Advances in Biological Chemistry
Vol.4 No.4(2014), Article
ID:47310,13
pages
DOI:10.4236/abc.2014.44032
IL-10 Inhibits LPS-Induced Expression of miR-147 in Murine Macrophages
Leah N. Cardwell, Brian K. Weaver*
Department of Biology, Missouri State University, Springfield, USA
Email: *brianweaver@missouristate.edu
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 20 April 2014; revised 31 May 2014; accepted 17 June 2014
ABSTRACT
Interleukin-10 (IL-10) mediates an anti-inflammatory response that constrains immune responses and limits inflammation-associated pathology. IL-10 does so, in part, by selectively inhibiting proinflammatory cytokine and chemokine expression induced in macrophages in response to Tolllike receptor (TLR) signaling. The IL-10-mediated anti-inflammatory response is executed through the activation of STAT3 leading to induction of target genes referred to as IL-10-induced genes. As miRNAs have emerged as important negative regulators of gene expression in various systems, we sought to address whether the IL-10-mediated anti-inflammatory response acts through regulated expression of miRNA genes. Using quantitative PCR-based arrays, we examined 140 miRNA genes with putative roles in inflammation for changes in expression in response to IL-10 and lipopolysaccharide (LPS) in primary mouse macrophages. IL-10 stimulation resulted in the inhibition of miR-147 expression induced in response to LPS, while having a potentiating effect on the induction of miR-455. miR-147 is the second TLR-induced miRNA, in addition to miR-155, identified to be counter-regulated by IL-10. Its suppression by IL-10 suggests that miR-147 may have an unknown pro-inflammatory function in TLR-triggered macrophages. The results extend the notion that IL-10 selectively regulates expression of miRNA genes, and that miRNA-mediated pathways are a component of the IL-10-mediated anti-inflammatory response.
Keywords:IL-10, Lipopolysaccharide, Macrophage, Inflammation, MicroRNA

1. Introduction
Cells of the innate immune system—including macrophages, dendritic cells, and neutrophils—recognize and respond to microbial pathogens using an array of germ-line encoded pattern recognition receptors (PRRs) [1] -[3] . PRRs are a diverse group of danger-signal sensors that fall into three major protein families: the membranebound Toll-like receptors (TLRs), the cytosolic NOD-like receptors (NLRs), and the cytosolic RIG-I-like receptors (RLRs). PRRs bind to specific cognate pathogen-associated molecular patterns (PAMPs) and signal for the activation of transcription factors that induce expression of numerous pro-inflammatory genes, including cytokines and chemokines, that promote the host inflammatory response [4] -[6] . This is exemplified by Toll-like Receptor 4 (TLR4) and its recognition of the prototypical PAMP lipopolysaccharide (LPS) produced by gramnegative bacteria [7] [8] . Although integral in host defenses, unconstrained inflammation can result in pathological and even life-threatening inflammatory conditions such as septic shock. Therefore, several anti-inflammatory pathways have evolved to impart negative regulation on PRR response pathways [9] [10] . The anti-inflammatory cytokine interleukin-10 (IL-10) has become recognized as a central player in this regulation and resolution of inflammation.
IL-10 acts on host cells by binding to the membrane-bound IL-10 receptor, a heterodimer of two polypeptide chains, IL-10R1 and IL-10R2, that form a tetrameric receptor when bound to cytokine [11] -[13] . IL-10 receptor engagement leads to trans-phosphorylation and activation of two JAK family receptor-associated tyrosine kinases, JAK1 and TYK2, which phosphorylate specific tyrosine residues in the IL-10R1 cytoplasmic domain. These phosphorylation events promote the recruitment, tyrosine-phosphorylation, and activation of the latent transcription factor STAT3 [14] [15] . The IL-10/STAT3-dependent activation of target genes (referred to as IL-10-induced genes) results in the execution of the IL-10-mediated anti-inflammatory response [16] -[18] . The importance of the IL-10/STAT3 signaling axis in constraining inflammation has been demonstrated by studies documenting the development of exacerbated inflammatory responses and autoimmune disease, particularly enterocolitis akin to human Crohn’s disease, in IL-10-, IL-10 receptor-, and STAT3-knockout mice [19] -[21] . Consistent with this, genetic alterations leading to impaired IL-10 responses have been linked with human diseases, including Crohn’s disease (and other inflammatory bowel diseases), arthritis, asthma, and psoriasis [13] [22] -[26] . Further studies utilizing tissue-specific knockout mice have revealed that macrophages are important cellular targets of IL-10’s actions in vivo [21] [27] [28] .
IL-10 acts on macrophages to suppress TLR responses that lead to the induction of pro-inflammatory gene expression. However, IL-10 does not work in an encompassing manner to block all signal transduction emanating from TLRs, nor does it act to block the expression of all TLR-induced pro-inflammatory genes. Instead, IL-10/STAT3 signaling works in a selective manner to inhibit expression of specific TLR-induced pro-inflammatory genes (e.g. TNF, IL-6, and IL-12), while leaving other genes unaffected (e.g. A20, IκBα, and IRF1) [10] [17] . TLR-induced genes that are inhibited by IL-10 stimulation have been referred to as “IL-10 counter-regulated genes”. IL-10 mediates the selective inhibition of pro-inflammatory gene expression via multiple mechanisms, including transcriptional repression, mRNA destabilization, and translational inhibition [16] [29] -[33] . In order to understand the mechanisms that underlie the anti-inflammatory response, researchers have worked to identify and characterize the genes induced by the IL-10/STAT3 pathway [34] -[39] . Hundreds of IL-10-induced genes have been identified, and a number of these, including SOCS3, BCL3, NFIL3, DUSP1, TTP, and TNIP3, have been shown to play specific roles in the anti-inflammatory response. An interesting phenomenon revealed by these studies, that further illustrates the selective nature of IL-10’s effects on TLR responses, is that while certain anti-inflammatory genes can be induced by IL-10 stimulation alone, many others are only up-regulated by IL-10 when cells are co-stimulated with a TLR agonist [36] -[38] . The term “IL-10 super-induced gene” was coined to describe these genes because IL-10 induces their expression synergistically along with TLR stimulation [37] .
Over the past decade, genes encoding small regulatory RNAs known as microRNAs (miRNAs) have been characterized as important regulators of animal development and homeostasis, including those with roles in development of the immune system and the immune response [40] -[44] . MicroRNAs act to negatively regulate the expression of specific protein-encoding genes largely through post-transcriptional mechanisms [45] -[48] . Partial or exact complementary base pairing between miRNA and the 3’-UTR of a target mRNA results in either translational inhibition or mRNA destabilization [48] -[50] . Several miRNAs have been identified as being induced in response to TLR or cytokine signaling [43] . Some of these have been characterized as having the ability to negatively regulate TLR or cytokine responses through the down-regulation of signaling proteins or cytokine gene expression [43] [51] -[54] . Considering these emerging aspects of miRNA function together with the classically known ability of IL-10 to counter-regulate specific pro-inflammatory genes through post-transcriptional mechanisms, we hypothesized that IL-10 may do so, in part, through the activation of one or more miRNAs.
In this study, we set out to determine whether the IL-10-mediated anti-inflammatory response involves a miRNA-mediated mechanism of counter-regulation by first attempting to identify miRNAs that could be induced by IL-10 in either resting or LPS-triggered macrophages. Using low-density quantitative PCR (qPCR)- based arrays, we screened for miRNAs that could be up-regulated in macrophages responding to IL-10 and LPS. As an unintended result of our screen, we identified an LPS-induced miRNA, namely miR-147, that is targeted for counter-regulation by IL-10. Thus, miR-147 joins miR-155 as a second miRNA known to be negatively regulated by IL-10 in TLR-triggered macrophages [55] . miR-155 has been shown to have pro-inflammatory effects, and thus, it is logical that IL-10 would act to suppress its expression [43] [56] -[58] . By comparison, the physiological functions of miR-147 are less clear. Based on its expression pattern of being induced in response to LPS/TLR signaling and counter-regulated by IL-10, we postulate that miR-147 may also play a role in promoting inflammation.
2. Materials and Methods
2.1. Cell Culture
Cell cultures were maintained in a 37˚C incubator with a humidified 5% CO2 atmosphere. Mouse bone marrow-derived macrophages (BMM) were prepared and cultured as described previously [38] [59] . In brief, femurs from 8-week-old C57BL/6J mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and bone marrow was eluted and plated at a density of 5 × 106 cells per 100 mm Petri dish (Fisher) in DMEM culture medium, supplemented with 10% fetal bovine serum (Hyclone), 5% donor horse serum (Atlanta Biologicals), 20% L cell-conditioned media, 2 mM L-glutamine (Hyclone), 1 mM sodium pyruvate (Hyclone), 50 U/ml penicillin (Hyclone), 50 μg/ml streptomycin (Hyclone), and 1 ng/ml M-CSF (eBioscience). After 7 days of culture, adherent macrophages (6 - 8 × 106 cells per dish) were harvested and re-plated for experiments into tissue culture plates (Corning) and fed with resting media (lacking 20% L cell-conditioned media) to allow the cells to become quiescent.
2.2. Cell Treatments and RNA Isolation
BMM were plated at a density of 5 × 106 cells per 60 mm plate and incubated in resting media for 24 h prior to treatments. Cells were treated with one or more of the following reagents alone or in combination: recombinant mouse IL-10 (eBioscience), rat anti-mouse IL-10R1 neutralizing monoclonal antibody (clone 1B1.3a) (BD Biosciences), and LPS derived from Escherichia coli O127:B8 (Sigma). At specific times post-stimulation (as indicated in the Results and Figures), cells were collected on ice and total RNA, including microRNAs, was purified using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. During RNA purification, genomic DNA was removed using an on-column DNase treatment (Qiagen).
2.3. Gene Expression Analysis for mRNA-Encoding Genes
First-strand cDNA was made from 0.5 μg of DNase-treated RNA using the miScript II RT Kit (Qiagen) according to the manufacturer’s protocol. In brief, RNA samples were subjected to reverse transcription at 37˚C for 60 min using the miScript HiFlex reaction buffer. Completed reactions were diluted with nuclease-free water to a volume of 250 μl (~2 ng/μl), and 10 μl (~20 ng cDNA) was used as template for PCR reactions. Relative gene expression was measured using SYBR Green-based qPCR. PCR reactions were performed in duplicate with a final reaction volume of 25 μl consisting of 12.5 μl GoTaq qPCR master mix (Promega), 10 μl cDNA template, PCR primers (final concentration of 200 nM each), and nuclease-free water. PCR was performed using an Mx3005P QPCR system (Agilent Technologies). PCR amplification was performed as follows: 95˚C (10 min), followed by 40 cycles of 95˚C (30 s), 60˚C (60 s), and a final dissociation curve segment to monitor the specificity of amplification. PCR primer pairs targeting mouse Tnip3, Bcl3, IL-6, and Gapdh were described previously [38] . Primer pairs were designed and tested based on the following parameters: primer length of 18 - 26 nt, primer melting temperature between 58˚C - 62˚C, amplicon length between 75 - 250 bp, hybridization to different exons spanning one or more introns of the target cDNA, PCR efficiency of >85% over a linear dynamic range of 6 log10, and correlation coefficient (r2) > 0.95. The Rpl13a primer pair was purchased as part of the Mouse Housekeeping Gene Primer Sets from RealTimePrimers.com. Primer sequences are available upon request. Relative expression values for genes of interest in cell samples were determined relative to mock-treated/ unstimulated cells using both the standard curve method (standard curves were made from serially diluted cDNA) and the ΔΔCt method of relative quantification. Expression values for genes of interest were normalized against the expression values of two endogenous reference genes: Gapdh and Rpl13a.
2.4. Gene Expression Analysis for miRNA Genes
The SYBR Green-based miScript miRNA PCR Mouse Immunopathology Array (Qiagen) and Mouse Inflammatory Response & Autoimmunity Array (Qiagen) were carried out according to the manufacturer’s protocol. In brief, 0.25 μg DNase-treated RNA was used to generate cDNA using the miScript II RT Kit (Qiagen) and the miScript HiSpec reaction buffer. Completed reactions were diluted with nuclease-free water to 200 μl (~1 ng/μl), and 1 ng cDNA template was used per PCR reaction. Confirmation and measurement of individual miRNA genes was performed using miScript Primer Assays (Qiagen) and the miScript SYBR Green PCR system (Qiagen). Reverse transcription reactions were performed with the miScript II RT Kit and the miScript HiSpec reaction buffer using 0.25 μg DNase-treated RNA and incubation at 37˚C for 60 min. Completed reactions were diluted with nuclease-free water to a final volume of 200 μl (~1 ng/μl), and 2.5 μl (~2.5 ng cDNA) was used as template for PCR reactions. PCR reactions were performed in duplicate with a final reaction volume of 25 μl consisting of 12.5 μl Quantitect SYBR Green master mix (Qiagen), 2.5 μl cDNA template, 2.5 μl of miScript Universal Primer, 2.5 μl miScript Primer Assay, and nuclease-free water. PCR amplification was performed as follows: 95˚C (15 min), followed by 40 cycles of 94˚C (15 s), 55˚C (30 s), 70˚C (30 s), and a final dissociation curve segment to monitor the specificity of amplification. Expression levels for miRNA genes of interest in cell samples were determined relative to mock-treated/unstimulated cells using the ΔΔCt method of relative quantification. Expression values for genes of interest were normalized against the expression levels of two endogenous reference genes: Snord68 and Snord72.
3. Results
3.1. Identification of MiRNAs Regulated in IL-10 and LPS Stimulated Mouse Macrophages
SYBR Green-based low-density miRNA qPCR arrays were used in an attempt to identify miRNAs that are upregulated by IL-10 in macrophages, and thus, that might play a role in the cytokine’s known anti-inflammatory effects. First, to ensure that cellular RNA samples were of suitable quality and to ensure that the macrophages had responded appropriately to the stimulations, we examined the expression of known IL-10- and LPS-induced protein-encoding genes: Bcl3 and Tnip3 [38] [60] . Primary mouse bone marrow-derived macrophages (BMM) were stimulated with IL-10, LPS, or the combination of IL-10 and LPS for 2, 8, and 16 h. When BMM were stimulated with IL-10 for 2 h, the induced expression of Bcl3 was observed as expected (Figure 1(a)). The induction of Bcl3 by IL-10 was blocked by the inclusion of a neutralizing monoclonal antibody against the IL-10R1 receptor chain. When cells were stimulated with LPS, Bcl3 expression was also induced. However, the induction by LPS was largely due to the autocrine effects of IL-10, as the inclusion of the neutralizing anti-IL-10R1 antibody blocked most of the effect. Cells stimulated with the combination of IL-10 and LPS displayed induction of Bcl3 comparable to that observed with either IL-10 or LPS stimulation. The second quality control gene examined, Tnip3, is known to have a different expression response pattern from that of Bcl3; Tnip3 had been identified as a gene that could be modestly induced by LPS alone, and not induced by IL-10 alone, but it could be synergistically induced by IL-10 in cells that were co-stimulated with LPS (i.e. an IL-10 super-induced gene) [38] . In accord, when BMM were stimulated with IL-10 for 2 h, Tnip3 expression was unchanged (Figure 1(a)). Stimulation with LPS led to an increase in Tnip3 expression, but this was largely due to the autocrine effects of IL-10 working in concert with the LPS/TLR4 signals, as shown by reduced induction when the neutralizing IL-10R1 antibody was added along with LPS. In agreement with previous reports, LPS stimulation in the absence of IL-10 induced modest levels of Tnip3 expression, and those levels were synergistically induced when IL-10 was combined with LPS (Figure 1(a)) [38] [61] . These results indicated that the reagents and cells were working properly and that the RNA samples were of appropriate quality to allow for the identification of novel IL-10-induced miRNA genes (akin to Bcl3) or IL-10 super-induced miRNA genes (akin to Tnip3).
As we were undertaking this project, another group reported on the identification of miRNAs induced in
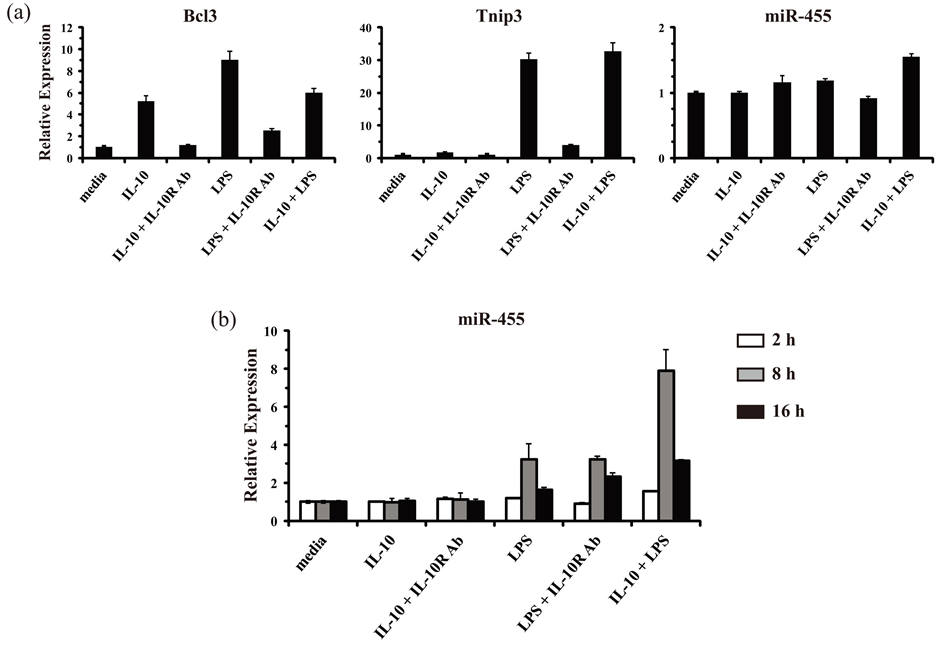
Figure 1. Validation of RNA/cDNA samples for the identification of miRNAs regulated by IL-10 and LPS in mouse macrophages. (a) Primary mouse bone marrow-derived macrophages (BMM) were either mock treated (media), or treated with IL-10 (10 ng/ml) alone, IL-10 plus an anti-IL-10R1-neutralizing antibody (Ab) (0.5 μg/ml), LPS (100 ng/ml) alone, LPS plus an anti-IL-10R1-neutralizing Ab, or IL-10 plus LPS together for 2 h. Total RNA was isolated and analyzed by RT-qPCR in duplicate. Relative expression values for genes of interest are presented as the mean ± s.d. of duplicate reactions after normalization against the reference genes Gapdh (in the case of Bcl3 and Tnip3) and Snord72 (in the case of miR-455). Results are representative of two independent experiments. (b) As described for panel (a) except that total RNA was analyzed from BMM stimulated for 2 h (white bars), 8 h (gray bars), and 16 h (black bars).
mouse macrophages in response to Candida albicans [62] . In addition to being regulated in response to C. albicans, that report also showed that miR-146b and miR-455 could be up-regulated by IL-10. In particular, miR- 455 was shown to be super-induced by IL-10 in LPS-triggered macrophages, in a manner analogous to the expression pattern observed with Tnip3. Therefore, we attempted to confirm that finding for miR-455 and use it as another positive control to verify the fidelity of our RNA samples. However, no induced expression was observed for miR-455 in BMM after 2 h of IL-10 and LPS stimulation (Figure 1(a)). Since inducible expression of miRNA genes in various systems often displays delayed kinetics, we examined miR-455 expression at later time points of IL-10 and LPS stimulation. Indeed, miR-455 was observed to be induced by LPS alone and super-induced by IL-10 in concert with LPS in cells at 8 h post-stimulation (Figure 1(b)). Therefore, an 8 h stimulation time was selected for use in the qPCR arrays aimed at identifying other IL-10-regulated miRNAs.
Results from the miRNA PCR arrays, which included 140 unique miRNAs with known or putative roles in the immune response, indicated a number of miRNAs as either induced or repressed ≥ 3-fold in IL-10 plus LPS stimulated macrophages relative to resting (media) macrophages (Figure 2(a)). Of note was the marked up-regulation observed for miR-155 (induced ~50-fold), which had been previously reported to be induced in both mouse and human monocytes/macrophages in response to TLR-stimulation, and also to be counter-regulated by IL-10 [55] [63] . In addition, miR-146b was also observed to be induced (~3-fold) by IL-10 and LPS, in accord with previous reports [62] [64] . A third miRNA found to be up-regulated in IL-10 plus LPS treated macrophages was miR-147 (induced ~4-fold). This miRNA had been previously reported to be

Figure 2. Identification of miRNAs regulated by IL-10 and LPS in mouse macrophages. (a) RNA/cDNA samples derived from BMM that were either mock treated (media) or treated with IL-10 (10 ng/ml) plus LPS (100 ng/ml) together for 8 h were analyzed using low-density SYBR Green-based miScript miRNA PCR arrays: Immunopathology Array (left panel) and Inflammatory Response & Autoimmunity Array (right panel). Relative expression values are presented as arbitrary units on a log scale after normalization against the six included reference genes: Snord61, Snord68, Snord72, Snord95, Snord96A, and RNU6-2. Each dot represents the relative expression level for an individual miRNA gene. Results are representative of two independent experiments. (b) BMM were either mock treated (media), or treated with IL-10 (10 ng/ml) alone, IL-10 plus an anti-IL-10R1-neutralizing antibody (Ab) (0.5 μg/ml), LPS (100 ng/ml) alone, LPS plus an anti-IL-10R1-neutralizing Ab, or IL-10 plus LPS together for 8 h. Total RNA was isolated and analyzed by RT-qPCR in duplicate for the expression of miR-455 and miR-147. Relative expression values are presented as mean ± s.d. of duplicate reactions after normalization against two reference genes: Snord68 and Snord72. Results are representative of three independent experiments.
induced by TLR stimulation, however whether or not it could be regulated by IL-10 remained unknown [65] . Of the 10 other miRNAs observed to be induced ≥3-fold using the arrays, none of them could be reproducibly confirmed to be induced by LPS, IL-10, or the combination of LPS and IL-10 in independent experiments using individual miRNA-targeted qPCR assays (data not shown).
During our analyses, miR-187 was reported to be induced by IL-10 in a speciesor cell-type-specific manner: induced in human monocytes but not in mouse macrophages [66] . In agreement, no increase was observed for miR-187 in our array results using mouse macrophages. One miRNA, namely miR-200c, appeared to be markedly repressed in IL-10 and LPS treated cells (Figure 2(a)); however, this was not found to be a reproducible result in independent experiments using individual miRNA-targeted qPCR assays (data not shown). The only miRNA, other than miR-155, identified in the array experiments that could be reproducibly observed to be induced in IL-10 and LPS treated macrophages was miR-147. The effects of LPS, IL-10, or the combination of IL-10 and LPS stimulation for 8 h on miR-147 expression are presented in Figure 2(b). For comparison, miR-455 expression could be observed to be modestly induced in response to LPS alone, unaffected by IL-10 alone, but super-induced by IL-10 acting in concert with LPS (Figure 2(b)). miR-147 was induced by LPS stimulation in a manner that was independent of the autocrine effects of IL-10. IL-10 stimulation alone or in concert with LPS did not induce the expression of miR-147. Instead, IL-10 appeared to partially repress the induction of miR-147 in response to LPS (Figure 2(b)). These results suggested that miR-147 may be a novel IL-10 counter-regulated miRNA gene. In this respect, it would join a list of specific pro-inflammatory genes known to be counter-regulated by IL-10, including the cytokines IL-1, IL-6, IL-12, TNF, and the one other miRNA miR-155.
3.2. Temporal Analysis of MiR-147 in LPS-Treated Macrophages and Its Counter-Regulation by IL-10
In order to confirm and extend the result that LPS-induced miR-147 expression could be partially inhibited by IL-10 observed at 8 h of stimulation (Figure 2(b)), we performed a time-course analysis of mouse macrophages stimulated from 0 to 36 h (Figure 3). The extended temporal analysis confirmed that miR-455 behaves as an IL-10 super-induced miRNA gene with somewhat delayed induction kinetics, whose expression is induced by LPS alone and potentiated by IL-10 in the presence of LPS. In this respect, miR-455 behaves similarly to the known IL-10 super-induced gene Tnip3 (Figure 3, left panels). In contrast, miR-147 was induced robustly by LPS alone with relatively delayed kinetics, continuing to increase in expression up to 36 h post-stimulation. IL-10 stimulation alone had no effect on miR-147. But, IL-10 markedly repressed the LPS-induced expression of miR-147 over the extended time-course. For comparison, induction of a known IL-10 counter-regulated cyto-
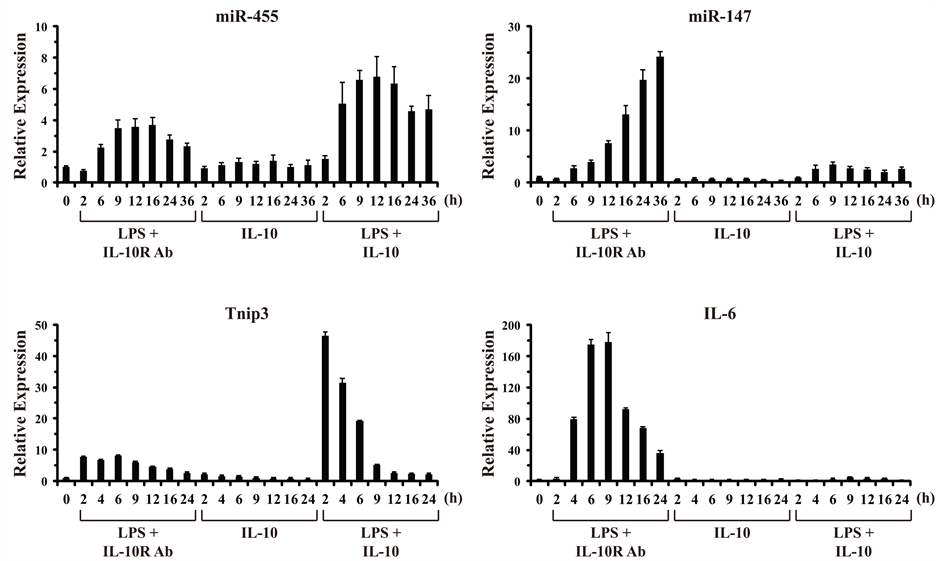
Figure 3. IL-10 inhibits LPS-induced expression of miR-147. BMM were stimulated over a time-course from 0 to 36 h with LPS (100 ng/ml) in the presence of an anti-IL-10R1-neutralizing Ab (0.5 μg/ml), IL-10 (10 ng/ml), or LPS plus IL-10 together. Total RNA was harvested from cells at the indicated times (h), and analyzed by RT-qPCR in duplicate. Relative expression values are presented as mean ± s.d. of duplicate reactions after normalization to two reference genes: Gapdh and Rpl13a (in the case of Tnip3 and IL-6), and Snord68 and Snord72 (in the case of miR-455 and miR-147). Results are representative of two independent experiments.
kine gene, IL-6, was also observed to be markedly inhibited by IL-10 (Figure 3, right panels). Thus, the LPS/ TLR-induced expression of miR-147 is counter-regulated by the anti-inflammatory cytokine IL-10.
4. Discussion
In this study, we sought to address the hypothesis that IL-10 could induce the expression of one or more miRNAs that would then act in an anti-inflammatory manner to selectively suppress expression of certain pro-inflammatory genes. Although we were unable to identify a novel IL-10-induced miRNA, studies reported by others in recent years have begun to support this hypothesis. First, miR-455, miR-146a, and miR-146b were identified as genes that could be induced in mouse macrophages in response to Candida albicans or LPS, and also induced/super-induced by IL-10 [62] . At that time, the roles that these miRNAs might play in the IL-10-mediated anti-inflammatory response remained unknown. A subsequent report identified a functional role for miR-146b in the IL-10-mediated anti-inflammatory response in human monocytes, in which its induction suppressed LPSdependent expression of several cytokines and chemokines [64] . Secondly, miR-187 was reported as being induced in response to IL-10 alone, and also super-induced by IL-10 acting in concert with LPS in human monocytes [66] . The induction of miR-187 by IL-10 appeared to be species-specific, occurring in human monocytes but not in mouse macrophages. miR-187 was shown to have an anti-inflammatory activity, as it negatively regulated the expression of the cytokines TNF, IL-6, and IL-12 [66] . We were able to confirm that IL-10 induces the expression of miR-455 and miR-146b, but not miR-187 in mouse macrophages (Figure 2). It remains to be determined whether miR-455, miR-146b, or other as yet unidentified miRNAs, play a functional role in the IL-10-mediated anti-inflammatory response in the mouse system, as has been suggested for the human system.
As an unplanned result of our exploration into IL-10-induced miRNAs, we found that the known LPS-induced miR-147 is counter-regulated by IL-10. In this respect, it joins miR-155, a well studied miRNA with established roles in lymphoid and myeloid oncogenesis, as well as in the inflammatory response [41] [58] [67] . TLR signaling induces miR-155 expression in macrophages and IL-10 is able to counter-regulate that induction [55] [68] . Thus, IL-10 constrains the pro-inflammatory actions of miR-155, which include: increasing TNF production, enhancing PI3K-dependent activation of NF-κB and MAPKs, enhancing cytokine responses through JAK-STAT signaling, and potentiating Th1/Th17 cell polarization [41] [53] [55] [58] [69] . Compared with miR-155, little is known about miR-147 and its role(s) in the immune response.
miR-147 was originally identified as a miRNA that could be induced in macrophages in response to TLR stimulation [65] . The miR-147 precursor transcript is expressed as part of the NMES1 (normal mucosa esophagus specific 1) gene primary transcript, from which it is processed into the mature miRNA. TLR signals were found to induce the expression of the miR-147/NMES1 primary transcript via a transcriptional mechanism that depends on both NF-κB and IRF3 activation [65] . Use of miR-147 mimics and inhibitors indicated that this miRNA can act to suppress the production of certain pro-inflammatory cytokines, including TNF and IL-6 [65] . The mechanism by which miR-147 acts to limit the production of these cytokines remains unknown, and thus far, no TLR signaling proteins have been identified as miR-147 targets. Since miR-147 is expressed with delayed and prolonged kinetics, it has been proposed to be more likely important in suppressing inflammatory cytokine production at late time points after TLR stimulation [65] . If miR-147 is indeed acting as a TLR-induced negative feedback regulator of inflammatory cytokine production, it would be uncharacteristic of IL-10 to target it for down-regulation as observed herein (Figure 3). Therefore, it is reasonable to hypothesize that miR-147 may have some presently unknown pro-inflammatory function(s). Alternatively, as miR-147 is up-regulated by TLR signaling at the transcriptional level along with NMES1, it is possible that IL-10 is actually aimed at targeting NMES1 for down-regulation.
As with miR-147, a function for NMES1 in the immune response is not presently known. NMES1 was first identified as a gene down-regulated in human esophageal squamous cell carcinoma, suggesting that it may act as a tumor suppressor [70] . Recently, suggestive of a role in the immune system, NMES1 was identified as a gene expressed highly in dendritic cells with an activated/effector phenotype, but not in ones with a regulatory/tolerogenic phenotype [71] . Consistent with the aforementioned result regarding NMES1 gene expression being modulated in cancer, miRNA profiling experiments have revealed miR-147 to be up-regulated in human gastric cancer cells [72] . Another recent study corroborated this proposed role for miR-147 as a tumor suppressor, as it was found to down-regulate expression of proteins in the EGFR/cell-cycle network, thereby inhibiting G1/S transition and proliferation of breast cancer cells [73] . These results were followed by others showing that expression of miR-147 could reverse epithelial-to-mesenchymal transition, and inhibit both colon and lung cancer cell proliferation [74] . Given the links between chronic inflammation and cancer, a persistence of IL-10 leading to a reduction of miR-147 (and NMES1) over a long period of time may act to promote oncogenesis. Likewise, miR-147 counter-regulation may play a role in the known effect of IL-10 in promoting B cell proliferation and lymphoma [11] [13] [75] .
In addition to ascertaining the physiological role miR-147 might play in the inflammatory response and/or in the IL-10-mediated anti-inflammatory response, future research should be aimed at determining the mechanism by which IL-10 inhibits LPS/TLR-induced miR-147 expression. For several decades there has been an active research focus on understanding how IL-10 selectively counter-regulates TLR-induced protein-encoding genes. Although the complete picture continues to be elucidated, IL-10 is known to act through multiple IL-10-induced gene products that work in specific ways to selectively inhibit pro-inflammatory gene expression at both transcriptional and post-transcriptional levels [10] [13] [17] . One might expect the mechanisms that IL-10 employs to counter-regulate miRNA induction to be comparably complex. This question has begun to be addressed with miR-155. First, the mechanism by which IL-10 inhibits TLR-induced miR-155 expression was shown to be mediated at the level of transcriptional inhibition [55] . This result was followed by work showing that IL-10 counter-regulates miR-155 by first down-regulating the expression of the Ets2 transcription factor, which is required for the LPS-induced transcription of miR-155 [76] . Reminiscent of the complexities regarding how IL-10 counter-regulates protein-encoding genes, another group reported that IL-10’s inhibitory effect on TLR-induced miR-155 was mediated at a post-transcriptional level via destabilizing the pri-miR-155 and pre-miR-155 transcripts [68] . It is certainly possible that IL-10 is acting on miR-155 at multiple levels, as has been shown for the mechanisms by which IL-10 can inhibit cytokine and chemokine gene expression [30] -[32] [60] [77] . The regulation of the cytokine TNF is a good example, as IL-10 antagonizes induction of this gene at the levels of transcriptional repression, mRNA destabilization, and translational repression. It remains to be seen whether IL-10 is acting to inhibit miR-147 at a transcriptional and/or post-transcriptional level. Furthermore, it will be important to identify the IL-10-induced gene(s) responsible for suppressing miR-147, and for that matter, the gene(s) responsible for suppressing miR-155.
5. Conclusion
In conclusion, we have shown that miR-147 is an LPS/TLR-induced gene that is counter-regulated by IL-10 in mouse macrophages. Despite its being reported as an anti-inflammatory miRNA, its suppression by IL-10 suggests that it may play additional unidentified roles in promoting TLR responses and inflammation. A better mechanistic understanding of how IL-10 counter-regulates miR-147 may help to elucidate the physiological consequences to the inflammatory response if this regulatory mechanism is abrogated. For example, if mice could be engineered so that IL-10 was specifically unable to suppress miR-147, the effect of overproduction of this miRNA in response to TLRs could be examined to test for a promotion of inflammation and associated exacerbated pathology. Given the putative role this miRNA plays in cancer, such an animal model system could also be used to test whether chronic overproduction of miR-147 might promote oncogenesis in models of inflammation-induced cancer. Finally, as miR-147 is only the second miRNA known to be counter-regulated by IL-10, it will be important to extend this work by using whole miRNome-based array platforms to characterize the full extent to which miRNA genes are counter-regulated by IL-10.
Acknowledgements
We thank all of the members of the Weaver laboratory for helpful discussions in the design and execution of experiments, particularly, Srinivas Reddy Doma for technical assistance in the performance of the miRNA qPCR arrays. We also thank Courtney Coleman, John Steiert, and Paula Steiert for advice and critical reading of the manuscript. This work was supported by funds awarded by the Biology Department and the Graduate College of Missouri State University and the Missouri State University Foundation.
References
- Broz, P. and Monack, D.M. (2013) Newly Described Pattern Recognition Receptors Team up against Intracellular Pathogens. Nature Reviews Immunology, 13, 551-565. http://dx.doi.org/10.1038/nri3479
- Medzhitov, R. (2008) Origin and Physiological Roles of Inflammation. Nature, 454, 428-435. http://dx.doi.org/10.1038/nature07201
- Takeuchi, O. and Akira, S. (2010) Pattern Recognition Receptors and Inflammation. Cell, 140, 805-820. http://dx.doi.org/10.1016/j.cell.2010.01.022
- O’Neill, L.A. and Bowie, A.G. (2007) The Family of Five: TIR-Domain-Containing Adaptors in Toll-Like Receptor Signalling. Nature Reviews Immunology, 7, 353-364. http://dx.doi.org/10.1038/nri2079
- Takeda, K. and Akira, S. (2005) Toll-Like Receptors in Innate Immunity. International Immunology, 17, 1-14. http://dx.doi.org/10.1093/intimm/dxh186
- Yamamoto, M. and Akira, S. (2005) TIR Domain-Containing Adaptors Regulate TLR Signaling Pathways. Advances in Experimental Medicine and Biology, 560, 1-9. http://dx.doi.org/10.1007/0-387-24180-9_1
- Poltorak, A., He, X., Smirnova, I., Liu, M.Y., Van Huffel, C., Du, X., Birdwell, D., Alejos, E., Silva, M., Galanos, C., Freudenberg, M., Ricciardi-Castagnoli, P., Layton, B. and Beutler, B. (1998) Defective LPS Signaling in C3H/HeJ and C57BL/10ScCr Mice: Mutations in Tlr4 Gene. Science, 282, 2085-2088. http://dx.doi.org/10.1126/science.282.5396.2085
- Rock, F.L., Hardiman, G., Timans, J.C., Kastelein, R.A. and Bazan, J.F. (1998) A Family of Human Receptors Structurally Related to Drosophila Toll. Proceedings of the National Academy of Sciences of the United States of America, 95, 588-593. http://dx.doi.org/10.1073/pnas.95.2.588
- Kondo, T., Kawai, T. and Akira, S. (2012) Dissecting Negative Regulation of Toll-Like Receptor Signaling. Trends in Immunology, 33, 449-458. http://dx.doi.org/10.1016/j.it.2012.05.002
- Murray, P. J. and Smale, S.T. (2012) Restraint of Inflammatory Signaling by Interdependent Strata of Negative Regulatory Pathways. Nature Immunology, 13, 916-924. http://dx.doi.org/10.1038/ni.2391
- Moore, K. W., de Waal Malefyt, R., Coffman, R. L., and O’Garra, A. (2001) Interleukin-10 and the interleukin-10 receptor, Annual Review of Immunology, 19, 683-765. http://dx.doi.org/10.1146/annurev.immunol.19.1.683
- Pestka, S., Krause, C.D., Sarkar, D., Walter, M.R., Shi, Y. and Fisher, P.B. (2004) Interleukin-10 and Related Cytokines and Receptors. Annual Review of Immunology, 22, 929-979. http://dx.doi.org/10.1146/annurev.immunol.22.012703.104622
- Sabat, R., Grutz, G., Warszawska, K., Kirsch, S., Witte, E., Wolk, K. and Geginat, J. (2010) Biology of Interleukin-10. Cytokine & Growth Factor Reviews, 21, 331-344. http://dx.doi.org/10.1016/j.cytogfr.2010.09.002
- Donnelly, R.P., Dickensheets, H. and Finbloom, D.S. (1999) The Interleukin-10 Signal Transduction Pathway and Regulation of Gene Expression in Mononuclear Phagocytes. Journal of Interferon & Cytokine Research, 19, 563-573. http://dx.doi.org/10.1089/107999099313695
- Weber-Nordt, R.M., Riley, J.K., Greenlund, A.C., Moore, K.W., Darnell, J.E. and Schreiber, R.D. (1996) Stat3 Recruitment by Two Distinct Ligand-Induced, Tyrosine-Phosphorylated Docking Sites in the Interleukin-10 Receptor Intracellular Domain. Journal of Biological Chemistry, 271, 27954-27961. http://dx.doi.org/10.1074/jbc.271.44.27954
- Grutz, G. (2005) New Insights into the Molecular Mechanism of Interleukin-10-Mediated Immunosuppression. Journal of Leukocyte Biology, 77, 3-15.
- Hutchins, A.P., Diez, D. and Miranda-Saavedra, D. (2013) The IL-10/STAT3-Mediated Anti-Inflammatory Response: Recent Developments and Future Challenges. Briefings in Functional Genomics, 12, 489-498. http://dx.doi.org/10.1093/bfgp/elt028
- Murray, P.J. (2006) Understanding and Exploiting the Endogenous Interleukin-10/STAT3-Mediated Anti-Inflammatory Response. Current Opinion in Pharmacology, 6, 379-386. http://dx.doi.org/10.1016/j.coph.2006.01.010
- Kühn, R., Löhler, J., Rennick, D., Rajewsky, K. and Müller, W. (1993) Interleukin-10-Deficient Mice Develop Chronic Enterocolitis. Cell, 75, 263-274. http://dx.doi.org/10.1016/0092-8674(93)80068-P
- Spencer, S.D., Di Marco, F., Hooley, J., Pitts-Meek, S., Bauer, M., Ryan, A.M., Sordat, B., Gibbs, V.C. and Aguet, M. (1998) The Orphan Receptor CRF2-4 Is an Essential Subunit of the Interleukin 10 Receptor. Journal of Experimental Medicine, 187, 571-578. http://dx.doi.org/10.1084/jem.187.4.571
- Takeda, K., Clausen, B.E., Kaisho, T., Tsujimura, T., Terada, N., Forster, I. and Akira, S. (1999) Enhanced Th1 Activity and Development of Chronic Enterocolitis in Mice Devoid of Stat3 in Macrophages and Neutrophils. Immunity, 10, 39-49. http://dx.doi.org/10.1016/S1074-7613(00)80005-9
- Asadullah, K., Sabat, R., Friedrich, M., Volk, H.D. and Sterry, W. (2004) Interleukin-10: An Important Immunoregulatory Cytokine with Major Impact on Psoriasis. Current Drug Targets-Inflammation & Allergy, 3, 185-192. http://dx.doi.org/10.2174/1568010043343886
- Correa, I., Veny, M., Esteller, M., Pique, J.M., Yague, J., Panes, J. and Salas, A. (2009) Defective IL-10 Production in Severe Phenotypes of Crohn’s Disease. Journal of Leukocyte Biology, 85, 896-903. http://dx.doi.org/10.1189/jlb.1108698
- Glocker, E.O., Kotlarz, D., Klein, C., Shah, N. and Grimbacher, B. (2011) IL-10 and IL-10 Receptor Defects in Humans. Annals of the New York Academy of Sciences, 1246, 102-107. http://dx.doi.org/10.1111/j.1749-6632.2011.06339.x
- Louis, E., Libioulle, C., Reenaers, C., Belaiche, J. and Georges, M. (2009) Genetics of Ulcerative Colitis: The Come-Back of Interleukin 10. Gut, 58, 1173-1176. http://dx.doi.org/10.1136/gut.2008.169235
- Shah, N., Kammermeier, J., Elawad, M. and Glocker, E.O. (2012) Interleukin-10 and Interleukin-10-Receptor Defects in Inflammatory Bowel Disease. Current Allergy & Asthma Reports, 12, 373-379. http://dx.doi.org/10.1007/s11882-012-0286-z
- Pils, M.C., Pisano, F., Fasnacht, N., Heinrich, J.M., Groebe, L., Schippers, A., Rozell, B., Jack, R.S. and Müller, W. (2010) Monocytes/Macrophages and/or Neutrophils Are the Target of IL-10 in the LPS Endotoxemia Model. European Journal of Immunology, 40, 443-448. http://dx.doi.org/10.1002/eji.200939592
- Welte, T., Zhang, S.S., Wang, T., Zhang, Z., Hesslein, D.G., Yin, Z., Kano, A., Iwamoto, Y., Li, E., Craft, J.E., Bothwell, A.L., Fikrig, E., Koni, P.A., Flavell, R.A. and Fu, X.Y. (2003) STAT3 Deletion during Hematopoiesis Causes Crohn’s Disease-Like Pathogenesis and Lethality: A Critical Role of STAT3 in Innate Immunity. Proceedings of the National Academy of Sciences of the United States of America, 100, 1879-1884. http://dx.doi.org/10.1073/pnas.0237137100
- Biswas, R., Datta, S., Gupta, J.D., Novotny, M., Tebo, J. and Hamilton, T.A. (2003) Regulation of Chemokine mRNA Stability by Lipopolysaccharide and IL-10. The Journal of Immunology, 170, 6202-6208. http://dx.doi.org/10.4049/jimmunol.170.12.6202
- Kim, H.S., Armstrong, D., Hamilton, T.A. and Tebo, J.M. (1998) IL-10 Suppresses LPS-Induced KC mRNA Expression via a Translation-Dependent Decrease in mRNA Stability. Journal of Leukocyte Biology, 64, 33-39.
- Kontoyiannis, D., Kotlyarov, A., Carballo, E., Alexopoulou, L., Blackshear, P.J., Gaestel, M., Davis, R., Flavell, R. and Kollias, G. (2001) Interleukin-10 Targets p38 MAPK to Modulate ARE-Dependent TNF mRNA Translation and Limit Intestinal Pathology. EMBO Journal, 20, 3760-3770.
- Murray, P.J. (2005) The Primary Mechanism of the IL-10-Regulated Antiinflammatory Response Is to Selectively Inhibit Transcription. Proceedings of the National Academy of Sciences of the United States of America, 102, 8686-8691. http://dx.doi.org/10.1073/pnas.0500419102
- Smallie, T., Ricchetti, G., Horwood, N.J., Feldmann, M., Clark, A.R. and Williams, L.M. (2010) IL-10 Inhibits Transcription Elongation of the Human TNF Gene in Primary Macrophages. Journal of Experimental Medicine, 207, 2081-2088. http://dx.doi.org/10.1084/jem.20100414
- Antoniv, T.T., Park-Min, K.H. and Ivashkiv, L.B. (2005) Kinetics of IL-10-Induced Gene Expression in Human Macrophages. Immunobiology, 210, 87-95. http://dx.doi.org/10.1016/j.imbio.2005.05.003
- Jung, M., Sabat, R., Krätzschmar, J., Seidel, H., Wolk, K., Schönbein, C., Schutt, S., Friedrich, M., Docke, W.D., Asadullah, K., Volk, H.D. and Grütz, G. (2004) Expression Profiling of IL-10-Regulated Genes in Human Monocytes and Peripheral Blood Mononuclear Cells from Psoriatic Patients during IL-10 Therapy. European Journal of Immunology, 34, 481-493. http://dx.doi.org/10.1002/eji.200324323
- Lang, R., Patel, D., Morris, J.J., Rutschman, R.L. and Murray, P.J. (2002) Shaping Gene Expression in Activated and Resting Primary Macrophages by IL-10. The Journal of Immunology, 169, 2253-2263. http://dx.doi.org/10.4049/jimmunol.169.5.2253
- Perrier, P., Martinez, F.O., Locati, M., Bianchi, G., Nebuloni, M., Vago, G., Bazzoni, F., Sozzani, S., Allavena, P. and Mantovani, A. (2004) Distinct Transcriptional Programs Activated by Interleukin-10 with or without Lipopolysaccharide in Dendritic Cells: Induction of the B Cell-Activating Chemokine, CXC Chemokine Ligand 13. The Journal of Immunology, 172, 7031-7042. http://dx.doi.org/10.4049/jimmunol.172.11.7031
- Weaver, B.K., Bohn, E., Judd, B.A., Gil, M.P. and Schreiber, R.D. (2007) ABIN-3: A Molecular Basis for Species Divergence in Interleukin-10-Induced Anti-Inflammatory Actions. Molecular and Cellular Biology, 27, 4603-4616. http://dx.doi.org/10.1128/MCB.00223-07
- Williams, L., Jarai, G., Smith, A. and Finan, P. (2002) IL-10 Ex-pression Profiling in Human Monocytes. Journal of Leukocyte Biology, 72, 800-809.
- Bushati, N. and Cohen, S.M. (2007) MicroRNA Functions. Annual Review of Cell and Developmental Biology, 23, 175-205. http://dx.doi.org/10.1146/annurev.cellbio.23.090506.123406
- Xiao, C. and Rajewsky, K. (2009) MicroRNA Control in the Immune System: Basic Principles. Cell, 136, 26-36. http://dx.doi.org/10.1016/j.cell.2008.12.027
- O’Connell, R.M., Rao, D.S., Chaudhuri, A.A. and Baltimore, D. (2010) Physiological and Pathological Roles for MicroRNAs in the Immune System. Nature Reviews Immunology, 10, 111-122. http://dx.doi.org/10.1038/nri2708
- O’Connell, R.M., Rao, D.S. and Baltimore, D. (2012) MicroRNA Regulation of Inflammatory Responses. Annual Review of Immunology, 30, 295-312. http://dx.doi.org/10.1146/annurev-immunol-020711-075013
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W. and Tuschl, T. (2001) Identification of Novel Genes Coding for Small Expressed RNAs. Science, 294, 853-858.
- Jing, Q., Huang, S., Guth, S., Zarubin, T., Motoyama, A., Chen, J., Di Padova, F., Lin, S.C., Gram, H. and Han, J. (2005) Involvement of MicroRNA in AU-Rich Element-Mediated mRNA Instability. Cell, 120, 623-634. http://dx.doi.org/10.1016/j.cell.2004.12.038
- Baek, D., Villén, J., Shin, C., Camargo, F.D., Gygi, S.P. and Bartel, D.P. (2008) The Impact of MicroRNAs on Protein Output. Nature, 455, 64-71. http://dx.doi.org/10.1038/nature07242
- Selbach, M., Schwänhausser, B., Thierfelder, N., Fang, Z., Khanin, R. and Rajewsky, N. (2008) Widespread Changes in Protein Synthesis Induced by MicroRNAs. Nature, 455, 58-63. http://dx.doi.org/10.1038/nature07228
- Guo, H., Ingolia, N.T., Weissman, J.S. and Bartel, D.P. (2010) Mammalian MicroRNAs Predominantly Act to Decrease Target mRNA Levels. Nature, 466, 835-840. http://dx.doi.org/10.1038/nature09267
- Valencia-Sanchez, M.A., Liu, J., Hannon, G.J. and Parker, R. (2006) Control of Translation and mRNA Degradation by miRNAs and siRNAs. Genes & Development, 20, 515-524. http://dx.doi.org/10.1101/gad.1399806
- Huntzinger, E. and Izaurralde, E. (2011) Gene Silencing by MicroRNAs: Contributions of Translational Repression and mRNA Decay. Nature Reviews Genetics, 12, 99-110. http://dx.doi.org/10.1038/nrg2936
- McCoy, C.E. (2011) The Role of miRNAs in Cytokine Signaling. Frontiers in Bioscience, 16, 2161-2171. http://dx.doi.org/10.2741/3845
- O’Neill, L.A., Sheedy, F.J. and McCoy, C.E. (2011) MicroRNAs: The Fine-Tuners of Toll-Like Receptor Signaling. Nature Reviews Immunology, 11, 163-175. http://dx.doi.org/10.1038/nri2957
- Kohanbash, G. and Okada, H. (2012) MicroRNAs and STAT Interplay. Seminars in Cancer Biology, 22, 70-75. http://dx.doi.org/10.1016/j.semcancer.2011.12.010
- Li, Y. and Shi, X. (2013) MicroRNAs in the Regulation of TLR and RIG-I Pathways. Cellular & Molecular Immunology, 10, 65-71. http://dx.doi.org/10.1038/cmi.2012.55
- McCoy, C.E., Sheedy, F.J., Qualls, J.E., Doyle, S.L., Quinn, S.R., Murray, P.J. and O’Neill, L.A. (2010) IL-10 Inhibits miR-155 Induction by Toll-Like Receptors. The Journal of Biological Chemistry, 285, 20492-20498. http://dx.doi.org/10.1074/jbc.M110.102111
- O’Connell, R.M., Kahn, D., Gibson, W.S., Round, J.L., Scholz, R.L., Chaudhuri, A.A., Kahn, M.E., Rao, D.S. and Baltimore, D. (2010) MicroRNA-155 Promotes Autoimmune Inflammation by Enhancing Inflammatory T Cell Development. Immunity, 33, 607-619. http://dx.doi.org/10.1016/j.immuni.2010.09.009
- Blüml, S., Bonelli, M., Niederreiter, B., Puchner, A., Mayr, G., Hayer, S., Koenders, M.I., van den Berg, W.B., Smolen, J. and Redlich, K. (2011) Essential Role of MicroRNA-155 in the Pathogenesis of Autoimmune Arthritis in Mice. Arthritis & Rheumatology, 63, 1281-1288. http://dx.doi.org/10.1002/art.30281
- Liu, G. and Abraham, E. (2013) ATVB in Focus: MicroRNAs: From Basic Mechanisms to Clinical Application in Cardiovascular Medicine. MicroRNAs in Immune Response and Macrophage Polarization. Arteriosclerosis, Thrombosis, and Vascular Biology, 33, 170-177. http://dx.doi.org/10.1161/ATVBAHA.112.300068
- Celada, A., Gray, P.W., Rinderknecht, E. and Schreiber, R.D. (1984) Evidence for a Gamma-Interferon Receptor That Regulates Macrophage Tumoricidal Activity. Journal of Experimental Medicine, 160, 55-74. http://dx.doi.org/10.1084/jem.160.1.55
- Kuwata, H., Watanabe, Y., Miyoshi, H., Yamamoto, M., Kaisho, T., Takeda, K. and Akira, S. (2003) IL-10-Inducible Bcl-3 Negatively Regulates LPS-Induced TNF-Alpha Production in Macrophages. Blood, 102, 4123-4129. http://dx.doi.org/10.1182/blood-2003-04-1228
- Wullaert, A., Verstrepen, L., Van Huffel, S., Adib-Conquy, M., Cornelis, S., Kreike, M., Haegman, M., El Bakkouri, K., Sanders, M., Verhelst, K., Carpentier, I., Cavaillon, J.M., Heyninck, K. and Beyaert, R. (2007) LIND/ABIN-3 Is a Novel Lipopolysaccharide-Inducible Inhibitor of NF-kappaB Activation. The Journal of Biological Chemistry, 282, 81-90. http://dx.doi.org/10.1074/jbc.M607481200
- Monk, C.E., Hutvagner, G. and Arthur, J.S. (2010) Regulation of miRNA Transcription in Macrophages in Response to Candida albicans. PLoS ONE, 5, e13669. http://dx.doi.org/10.1371/journal.pone.0013669
- O’Connell, R.M., Taganov, K.D., Boldin, M.P., Cheng, G. and Baltimore, D. (2007) MicroRNA-155 Is Induced during the Macrophage Inflammatory Response. Proceedings of the National Academy of Sciences of the United States of America, 104, 1604-1609. http://dx.doi.org/10.1073/pnas.0610731104
- Curtale, G., Mirolo, M., Renzi, T.A., Rossato, M., Bazzoni, F. and Locati, M. (2013) Negative Regulation of Toll-Like Receptor 4 Signaling by IL-10-Dependent MicroRNA-146b. Proceedings of the National Academy of Sciences of the United States of America, 110, 11499-11504. http://dx.doi.org/10.1073/pnas.1219852110
- Liu, G., Friggeri, A., Yang, Y., Park, Y.J., Tsuruta, Y. and Abraham, E. (2009) MiR-147, a MicroRNA That Is Induced upon Toll-Like Receptor Stimulation, Regulates Murine Macrophage Inflammatory Responses. Proceedings of the National Academy of Sciences of the United States of America, 106, 15819-15824. http://dx.doi.org/10.1073/pnas.0901216106
- Rossato, M., Curtale, G., Tamassia, N., Castellucci, M., Mori, L., Gasperini, S., Mariotti, B., De Luca, M., Mirolo, M., Cassatella, M.A., Locati, M. and Bazzoni, F. (2012) IL-10-Induced MicroRNA-187 Negatively Regulates TNF-Alpha, IL-6, and IL-12p40 Production in TLR4-Stimulated Monocytes. Proceedings of the National Academy of Sciences of the United States of America, 109, E3101-3110. http://dx.doi.org/10.1073/pnas.1209100109
- Vigorito, E., Kohlhaas, S., Lu, D. and Leyland, R. (2013) MiR-155: An Ancient Regulator of the Immune System. Immunological Reviews, 253, 146-157. http://dx.doi.org/10.1111/imr.12057
- Cheung, S.T., So, E.Y., Chang, D., Ming-Lum, A. and Mui, A.L. (2013) Interleukin-10 Inhibits Lipopolysaccharide Induced MiR-155 Precursor Stability and Maturation. PLoS ONE, 8, e71336.
- Quinn, S.R. and O’Neill, L.A. (2011) A Trio of MicroRNAs That Control Toll-Like Receptor Signaling. International Immunology, 23, 421-425. http://dx.doi.org/10.1093/intimm/dxr034
- Zhou, J., Wang, H., Lu, A., Hu, G., Luo, A., Ding, F., Zhang, J., Wang, X., Wu, M. and Liu, Z. (2002) A Novel Gene, NMES1, Downregulated in Human Esophageal Squamous Cell Carcinoma. International Journal of Cancer, 101, 311-316. http://dx.doi.org/10.1002/ijc.10600
- Zimmer, A., Bouley, J., Le Mignon, M., Pliquet, E., Horiot, S., Turfkruyer, M., Baron-Bodo, V., Horak, F., Nony, E., Louise, A., Moussu, H., Mascarell, L. and Moingeon, P. (2012) A Regulatory Dendritic Cell Signature Correlates with the Clinical Efficacy of Allergen-Specific Sublingual Immuno-therapy. Journal of Allergy and Clinical Immunology, 129, 1020-1030. http://dx.doi.org/10.1016/j.jaci.2012.02.014
- Yao, Y., Suo, A.L., Li, Z.F., Liu, L.Y., Tian, T., Ni, L., Zhang, W.G., Nan, K.J., Song, T.S. and Huang, C. (2009) MicroRNA Profiling of Human Gastric Cancer. Molecular Medicine Reports, 2, 963-970. http://dx.doi.org/10.3892/mmr_00000199
- Uhlmann, S., Mannsperger, H., Zhang, J.D., Horvat, E.A., Schmidt, C., Kublbeck, M., Henjes, F., Ward, A., Tschulena, U., Zweig, K., Korf, U., Wiemann, S. and Sahin, O. (2012) Global MicroRNA Level Regulation of EGFR-Driven Cell-Cycle Protein Network in Breast Cancer. Molecular Systems Biology, 8, 570. http://dx.doi.org/10.1038/msb.2011.100
- Lee, C.G., McCarthy, S., Gruidl, M., Timme, C. and Yeatman, T.J. (2014) MicroRNA-147 Induces a Mesenchymal-to-Epithelial Transition (MET) and Reverses EGFR Inhibitor Resistance. PLoS ONE, 9, e84597. http://dx.doi.org/10.1371/journal.pone.0084597
- Klein, B., Lu, Z.Y., Gu, Z.J., Costes, V., Jourdan, M., and Rossi, J.F. (1999) Interleukin-10 and Gp130 Cytokines in Human Multiple Myeloma. Leukemia & Lymphoma, 34, 63-70.
- Quinn, S.R., Mangan, N.E., Caffrey, B.E., Gantier, M.P., Williams, B.R., Hertzog, P.J., McCoy, C.E. and O’Neill, L.A. (2013) The Role of Ets2 Transcription Factor in the Induction of MicroRNA-155 by LPS, and Its Targeting by IL-10. Journal of Biological Chemistry, 289, 4316-4325. http://dx.doi.org/10.1074/jbc.M113.522730
- Kishore, R., Tebo, J.M., Kolosov, M. and Hamilton, T.A. (1999) Cutting Edge: Clustered AU-Rich Elements Are the Target of IL-10-Mediated mRNA Destabilization in Mouse Macrophages. Journal of Immunology, 162, 2457-2461.
NOTES

*Corresponding author.

