assays need to be addressed.
To explore the possibility of wide applicability and potentials for the routine use in the hospitals and rural clinics of provincial and municipal regions, in the present study, an improved RT-LAMP assay using a commercial loopamp RNA amplification kit and a fluorescent detection reagent (Eiken Chemical Co., Ltd., Tokyo, Japan) was evaluated for the detection of EV71 and CVA16. RT-LAMP assay was examined with the specificity and sensitivity for EV71 and CVA16, respectively. The clinical reliability of RT-LAMP assays for the detection of EV71 and CVA16 was conducted using a total of 515 clinical samples. The results were compared with those obtained from commercial qRT-PCR assays, and the discrepancies from two methods were further clarified by nRT-PCR and sequencing.
2. Material and Methods
2.1. Viral RNA and Specimen
Field isolates of human enterovirus known to be genetically related to HFMD were obtained from the National Laboratory for Poliomyelitis, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention. These isolates were used as control viruses to evaluate the specificity of the RT-LAMP assay for EV71 and CVA16. The control viruses included Coxsackievirus A (CVA 2, 4, 5, 7, 9, 10, 14, and 24), Coxsackievirus B (CVB 1, 2, 3, 4, and 5) and ECHO viruses (ECHO 3, 6, 11, and 19). All isolates had been verified previously by RT-PCR and sequencing. The EV71 subgenotype C4 isolate (Strain FY17.08/ AN/CHN/2008, GenBank accession no. EU703812) and CVA16 isolate (Strain FY18/AN/CHN/2008, GenBank accession No. EU812514) from the National Laboratory for Poliomyelitis were used as reference viruses in this study; both reference viruses had an infectivity titer of 106.5 50% tissue culture infective doses (TCID50)/ml on human rhabdomyosarcoma (RD) cells. One EV71—Negative stool sample collected from other HFMD patients was used as a negative control. A total of 515 clinical specimens (445 fecal samples, 60 anal swabs, 1 cerebrospinal fluid, 9 nasopharyngeal swabs)(Table 1) from suspicious patients with HFMD between 1 month and 10 years old enrolled in Changsha, Hunan, China in 2011 were collected. This study was approved by the local ethics committee. Sample collection was agreed by child’s parents or grandparents with informed consent.
2.2. Extraction of Viral RNA
Viral RNA was extracted from 140 µL of 515 clinical specimens, various control viruses and reference viruses stocks (106.5 TCID50) by using the QIAampViral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The RNA was eluted from the QIAspin column in a final volume of 60 µL of RNase-free water and stored at –80˚C until further analysis.
2.3. Primer Design
As described in our previous study [22], the VP1 gene of enterovirus was used to distinguish enterovirus serotypes and the primers were designed based on sequence information obtained from GenBank. The VP1 nucleotide sequences of EV71 strains circulating in mainland China since 1998 (n = 422) and CVA16 strains (n = 175) in GenBank were all downloaded and aligned. The highly conserved segment within the VP1 gene of EV71 and CVA16 were selected as target sequences for RT-LAMP assay primer design by using online software program PrimerExplorer V4 (http://primerexplorer.jp/e/). The primers for CVA16 were re-designed intended to increase the sensitivity based on our pre-experimental data. All the primers were HPLC purified and synthesized by Invitrogen. The primers for RT-LAMP assay and nRT-PCR [23] are listed in Tables 2 and 3, respectively.
2.4. Reaction Protocol for RT-LAMP Assay
LAMP was performed in a total of 25 μL of reaction mix prepared as follows: 2 µL of template RNA, 12.5 µL of the RNA amplification reaction mix, 1 µL of each primer of EV71 or CVA16 (F3 and B3: 5 pmol/µL; BIP and FIP: 40 pmol/µL; Loop-1 and Loop-2: 20 pmol/µL), 1 µL of the enzyme mix, 1 µL of the Fluorescent Detection Reagent (FDR) (Eiken Chemical Co., Ltd., Tokyo, Japan)
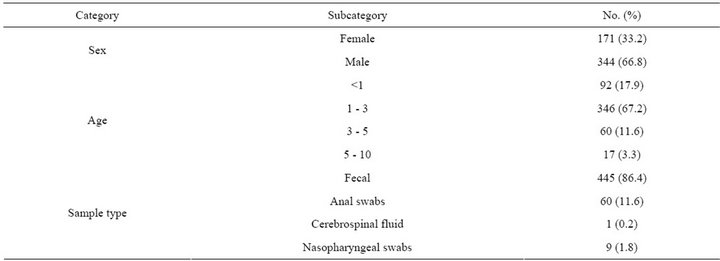
Table 1. The epidemiological characteristics of specimens (n = 515) in this study.
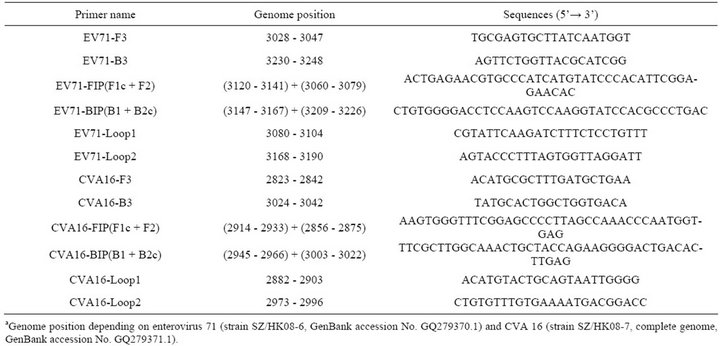
Table 2. RT-LAMP primers designed for the detection of the VP1 gene sequences of human enterovirus 71 and Coxsackievirus A16.
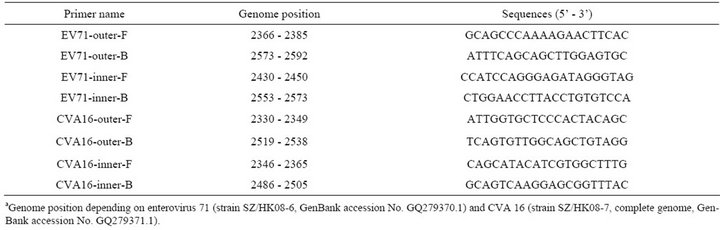
Table 3. Nested RT-PCR primers for the detection of human enterovirus 71 and Coxsackievirus A16.
and 2.5 µL of dH2O. The RT-LAMP reaction was incubated in a Loopamp turbidimeter (LA: 32˚C; Teramecs, Japan) for real-time monitoring of the amplification at 65˚C for 45 min for EV71 detection and 35 min for CVA16 detection, followed by heating at 85˚C for 3 min to terminate the reaction. The turbidities of the corresponding LAMP products were detected using a Loopamp turbidimeter LA: 32˚C. Positive reactions were defined as those samples having a threshold value of greater than 0.2 or a color change from faint orange to yellowish green. Positive and negative controls were included in each run, as described previously [22].
2.5. Specificity of the RT-LAMP Assay
To establish the specificity of the RT-LAMP assays for EV71 and CVA16, the extracted RNAs from various control viruses (CVA 2, 4, 5, 7, 9, 10, 14, and 24; CVB 1, 2, 3, 4, and 5; ECHO 3, 6, 11, and 19) and the reference viruses (EV71 Strain FY17.08 and CVA16 Strain FY18) were used as templates, respectively. RT-LAMP assay was performed as described in above reaction protocol. Positive and negative controls were included in each run.
2.6. Sensitivity of the RT-LAMP Assay
The EV71 reference viral stock obtained from infected human RD cells was titrated, and 10—Fold serial dilutions were made. A panel of RNAs with concentrations ranging from 104 to 10–2 TCID50 per assay was prepared. RT-LAMP assay was performed as described in above reaction protocol. The CVA16 detection limit of the RTLAMP assay was determined similarly with CVA16 reference virus. The reaction at each template concentration was repeated three times, and similar results were obtained.
2.7. Evaluation of RT-LAMP Assay with Clinical Specimens and Comparison with qRT-PCR Assay
A total of 515 clinical specimens from suspicious patients with HFMD were analyzed by RT-LAMP assay for the detection of EV71 and CVA16, respectively, as described in above reaction protocol. The reaction time was set up to 45 min for EV71 or 35 min for CVA16. In comparison, qRT-PCR assay was carried out in an ABI RealTime System 7300 device (Applied Biosystems, USA) with same amount of viral RNA used in RT-LAMP assay using commercial qRT-PCR Diagnostic Kits (PCRFluorescence Probing) for EV71, CVA16 and panenterovirus RNA (DaAn Gene, China) approved by the State Food and Drug Administration of China according to the manufacturer’s instructions.
2.8. nRT-PCR Assays for EV71 and CVA16 and Sequencing
The clinical samples with discrepant detection results from RT-LAMP and qRT-PCR were further verified using nRT-PCR assays, which are recommended methods for HFMD pathogens surveillance used in CDC of provincial and municipal regions in China, as described elsewhere [2,23]. The resulting nRT-PCR products were subjected to sequencing for confirmation with an ABI 3730 automated DNA sequencer (Applied Biosystems, USA) [23].
3. Results
3.1. Specificity of RT-LAMP Assay
The specificity of the RT-LAMP assays for EV71and CVA16 was analyzed using extracted RNAs from various control viruses and the reference viruses as templates, respectively. Positive reactions were defined as those samples having a turbidity threshold value of greater than 0.2 or a color change. The original faint orange color of FDR changed to yellowish green in the case of positive amplification, whereas the original color was retained for negative reactions without amplification (not shown). Turbidity increase was only observed in the preparation of the EV71 or CVA16 reference virus, whereas none of the control viruses showed turbidity increase for the detection of EV71 (Figure 1(a)) or CVA16 (Figure 1(b)).
3.2. Sensitivity of RT-LAMP Assay
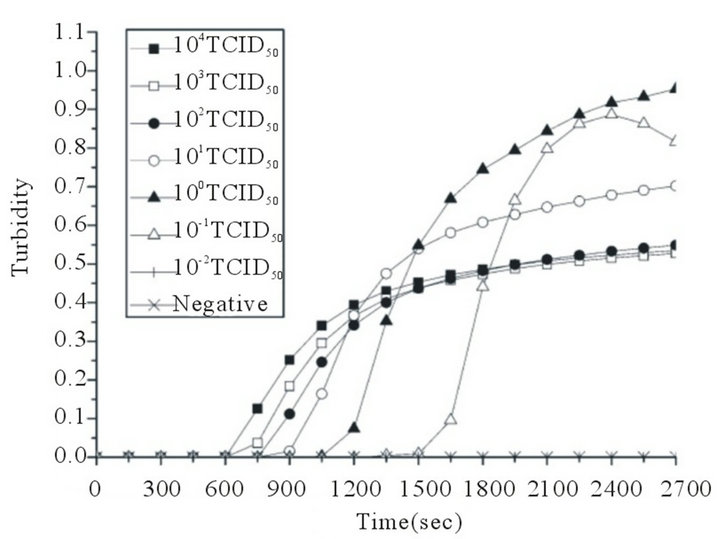
A serial of 10—Fold dilutions of titrated reference EV71 or CVA16 stock was used to examine the sensitivity of RT-LAMP assay. The EV71 detection limit of the RT-LAMP assay was 0.1 TCID50 per reaction. Positive reactions were also indicated by the observation by the naked eye of a color change from faint orange to yellowish green (Figures 2(a) and (b)). The CVA16 detection limit of the RT-LAMP assay was determined similarly with CVA16 reference virus, and the sensitivity was also 0.1 TCID50 per reaction. Positive reactions were also indicated by the observation of a color change from faint orange to yellowish green (Figures 2(c) and (d)). The reaction at each template concentration was repeated three times, and similar results were obtained. Compared with the sensitivities of RT-LAMP assays using homemade reagent previously reported by our research group [22], the detection limit of the RT-LAMP assays for EV71 and CVA16 increased up to 0.1 TCID50 per reaction.
3.3. Evaluation of RT-LAMP Assay with Clinical Specimens and Comparison with qRT-PCR Assay
The RT-LAMP assays for EV71 and CVA16 were
 (a)
(a)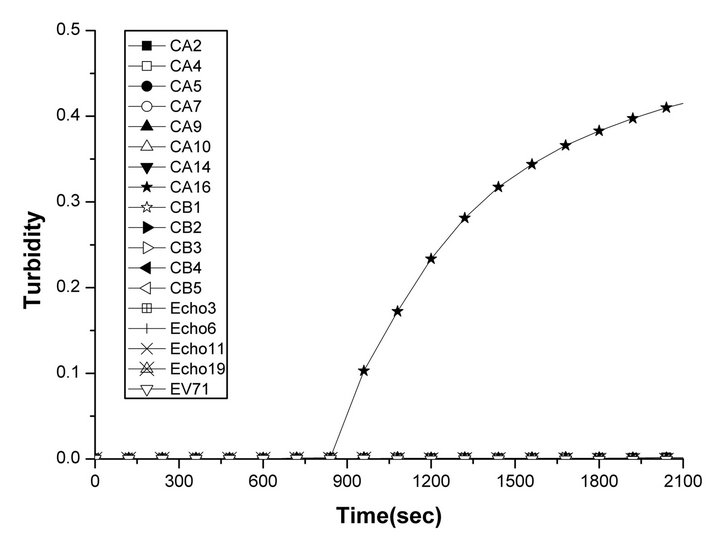 (b)
(b)
Figure 1. Specificities of the RT-LAMP assays for the detection of human EV71 and CVA16. Specificities of the RT-LAMP assays were monitored by real-time measurement of turbidity. Positive reactions were defined as those samples having a threshold value of greater than 0.2 within 60 minutes. Positive reactions were only observed in (a) the EV71 reference virus (cross) and (b) the CVA16 reference virus (pentagram). None of the control viruses (Coxsackievirus A viruses 2, 4, 5, 7, 9, 10, 14, 24; Coxsackievirus B viruses 1, 2, 3, 4, 5; or ECHO viruses 3, 6, 11, 19) showed an increase in turbidity.
 (a)
(a) (b)
(b)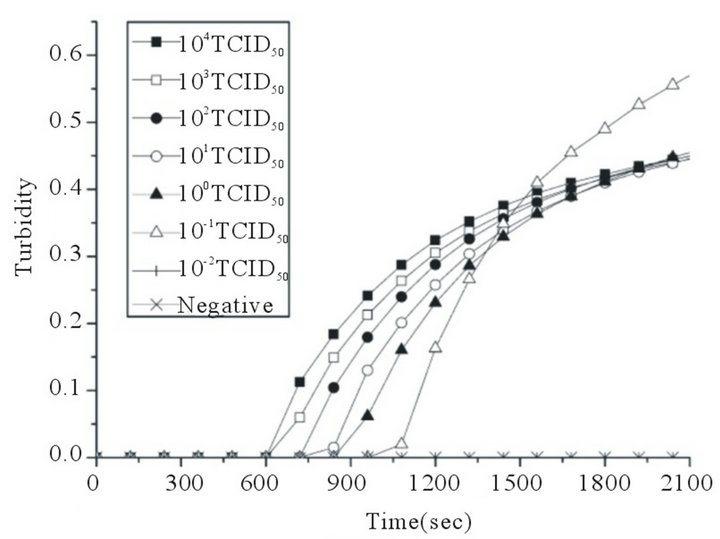 (c)
(c)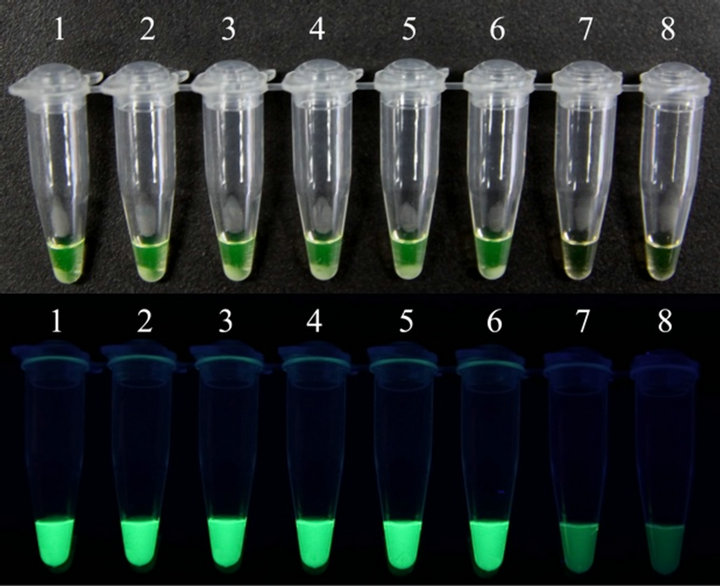 (d)
(d)
Figure 2. Sensitivities of the RT-LAMP assays for the detection of human EV71 and CVA16. RT-LAMP was carried out using different concentrations of RNA from a titrated EV71 subgenotype C4 isolate (Strain FY17.08/AN/CHN/2008, GenBank accession No. EU703812) or CVA16 isolate (Strain FY18/AN/CHN/2008, GenBank accession No. EU812514). Sensitivities of the RT-LAMP assays for EV71 (a) and CVA16 (c) were monitored by real-time measurement of turbidity. Sensitivities of the RT-LAMP assays for EV71 (b) and CVA16 (d) were also indicated by the observation of a color change from faint orange to yellowish green (anes 1 to 8, 104 to 10–2 TCID50 and negative control). The tubes shown from left to right have decreasing concentrations of RNA, correspondingly. The detection limits for both EV71 and CVA16 were 0.1 TCID50 per reaction.
evaluated and compared with commercial qRT-PCR diagnostic kits (DaAn Gene, China) using 515 clinical specimens from suspicious patients with HFMD. The qRT-PCR results were defined as the positive for Ct not higher than 34.8. As shown in Table 4, the results from the RT-LAMP and the qRT-PCR were in complete agreement for 513/515 (99.6%) of the specimens. Of the 515 samples, 233 (45.2%) were found to be positive for EV71, 105 (20.4%) positive for CVA16, 107 (20.8%) positive for pan-enterovirus and 70 (13.6%) negative by qRT-PCR assays, while 233 (45.2%) were found to be positive for EV71, 103 (20%) positive for CVA16 and 179/515 (34.8%) negative by RT-LAMP assays. Two negative samples (fecal samples) detected by RT-LAMP were found to be CVA16 positive with high Ct values (>33) by qRT-PCR. Mixed infection of EV71 and CVA16 was not observed in the detection of 515 clinical samples.
3.4. nRT-PCR Assay for EV71 and CVA16 and Sequencing
Two samples with discrepant detection results from RTLAMP assay and qRT-PCR assay were further clarified by nRT-PCR and sequencing to be true positives for CVA16 (data not shown).
4. Discussion
Hand-Foot-and-Mouth Disease (HFMD), mainly caused by
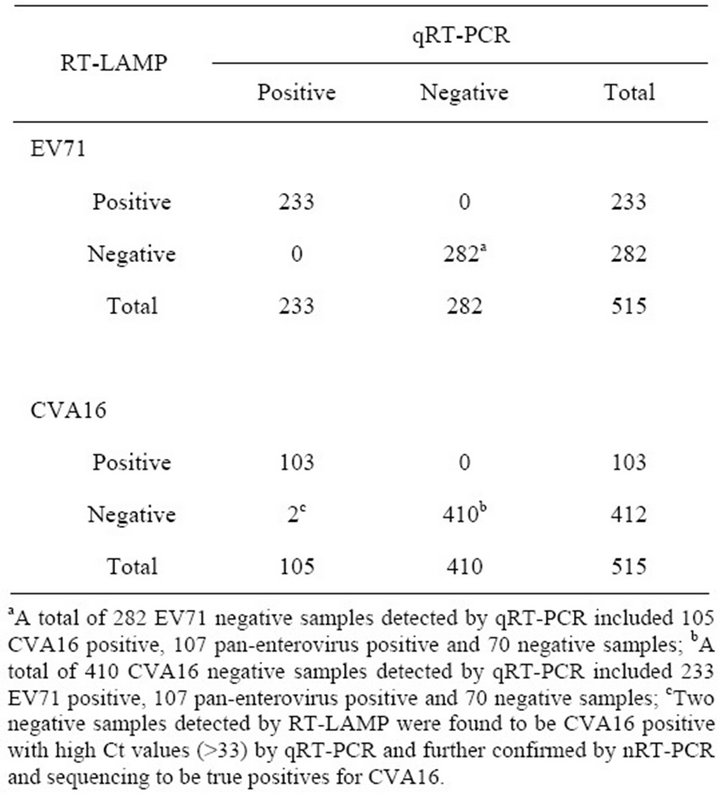
Table 4. Comparison of results for RT-LAMP assay and qRT-PCR obtained from 515 clinical specimens.
enterovirus 71 (EV71) and coxsackievirus A16 (CVA16), is a common acute enteroviral infectious disease, which was transmitted from person to person by direct contact with nose and throat discharges or by fecal oral routes. Individual cases of HFMD occur worldwide, and 1.77 million cases were reported in China, 2010. Thus, accurate and rapid detection of EV71 and CVA16 is required to enable measures to be implemented to prevent widespread transmission.
Routine methods for EV71 and CVA16 detection are virus isolation, neutralization, and RT-PCR (nRT-PCR or qRT-PCR). Especially the RT-PCR, which is the most common method for HFMD pathogens surveillance, has been widely used in CDC of provincial and municipal regions in China. There are a few qRT-PCR diagnostic kits for EV71, CVA16 and pan-enterovirus RNA commercially available and approved by the State Food and Drug Administration of China.
However, these methods either are with low specificity and sensitivity (virus isolation and neutralization) or need large expense for equipments and a relative long-time reaction to achieve the goal of detection (RT-PCR). LAMP was first described in 2000 and proved to be a simple, rapid, cost-effective, highly specific and sensitive nucleic acid amplification method for virus detection. No PCR machine is needed in practical application and only an ordinary water bath is required to perform the LAMP reaction. Thus, RT-LAMP for EV71 and CVA16 detection should be superior to those routine ones.
When this manuscript was in preparation, RT-LAMP assay for EV71 has been reported in four publications [22,24-26] including the one developed in our research group using HNB dye and home-made reagents [22]. As illustrated in previous publications, different target genes were chosen for LAMP primer design and comparable sensitivity of EV71 was achieved. Unlike others reports, using limited number of phylogenetically related enteroviruses to examine the specificity and less samples to evaluate the clinical application of RT-LAMP assay, a total of 17 related enteroviruses including Coxsackievirus A (CVA) viruses (CVA 2, 4, 5, 7, 9, 10, 14, and 25), Coxsackievirus B (CVB) viruses (CVB 1, 2, 3, 4, and 5) and ECHO viruses (ECHO 3, 6, 11, and 19) was used and a total of 515 clinical samples was tested by RT-LAMP assays in this study with the parallel analysis by commercial qRT-PCR diagnostic kits.
In the present study, a RT-LAMP assay for EV71 and CVA16 was developed, respectively, using a Loopamp RNA Amplification Kit and a fluorescent detection reagent (Eiken Chemical Co., Ltd., Tokyo, Japan). Compared with 0.33 TCID50 for EV71 and 1.58 TCID50 for CVA16 in our previous report [22], the specificities of the RT-LAMP assays for EV71 and CVA16 were retained, while the sensitivities of both EV71 and CVA16 increased up to 0.1 TCID50 per reaction. In addition, the reaction time was shortened from 60 min for EV71 and CVA16 [22] to 45 min for EV71 and 35 min for CVA16 in this study, suggesting the commercial reaction mixture is more sensitive and stable than home-made reaction mixture.
To be further, the reliability of RT-PCR assay in clinical performance was evaluated using a large number of clinical samples (totally 515 samples, of which, 445 fecal samples, 60 anal swabs, 1 cerebrospinal fluid and 9 nasopharyngeal swabs). In comparison with the results obtained from commercial qRT-PCR diagnostic kits (DaAn Gene, China), the results from RT-PCR-kit assay showed a 99.6% (513/515) rate of coincidence. Two negative samples detected by RT-LAMP assay were found to be CVA16 positive with high Ct values (>33) by qRT-PCR and further confirmed by nRT-PCR and sequencing to be true positives for CVA16, indicating that RT-LAMP assay for CVA16 detection was slightly less sensitive than qRT-PCR. Compared with qRT-PCR diagnostic kits, the sensitivity of RT-LAMP assays for EV71 and CVA16 was 100% and 98.09%, respectively, and the specificity was 100% and 100%, respectively. It was noticeable that the detection rate of EV71 and CVA 16 using RT-LAMP varied depending on the origin of the collected samples (unpublished).
By using a fluorescent detection reagent, the change of color from faint orange to yellowish green can be observed visually thereby no electrophoresis instrument is needed to determine the positive amplification, making this improved RT-LAMP assay in this study more appropriate for rapid detection in hospitals and clinics especially those resource-limit ones.
5. Conclusion
Improved RT-LAMP assays were developed and further evaluated with large number of clinical samples. Our results demonstrate that the RT-LAMP assays are sensitive and specific methods and have a great potential for the rapid and visual screening of EV71 and CVA16 in China, especially in those resource-limited hospitals and rural clinics of provincial and municipal regions.
6. Conflict of Interests
Two Chinese patents (application numbers: 20101029 4464.X, 201010567932.6) have been filed for the LAMP primers specific to EV71 and CVA16. The author Xuejun Ma is the inventor on the patent applications. The technology is available for research-only purposes.
7. Acknowledgements
We are grateful to Eiken Chemical Co., Ltd., Tokyo, Japan, for providing the fluorescent detection reagent, the Loopamp RNA Amplification kit and the Loopamp turbidimeter. This work was supported by the China Mega-Project for Infectious Disease (2011ZX10004-001, 2012ZX10004-215) and a research grant from the State Key Laboratory for Genetic Engineering and Molecular Virology.
8. Disclaimers
The opinions expressed by the authors contributing to this journal do not necessarily reflect the opinions of the Centers for Disease Control and Prevention or the institutions with which the authors are affiliated.
REFERENCES
- E. L. Tan, L. L. Yong, S. H. Quak, W. C. Yeo, V. T. Chow and C. L. Poh, “Rapid Detection of Enterovirus 71 by RealTime TaqMan RT-PCR,” Journal of Clinical Virology, Vol. 42, No. 2, 2008, pp. 203-206. doi:10.1016/j.jcv.2008.01.001
- Y. Zhang, X. J. Tan, H. Y. Wang, D. M. Yan, S. L. Zhu, D. Y. Wang, F. Ji, X. J. Wang, Y. J. Gao, L. Chen, H. Q. An, D. X. Li, S. W. Wang, A. Q. Xu, Z. J. Wang and W. B. Xu, “An Outbreak of Hand, Foot, and Mouth Disease Associated with Subgenotype C4 of Human Enterovirus 71 in Shandong, China,” Journal of Clinical Virology, Vol. 44, No. 4, 2009, pp. 262-267. doi:10.1016/j.jcv.2009.02.002
- L. W. Ang, B. K. Koh, K. P. Chan, L. T. Chua, L. James and K. T. Goh, “Epidemiology and Control of Hand, Foot and Mouth Disease in Singapore, 2001-2007,” Annals Academy of Medicine Singapore, Vol. 38, No. 2, 2009, pp. 106-112.
- S. Blomqvist, P. Klemola, S. Kaijalainen, A. Paananen, M. L. Simonen, T. Vuorinen and M. Roivainen, “CoCirculation of Coxsackieviruses A6 and A10 in Hand, Foot and Mouth Disease Outbreak in Finland,” Journal of Clinical Virology, Vol. 48, No. 1, 2010, pp. 49-54. doi:10.1016/j.jcv.2010.02.002
- Z. Zhu, W. B. Xu, A. Q. Xu, H. Y. Wang, Y. Zhang, L. Z. Song, H. L. Yang, Y. Li and F. Ji, “Molecular Epidemiological Analysis of Echovirus 19 Isolated from an Outbreak Associated with Hand, Foot, and Mouth Disease (HFMD) in Shandong Province of China,” Biomedical and Environmental Sciences, Vol. 20, No. 4, 2007, pp. 321- 328.
- F. Zhou, F. Kong, K. McPhie, M. Ratnamohan, L. Donovan, F. Zeng, G. L. Gilbert and D. E. Dwyer, “Identification of 20 Common Human Enterovirus Serotypes by Use of a Reverse Transcription-PCR-Based Reverse Line Blot Hybridization Assay,” Journal of Clinical Microbiology, Vol. 47, No. 9, 2009, pp. 2737-2743. doi:10.1128/JCM.00823-09
- T. Fujimoto, S. Yoshida, T. Munemura, K. Taniguchi, M. Shinohara, O. Nishio, M. Chikahira and N. Okabe, “Detection and Quantification of Enterovirus 71 Genome from Cerebrospinal Fluid of an Encephalitis Patient by PCR Applications,” Japanese Journal of Infectious Diseases, Vol. 61, No. 6, 2008, pp. 497-499.
- X. L. Xiao, Y. Q. He, Y. G. Yu, H. Yang, G. Chen, H. F. Li, J. W. Zhang, D. M. Liu, X. F. Li, X. Q. Yang and H. Wu, “Simultaneous Detection of Human Enterovirus 71 and Coxsackievirus A16 in Clinical Specimens by Multiplex Real-Time PCR with an Internal Amplification Control,” Archives of Virology, Vol. 154, No. 1, 2009, pp. 121-125. doi:10.1007/s00705-008-0266-8
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, “Loop-Mediated Isothermal Amplification of DNA,” Nucleic Acids Research, Vol. 28, No. 12, 2000, p. E63. doi:10.1093/nar/28.12.e63
- K. A. Curtis, D. L. Rudolph and S. M. Owen, “Rapid Detection of HIV-1 by Reverse-Transcription, Loop-Mediated Isothermal Amplification (RT-LAMP),” Journal of Virological Methods, Vol. 151, No. 2, 2008, pp. 264-270. doi:10.1016/j.jviromet.2008.04.011
- J. P. Dukes, D. P. King and S. Alexandersen, “Novel Reverse Transcription Loop-Mediated Isothermal Amplification for Rapid Detection of Foot-and-Mouth Disease Virus,” Archives of Virology, Vol. 151, No. 6, 2006, pp. 1093-1106. doi:10.1007/s00705-005-0708-5
- M. Goto, E. Honda, A. Ogura, A. Nomoto and K. Hanaki, “Colorimetric Detection of Loop-Mediated Isothermal Amplification Reaction by Using Hydroxy Naphthol Blue,” Biotechniques, Vol. 46, No. 3, 2009, pp. 167-172. doi:10.2144/000113072
- T. C. Hong, Q. L. Mai, D. V. Cuong, M. Parida, H. Minekawa, T. Notomi, F. Hasebe and K. Morita, “Development and Evaluation of a Novel Loop-Mediated Isothermal Amplification Method for Rapid Detection of Severe Acute Respiratory Syndrome Coronavirus,” Journal of Clinical Microbiology, Vol. 42, No. 5, 2004, pp. 1956- 1961. doi:10.1128/JCM.42.5.1956-1961.2004
- Y. Kurosaki, A. Takada, H. Ebihara, A. Grolla, N. Kamo, H. Feldmann, Y. Kawaoka and J. Yasuda, “Rapid and Simple Detection of Ebola Virus by Reverse Transcription-Loop-Mediated Isothermal Amplification,” Journal of Virological Methods, Vol. 141, No. 1, 2007, pp. 78-83. doi:10.1016/j.jviromet.2006.11.031
- Y. Misawa, A. Yoshida, R. Saito, H. Yoshida, K. Okuzumi, N. Ito, M. Okada, K. Moriya and K. Koike, “Application of Loop-Mediated Isothermal Amplification Technique to Rapid and Direct Detection of Methicillin-Resistant Staphylococcus Aureus (MRSA) in Blood Cultures,” Journal of Infection and Chemotherapy, Vol. 13, No. 3, 2007, pp. 134-140. doi:10.1007/s10156-007-0508-9
- R. Ohtsuki, K. Kawamoto, Y. Kato, M. M. Shah, T. Ezaki and S. I. Makino, “Rapid Detection of Brucella spp. by the Loop-Mediated Isothermal Amplification Method,” Journal of Applied Microbiology, Vol. 104, No. 6, 2008, pp. 1815-1823. doi:10.1111/j.1365-2672.2008.03732.x
- M. Parida, K. Horioke, H. Ishida, P. K. Dash, P. Saxena, A. M. Jana, M. A. Islam, S. Inoue, N. Hosaka and K. Morita, “Rapid Detection and Differentiation of Dengue Virus Serotypes by a Real-Time Reverse Transcription-LoopMediated Isothermal Amplification Assay,” Journal of Clinical Microbiology, Vol. 43, No. 6, 2005, pp. 2895- 2903. doi:10.1128/JCM.43.6.2895-2903.2005
- H. Toriniwa and T. Komiya, “Rapid Detection and Quantification of Japanese Encephalitis Virus by Real-Time Reverse Transcription Loop-Mediated Isothermal Amplification,” Microbiology and Immunology, Vol. 50, No. 5, 2006, pp. 379-387.
- T. Yoneyama, T. Kiyohara, N. Shimasaki, G. Kobayashi, Y. Ota, T. Notomi, A. Totsuka and T. Wakita, “Rapid and Real-Time Detection of Hepatitis A Virus by Reverse Transcription Loop-Mediated Isothermal Amplification Assay,” Journal of Virological Methods, Vol. 145, No. 2, 2007, pp. 162-168. doi:10.1016/j.jviromet.2007.05.023
- L. Luo, K. Nie, M. J. Yang, M. Wang, J. Li, C. Zhang, H. T. Liu and X. J. Ma, “Visual Detection of High-Risk Human Papillomavirus Genotypes 16, 18, 45, 52, and 58 by Loop-Mediated Isothermal Amplification with Hydroxynaphthol Blue Dye,” Journal of Clinical Microbiology, Vol. 49, No. 10, 2011, pp. 3545-3550. doi:10.1128/JCM.00930-11
- X. J. Ma, Y. L. Shu, K. Nie, M. Qin, D. Y. Wang, R. B. Gao, M. Wang, L. Y. Wen, F. Han, S. M. Zhou, X. Zhao, Y. H. Cheng, D. X. Li and X. P. Dong, “Visual Detection of Pandemic Influenza A H1N1 Virus 2009 by Reverse-Transcription Loop-Mediated Isothermal Amplification with Hydroxynaphthol Blue Dye,” Journal of Virological Methods, Vol. 167, No. 2, 2010, pp. 214-217. doi:10.1016/j.jviromet.2010.03.027
- K. Nie, Y. Zhang, L. Luo, M. J. Yang, X. M. Hu, M. Wang, S. L. Zhu, F. Han, W. B. Xu and X. J. Ma, “Visual Detection of Human Enterovirus 71 Subgenotype C4 and Coxsackievirus A16 by Reverse Transcription Loop-Mediated Isothermal Amplification with the Hydroxynaphthol Blue Dye,” Journal of Virological Methods, Vol. 175, No. 2, 2011, pp. 283-286. doi:10.1016/j.jviromet.2011.05.020
- J. Han, X. J. Ma, J. F. Wan, Y. H. Liu, Y. L. Han, C. Chen, C. Tian, C. Gao, M. Wang and X. P. Dong, “Long Persistence of EV71 Specific Nucleotides in Respiratory and Feces Samples of the Patients with Hand-Foot-Mouth Disease after Recovery,” BMC Infectious Diseases, Vol. 10, No. 2010, 2010, p. 178.
- T. Jiang, J. Liu, Y. Q. Deng, L. J. Xu, X. F. Li, J. F. Han, R. Y. Cao, E. D. Qin and C. F. Qin, “Development and Evaluation of a Reverse Transcription-Loop-Mediated Isothermal Amplification Assay for Rapid Detection of Enterovirus 71,” Journal of Clinical Microbiology, Vol. 49, No. 3, 2011, pp. 870-874. doi:10.1128/JCM.02045-10
- W. Shi, K. Li, Y. Ji, Q. Jiang, M. Shi and Z. Mi, “Development and Evaluation of Reverse Transcription-LoopMediated Isothermal Amplification Assay for Rapid Detection of Enterovirus 71,” BMC Infectious Diseases, Vol. 11, No. 2011, 2011, p. 197.
- J. F. Xia, X. F. Yan, H. Yu, D. Qu and J. E. Long, “Simple and Rapid Detection of Human Enterovirus 71 by Reverse-Transcription and Loop-Mediated Isothermal Amplification: Cryopreservation Affected the Detection Ability,” Diagnostic Microbiology and Infectious Disease, Vol. 71, No. 3, 2011, pp. 244-251. doi:10.1016/j.diagmicrobio.2011.07.014
Abbreviations
CVA16, Coxsackievirus A16; EV71, human enterovirus 71; FDR, fluorescent detection reagent; HEV, human enteroviruses; HFMD, hand-foot-and-mouth disease; HNB, hydroxyl naphthol blue dye; nRT-PCR, nested reverse transcription polymerase chain reaction; qRT-PCR, quantitative real-time polymerase chain reaction; RT-LAMP, reverse transcription loop-mediated isothermal amplification; TCID50, 50% tissue culture infective dose.
NOTES
*The first three authors contributed equally to this study.
#Corresponding authors.

