Stem Cell Discovery
Vol.3 No.2(2013), Article ID:30117,6 pages DOI:10.4236/scd.2013.32017
Distinct functions of Dnmt3a and Dnmt3b de novo DNA methyltransferases in ES cell proliferation and differentiation
![]()
Laboratory of Molecular Embryology, Department of Bioscience, Kitasato University School of Science, Sagamihara, Japan; *Corresponding Author: watanada@kitasato-u.ac.jp
Copyright © 2013 Yoshie Umehara et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 18 January 2013; revised 25 February 2013; accepted 25 March 2013
Keywords: DNA Methylation; Dnmt3a; Dnmt3b; Stem Cells; ES Cells; Differentiation; Ploliferation
ABSTRACT
Two de novo DNA methyltransferases, Dnmt3a and Dnmt3b, have been identified in humans and mice to contribute to the methylation of unmodified DNA. We recently showed a transition of de novo DNA methyltransferase expression from Dnmt3b to Dnmt3a during mouse embryogenesis and in tissue-specific stem cells, suggesting distinct functions of Dnmt3a and Dnmt3b during these processes. In this study, to characterize the functions of Dnmt3a and Dnmt3b in pluripotent stem cells, we exogenously transfected ES cells with Dnmt3a and Dnmt3b cDNAs linked to an internal ribosome entry site-green fluorescent protein gene, and then analyzed the effects of expression of these de novo DNA methyltransferases on ES cell growth and differentiation. ES cells expressing Dnmt3b showed specific downregulation of pluripotency marker genes such as Nanog and Oct 3/4. In addition, Dnmt3a-transfected ES cells showed a specific increase in mitotic index, while Dnmt3b-transfected ES cells showed a decrease in mitotic index. These results suggest that Dnmt3b has important physiological roles in the initial process of stem cell differentiation and that Dnmt3a has a function in stem cell proliferation.
1. INTRODUCTION
DNA methylation is an epigenetic modification that plays crucial roles in multiple biological processes, such as transcriptional repression of genes, genomic imprinting, X chromosome inactivation, and transposon silencing [1-3]. In addition, aberrant DNA methylation patterns have been reported in many tumor and diseases in humans [4]. In the mammalian genome, cytosine residues of CpG dinucleotide sequences are modified postreplication by DNA methylation; about 70% - 80% of CpG dinucleotides are methylated [5]. During mammalian development, the levels of methylation of genomic DNA undergo dynamic changes. De novo DNA methylation starts after implantation, following genome-wide demethylation after fertilization [6]. Three active DNA methyltransferases, Dnmt1, Dnmt3a and Dnmt3b, have been identified in humans and mice [7-10]. Dnmt1 preferentially acts on hemimethylated CpG dinucleotides and is necessary for the maintenance of specific methylation patterns during DNA replication, while Dnmt3a and Dnmt3b contribute to the methylation of unmodified DNA. These enzymes are essential for normal mouse development. Inactivation of Dnmt1 in mice leads to global genome DNA demethylation and embryonic lethality [11]. Dnmt3a null mice die 4 weeks after birth, while Dnmt3b null mice die in the post-gestation stage as a result of multiple developmental defects, including growth impairment and rostral neural tube defects [12]. Furthermore, conditional knockout of Dnmt3a in germ cells resulted in a lack of methylation at imprinted loci, impaired spermatogenesis and embryonic death of offspring from Dnmt3a conditional mutant females. However, disruption of Dnmt3b in germ cells did not result in any apparent phenotype [13].
Recently, we study the expression patterns of de novo methyltransferases during mouse embryogenesis and stem cell differentiation, and show that Dnmt3b is specifically expressed in pluripotent embryonic cells such as epiblasts and embryonic ectoderm, while Dnmt3a is significantly and ubiquitously expressed in embryos after E10.5 [14]. Dnm3b is also expressed in CD34-positive hematopoietic progenitor cells in fetal liver and dorsal aorta, type A spermatogonia cells in adult testis, and neural progenitor cells in the spinal cord, retina and olfactory epithelium cells. Furthermore, we observe the transition of expression of these de novo methyltransferases from Dnmt3b to Dnmt3a during the differentiation of stem/progenitor cells in these tissues [15,16]. These results imply important roles for Dnmt3b in stem/progenitor cell development and the transition of de novo methyltransferase expression from Dnmt3b to Dnmt3a suggests the existence of distinct functions of Dnmt3b and Dnmt3a during embryogenesis and ontogenesis. Dnmt3b but not Dnmt3a is highly expressed in the nuclei of ES cells [14,16-18]. During in vitro neural differentiation of ES cells, Dnmt3a is transiently expressed in differentiating immature neural cells, but not in undifferentiated ES cells or differentiated neurons [16]. Thus, the transition of the predominant de novo methyltransferase from Dnmt3b to Dnmt3a is also reproduced during in vitro differentiation of ES cells.
In this study, to explore the functions of Dnmt3a and Dnmt3b de novo methyltransferases, we exogenously transfect ES cells with Dnmt3b and Dnmt3a cDNAs linked to an internal ribosome entry site (IRES)—green fluorescent protein (GFP) gene, and then analyze the functions of these de novo DNA methyltransferases in the pluripotent stem cells. Exogenous Dnmt3b progress ES cell differentiation through the downregulation of Oct 3/4 and Nanog genes, while Dnmt3a specifically promote cell proliferation.
2. MATERIALS AND METHODS
2.1. ES Cell Culture
The ES cell line (R1) was grown on a 0.1% gelatin-coated cell culture dish with KnockOut™ D-MEM (GIBCO) supplemented with 15% KnockOut™ Serum Replacement (GIBCO), 0.1 mM non-essential amino acids (Invitrogen), 0.1 mM 2-mercaptoethanol, L-glutamine, and 1000 U/ml leukemia inhibitory factor (LIF; Nacalai Tesque). Cells were routinely propagated by trypsinization and re-plating every two days, with a split ratio of 1 in 10.
2.2. Plasmids Constructs and DNA Transfection
Dnmt3a or Dnmt3b full-length cDNAs and an IRES linked to the enhanced GFP gene sequence were cloned into pcDNA3 vector using the EcoRV cloning site. Plasmids were purified using an EndoFree Plasmid Maxi Kit (QIAGEN). Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Forty-eight hours after transfection, the ES cells were fixed with 4% paraformaldehyde/ PBS and then immunohistochemically stained. The numbers of anti-GFP-, Oct 3/4-, Nanogand phosphohistone H3-positive mitotic cells were counted in images acquired with a laser scanning confocal microscope (Carl Zeiss, LSM 510 Meta). Means ± standard error (SE) for three independent experiments were calculated.
2.3. Immunohistochemistry
After fixation with 4% paraformaldehyde/PBS, the cells were reacted with rabbit polyclonal anti-Dnmt3a (1:2000, a gift from Dr. S. Tajima), mouse monoclonal anti-Dnmt3b (1:800, IMGENEX), rabbit polyclonal antiOct 3/4 (1:1000, MBL), rabbit polyclonal anti-Nanog (1:5000, COSMO BIO), rabbit polyclonal anti-GFP (1:10,000, MBL), rat monoclonal anti-GFP (1:2000, Nacalai Tesque), or rabbit polyclonal anti-phospho-histone H3 antibody (1:5000, Abcam). They were then incubated with goat anti-mouse IgG conjugated with Alexa Fluor 568, goat anti-rabbit IgG conjugated with Alexa Fluor 488, goat anti-rabbit IgG conjugated with Alexa Fluor 568 (1:2000, Molecular Probes), or donkey anti-rat IgG conjugated with DyLight 488 as a secondary antibody (1:2000, Jackson ImmunoResearch Laboratories). Antibodies were diluted with Signal Enhancer HIKARI (Nacalai Tesque). The samples were counterstained with Hoechst 33258. Images were acquired using a laser scanning confocal microscope (Carl Zeiss, LSM 510 Meta).
3. RESULTS AND DISCUSSION
3.1. Specific Expression of Dnmt3a and Dnmt3b in GFP-Positive ES Cells
To elucidate the functions of Dnmt3a and Dnmt3b, we exogenously transfected ES cells with cDNA vectors encoding Dnmt3a or Dnmt3b by lipofection. To identify cells expressing Dnmt3a and Dnmt3b, each cDNA was subcloned upstream of the IRES-GFP gene. The vectors express bicistronic transcripts including Dnmt3a or Dnmt3b and EGFP under the control of the CMV promoter (Figure 1(A)). Forty-eight hours after transfection, expression of Dnmt3a and Dnmt3b was confirmed by immunohistochemistry. Dnmt3a was specifically expressed in the nuclei of GFP-positive ES cells transfected with Dnmt3a-IRES-GFP, but not in GFP-negative ES cells, or in control IRES-GFPand Dnmt3b-IRES-GFP transfected ES cells (Figures 1(B)-(D)). Dnmt3b is ubiquitously expressed in the nuclei of untransfected ES cells, and was specifically upregulated in GPF-positive ES cells transfected with Dnmt3b-IRES-GFP, but not in IRES-GFPor Dnmt3a-IRES-GFP transfected ES cells (Figures 1(E)-(G)). Thus, Dnmt3a and Dnmt3b were specifically upregulated in the nuclei of ES cells identified by GFP expression.

Figure 1. Dnmt3a and Dnmt3b expression in ES cells transfected with Dnmt3a-IRES-GFP and Dnmt3b-IRESGFP. Schematic representations of Dnmt-IRES-GFP vectors (A); Dnmt3a-IRES-GFP (C, F); Dnmt3b-IRESGFP (D, G) and control IRES-GFP (B, E) vectors were transfected into ES cells. After transfection, the ES cells were double-stained with anti-Dnmt3a (B, C, D; red) or anti-Dnmt3b (E, F, G; red) and GFP (green) antibodies. Dnmt3a and Dnmt3b protein levels were specifically enhanced in GFP-positive ES cells (C, G arrowheads). Scale bar: 10 μm.
3.2. Dnmt3b Expression Downregulates Nanog and Oct3/4 Expression in ES Cells
Nanog and Oct3/4 are the two most important genes for the self-renewal and maintenance of ES cell pluripotency, and are downregulated following the initiation of ES cell differentiation. To investigate the physiological functions of Dnmt3a and Dnmt3b in the maintenance of pluripotency or the differentiation of ES cells, we first examined the levels of Nanog and Oct 3/4 proteins in ES cells exogenously transfected with Dnmt3a-IRES-GFP or Dnmt3b-IRES-GFP (Figures 2(A)-(F)). Among GFPpositive ES cells expressing the Dnmt3b gene, the proportions of Nanogand Oct 3/4-positive cells were both specifically reduced to 47.7% and 78.4%, respectively, compared with control IRES-GFP transfected ES cells which showed 56.2% and 88.3% positivities for Nanog and Oct 3/4, respectively. Dnmt3a-IRES-GFP transfected ES cells showed 60.0% and 89.4% positivities, respectively, almost equal to the levels in IRES-GFP transfected ES cells. Conversely, the numbers of Nanogand Oct 3/4-negative cells were increased in Dnmt3b-IRESGFP transfected ES cells to 52.3% and 21.6%, respectively, compared with 43.8% and 11.7%, respectively, in control IRES-GFP transfected ES cells. Thus, the transient upregulation of Dnmt3b expression in ES cells specifically downregulates Oct 3/4 and Nanog expression
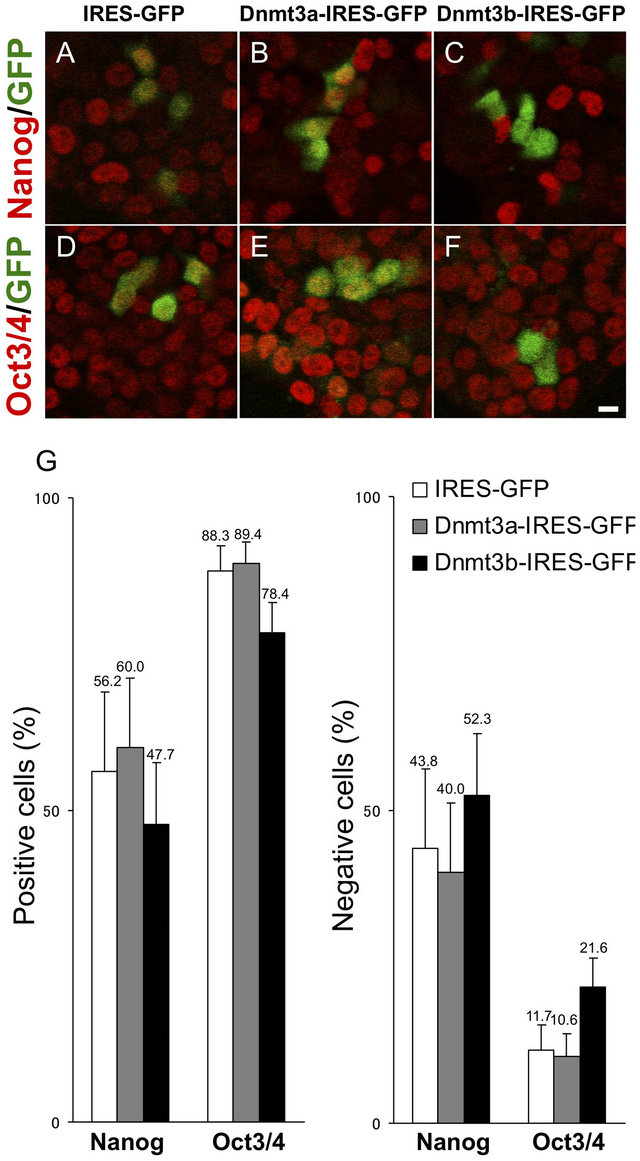
Figure 2. Nanog and Oct 3/4 expression in ES cells transfected with Dnmt3a-IRES-GFP or Dnmt3bIRES-GFP. ES cells transfected with Dnmt3aIRES-GFP (B, E), Dnmt3b-IRESGFP (C, F) and control IRES-GFP (A, D) are double-stained with anti-GFP (green) and anti-Nanog (A-C, red) or antiOct 3/4 (D-F, red) antibodies. Percentages of Nanogand Oct 3/4-positive or -negative cells in Dnmt3aor Dnmt3b-expressing GFP-positive ES cells. Error bars represent standard error (n = 3). Scale bar: 10 μm.
and promotes ES cell differentiation.
These results are consistent with a former report that Dnmt3b knockout ES cells failed to downregulate Oct3/4 and Nanog expression when differentiation was induced [19]. Dnmt3b may function to control ES cell differentiation through the de novo methylation of transcription factors required for the maintenance of pluripotency.
By immunohistochemical staining, Dnmt3b was heterogeneously expressed in ES cells and some ES cells express very low levels of Dnmt3b; however, Dnmt3b was transiently upregulated in ES cells in which differentiation was initiated by LIF withdraw or retinoic acid treatment, and then continuously downregulated during the differentiation process [14,16,18, data not shown].
Furthermore, a greater than 10-fold induction of Dnmt3b mRNA was reported in ES cells that in which differentiation had been induced [20]. During mouse embryogenesis, Dnmt3b is heterogeneously expressed in inner cell mass (ICM) cells, but is most abundantly expressed in epiblast cells [14,21]. These results strongly suggest that the differentiation of pluripotent stem cells is regulated by the transient upregulation of Dnmt3b that induses the downregulation of Oct 3/4 and Nanog through the de novo methylation of promoter sequences. Dnmt3b is specifically expressed in stem/progenitor cells and has very important roles in the initial steps of stem/ progenytor cell differentiation rather than in the maintenance of pluripotency.
3.3. Distinct Functions of Dnmt3a and Dnmt3b in ES Cell Proliferation
In Dnmt3a-IRES-GFP transfected ES cells, we observed a relatively higher number of clustered GFPpositive cells that proliferated from one transfected progenitor cell, compared with the number that proliferated from a single IRES-GFPor Dnmt3b-IRES-GFP transfected ES cell (Figures 3(A)-(C)). We then analyzed cell proliferation activity by counting the numbers of clustered GFP-positive cells expressing the Dnmt3a-IRESGFP or Dnmt3b-IRES-GFP gene at 48 hours post-transfection. Compared with IRES-GFP transfected ES cells, Dnmt3a-IRES-GFP transfected ES cells showed a greater average number of clustered GFP-positive cells (3.7 vs 2.2). Dnmt3b-IRES-GFP transfected ES cells produced an average of 2.4 clustered GFP-positive cells, almost equal to the number in IRES-GFP transfected ES cells (Figure 3(D)). To examine the possibility of a direct relationship between de novo methyltransferase expression and cell cycle progression in each ES cell, we next analyzed the mitotic index in Dnmt3b-IRES-GFPand Dnmt3a-IRES-GFP transfected ES cells. For this purpose, we doublestained cells with anti-phospho-histone H3 (a mitotic marker) and an anti-GFP antibody (Figures 4(A)-(C)). We then analyzed the mitotic index (phospho-histone H3-positive cells/GFP-positive cells) at 48 hours post-transfection. As expected, the mitotic index was also specifically increased in Dnmt3a-IRES-GFP transfected ES cells, which had a mitotic index of 6.5% compared with 5.6% in IRES-GFP transfected ES cells (Figure 4(D)). Thus, the expression of Dnmt3a specifically promotes the proliferation of ES cells as confirmed by the upregulation of the mitotic index and the GFP-positive cell number after transfection.
In adult mouse testis, Dnmt3a is specifically expressed in type B spermatogonia cells, which show high proliferation activities compared with type A spermatogonia cells [15]. In addition, Dnmt3a-deficient knockout mice show impaired mitotic proliferation of spermatogonia
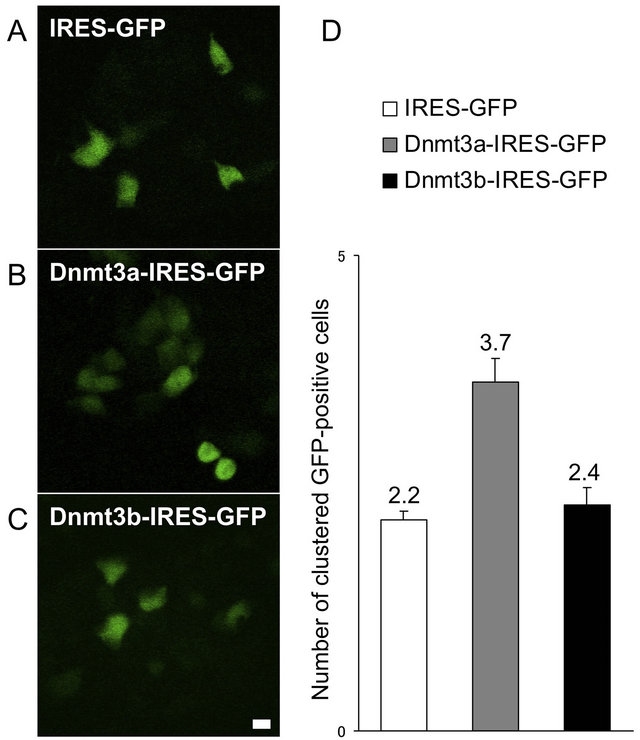
Figure 3. Cell proliferation activities in Dnmt3aand Dnmt3b-transfected ES cells. Dnmt3a-IRESGFP (B), Dnmt3b-IRES-GFP (C) and control IRES-GFP (A) vectors were transfected into ES cells, and the average numbers of GFP-positive colonies were counted after 48 hours (D). Error bars represent standard error (n = 3).

Figure 4. Mitotic index of Dnmt3a-IRES-GFP or Dnmt3b-IRES-GFP transfected ES cells. Dnmt3aIRES-GFP (B), Dnmt3b-IRES-GFP (C) and control IRES-GFP (A) vectors were transfected into ES cells, and the mitotic index (phospho-histone H3-positive cells/GFP-positive cells) was assessed (D). Error bars represent standard error (n = 3).
cells [13]. Thus, Dnmt3a seemed to have a general function in stem/progenitor cell proliferation. Furthermore, Dnmt3a begins to be expressed in the nuclei of all tissues from E10.5 mouse embryos, but its expression is decreased during postnatal mouse development, with almost undetectable levels in adult tissues except for some epithelial tissues and stem/progenitor cells including neural progenitor cells and type B spermatogonia cells [14-16]. Thus, Dnmt3a expression levels are well correlated with cell proliferation activities in many tissues after birth. During postnatal development, cell proliferation may be directly or indirectly regulated by the expression of Dnmt3a. Dnmt3a may have an important function in promoting cell proliferation, not only in pluripotent stem cells, but also in other somatic cells during embryogenesis or organogenesis via the inactivation of some genes that inhibit cell proliferation through de novo methylations of their promoter sequences.
In the present study, we detected significant downregulation of the mitotic index in Dnmt3b-transfected ES cells. Dnmt3b-transfected ES cells had a mitotic index of 3.8%, while IRES-GFP transfected ES cells had an index of 5.6% (Figure 4(D)). In contrast, the numbers of Dnmt3b-transfected ES cells did not differ from the numbers of IRES-GFP transfected ES cells (Figure 3(D)). It is possible that there is a delay in the Dnmt3bdependent suppression of cell proliferation, and that GFP-positive cells may proliferate before Dnmt3b activation. The mitotic index directly represents cell cycle progression in ES cells. Thus, the transient expression of Dnmt3b suppresses pluripotent stem cell proliferation. These results are consistent with former results and suggest novel functions of Dnmt3b that induse the initiation of stem/progenitor cell differentiation via downregulation of ES cell proliferation coordinated with the downregulation of Oct3/4 and Nanog genes.
4. ACKNOWLEDGEMENTS
We wish to thank Dr. Shoji Tajima (Osaka University) for providing the anti-Dnmt3a and anti-Dnmt3b anti-bodies. This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- Reik, W. and Walter, J. (2001) Genomic imprinting: Parental influence on the genome. Nature Reviews Genetics, 2, 21-32. doi:10.1038/35047554
- Bird, A. (2002) DNA methylation patterns and epigenetic memory. Genes & Development, 16, 6-21. doi:10.1101/gad.947102
- Bestor, T.H. (2005) Transposons reanimated in mice. Cell, 122, 322-235. doi:10.1016/j.cell.2005.07.024
- Jones, P.A. and Baylin, S.B. (2002) The fundamental role of epigenetic events in cancer. Nature Reviews Genetics, 3, 415-428.
- Ehrlich, M., Gama-Sosa, M.A., Huang, L.H., Midgett, R.M., Kuo, K.C., McCune, R.A. and Gehrke, C. (1982) Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Research, 10, 2709-2721. doi:10.1093/nar/10.8.2709
- Reik, W., Dean, W. and Walter, J. (2001) Epigenetic reprogramming in mammalian development. Science, 293, 1089-1093. doi:10.1126/science.1063443
- Bestor, T., Laudano, A., Mattaliano, R. and Ingram, V. (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells: The carboxylterminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. Journal of Molecular Biology, 203, 971-983. doi:10.1016/0022-2836(88)90122-2
- Yen, R.W., Vertino, P.M., Nelkin, B.D., Yu, J.J., El-Deiry, W., Cumaraswamy, A., Lennon, G.G., Trask, B.J., Celano, P. and Baylin, S.B. (1992) Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Research, 20, 2287-2291. doi:10.1093/nar/20.9.2287
- Okano, M., Xie, S. and Li, E. (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genetics, 19, 219-220. doi:10.1038/890
- Xie, S., Wang, Z., Okano, M., Nogami, M., Li, Y., He, W.W., Okumura, K. and Li, E. (1999) Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene, 236, 87-95. doi:10.1016/S0378-1119(99)00252-8
- Li, E., Bestor, T.H. and Jaenisch, R. (2002) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69, 915-926. doi:10.1016/0092-8674(92)90611-F
- Okano, M., Bell, D.W., Haber, D.A. and Li, E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247-257. doi:10.1016/S0092-8674(00)81656-6
- Kaneda, M., Okano, M., Hata, K., Sado, T., Tsujimoto, N., Li, E. and Sasaki, H. (2004) Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature, 429, 900-903. doi:10.1038/nature02633
- Watanabe, D., Suetake, I., Tada, T. and Tajima, S. (2002) Stageand cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mechanisms of Development, 118, 187-190. doi:10.1016/S0925-4773(02)00242-3
- Watanabe, D., Suetake, I., Tajima, S. and Hanaoka, K. (2004) Expression of Dnmt3b in mouse hematopoietic progenitor cells and spermatogonia at specific stages. Gene Expression Patterns, 5, 43-49. doi:10.1016/j.modgep.2004.06.008
- Watanabe, D., Uchiyama, K. and Hanaoka, K. (2006) Transition of mouse de novo methyltransferases expression from Dnmt3b to Dnmt3a during neural progenitor cell development. Neuroscience, 142, 727-737 doi:10.1016/j.neuroscience.2006.07.053
- Chen, T., Tsujimoto, N. and Li, E. (2004) The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Molecular and cellular biology, 20, 9048-9058. doi:10.1128/MCB.24.20.9048-9058.2004
- Lanner, F., Lee, K.L., Sohl, M., Holmborn, K., Yang, H., Wilbertz, J., Poellinger, L., Rossant, J. and Farnebo, F. (2010) Heparan sulfation-dependent fibroblast growth factor signaling maintains embryonic stem cells primed for differentiation in a heterogeneous state. Stem Cells, 28, 191-200.
- Li, J.Y., Pu, M.T., Hirasawa, R., Li, B.Z., Huang, Y.N., Zeng, R., Jing, N.H., Chen, T., Li, E., Sasaki, H. and Xu, G.L. (2007) Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in themethylation of Oct4 and Nanog. Molecular and cellular biology, 27, 8748- 8759. doi:10.1128/MCB.01380-07
- Thomson, M., Liu, S.J., Zou, L.N., Smith, Z., Meissner, A. and Ramanathan, S. (2011) Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell, 145, 875-889. doi:10.1016/j.cell.2011.05.017
- Hirasawa, R. and Sasaki, H. (2009) Dynamic transition of Dnmt3b expression in mouse preand early post-implantation embryos. Gene Expression Patterns, 9, 27-30. doi:10.1016/j.gep.2008.09.002

