American Journal of Molecular Biology
Vol.2 No.1(2012), Article ID:16546,11 pages DOI:10.4236/ajmb.2012.21006
Molecular docking investigation for Indonesian H274Y mutant neuraminidase type 1 with neuraminidase inhibitors
![]()
Department of Physics, Mathematics and Science Faculty, University of Indonesia, Depok, Indonesia
Email: *sigit.jaya.herlambang@gmail.com
Received 11 August 2011; revised 14 September 2011; accepted 21 September 2011
Keywords: Molecular Docking; Neuraminidase; Inhibitor; Resistance
ABSTRACT
The aim of this study is to get insight the interaction between Indonesian H274Y mutant neuraminidase with four inhibitors. Not only to seek preferable inhibitor to be used, but also to investigate the interaction occurred, especially hydrogen bonds formed. Hydrogen bonds analysis and its interaction energies calculation showed that zanamivir is the most preferable inhibitor with 13 hydrogen bonds formed and –439.96 kcal/mol. Laninamivir would be an alternative inhibitor since it has 10 hydrogen bonds and –307.19 kcal/mol. The investigation of ΔSAS showed almost all active site residues buried when interacted with inhibitors. Only a few residues have an increases ΔSAS. Lipinski rule analysis showed that zanamivir and laninamivir would be best taken by injection or inhalation.
1. INTRODUCTION
The avian influenza has damaged the economies of developing countries, such as Indonesia. The virus has infected poultry in 31 from 33 provinces, and 12 provinces were reported hundreds cases of human infection [1]. There were also 163 human cases with 135 deaths were reported until April 2010 [2]. The spread of infection with a variety of susceptible avian hosts has resulted in rapid viral adaption. The virus ability to shift and drift the antigen has increased the potential for these viruses to repeat influenza pandemic.
The high mortality of the viruses may be caused either by the high pathogenicity of the viruses itself or by the less effectiveness of the drugs inhibited the replication process. Recent antiviral medications which prevent the virus to replicate were divided by two classes: the M2 ion channel blocker (Adamantane and Rimantadine) and the neuraminidase inhibitor. Many studies show the resistance of M2 ion channel blockers [3-18] has brought the neuraminidase inhibitor research as the main stream of influenza virus investigation. Although several mutation in neuraminidase active site residues also causing the less attracted NA-inhibitor interaction [19-29], there are more improvement were made to produce a better neuraminidase inhibitor than in M2 ion channel blocker.
Neuraminidase inhibitors are produced mimicking sialic acid as its competitor. Neuraminidase inhibitors work in the last step of replication process. The matured virion budding in the edge of host cell and neuraminidase would enhance the cleavage activity to release them. If neuraminidase inhibitor interacted in neuraminidases active site, the matured virion will stay attached with host cell sialic acid [30]. This mechanism will prevent the matured virion to infect another host cell. Neuraminidase will clearly not going anywhere, and the infection was effectively terminated.
The mutations in neuraminidase active site residues could reduce the sensitivity to interact with neuraminidase inhibitors. One of a few well known active site residues mutations is at the position 274 which replaces histidine (H) into tyrosine (Y). It was observed from 2006 H5N1 NA (3CKZ) that the H274Y mutation has caused the reorientation of the adjacent E276 forcing it carboxyl group to move closer to binding site and decrease the binding affinity with oseltamivir [31,32].
The aim of this study is to investigate drugs attachment in mutated N1 neuraminidase influenza virus in order to search the best interaction between Indonesian H5N1 NA and several inhibitors. This is needed to provide the information about what drug is preferable to inhibit Indonesian H274Y mutated NA virus shed. The receptor-ligand docking was done to seek the probability of the attachment which correlated with non-bonded interactions. The observation was completed through structural and energetic investigation of the receptor-ligand interaction following NA-inhibitor interaction through hydrogen bonds formed, DSAS and interaction energies calculation. The Lipinski rule investigation also added as complementary information.
2. MATERIALS AND METHODS
2.1. Structures Preparation
The amino acid sequence of Indonesian mutant H274Y was obtained from influenza sequences database in NCBI [33]. The searching panel, provided by NCBI, was set with the keyword “Indonesia” in order to collect all the neuraminidase sequence from all cases which occurred in Indonesia. During that time, there are only 133 neuraminidase sequences found. The collecting years of sequences varies from 2003 to 2007 and has only one mutant neuraminidase in 274’s position. The (A/Indonesia/ 560H/2006(H5N1)) sequence with accession code ABW- 06159 obtained from influenza virus.
The searching of template structure which have a high percentage of similarity was done by BLASTP 2.2.16 [34] and InterProScan [35] provided by Swiss Expasy [36-40]. The template structure suggested from Swiss Expasy is 3CKZ which then obtained in Research Collaboratory for Structural Bioinformatics (RCSB) [41]. The template sequence is 97% similar with the sequence of ABW06159. Five homology models were produced using MODELER [42]. All models were assessed with PROCHECK [43-45], DOPE-HR [46] and 3D-Profiles scores [47,48] with Kabsch-Sander algorithm [49] for secondary structure in order to get the best model.
The structures of inhibitors were collected in the database of chemical molecules, PubChem [50] which is maintained by NCBI. At this moment, PubChem provide over 31 millions compounds, 75 millions substances and bioactivity results from 1644 high-throughput screening programs. Also, the information needed to check Lipinski Rules were obtained in PubChem. Until the time, there were 13 neuraminidase inhibitor compounds. Only four inhibitors were used in this experiment. All neuraminidase inhibitors compounds used in this experiment and its properties were listed in Table 1.
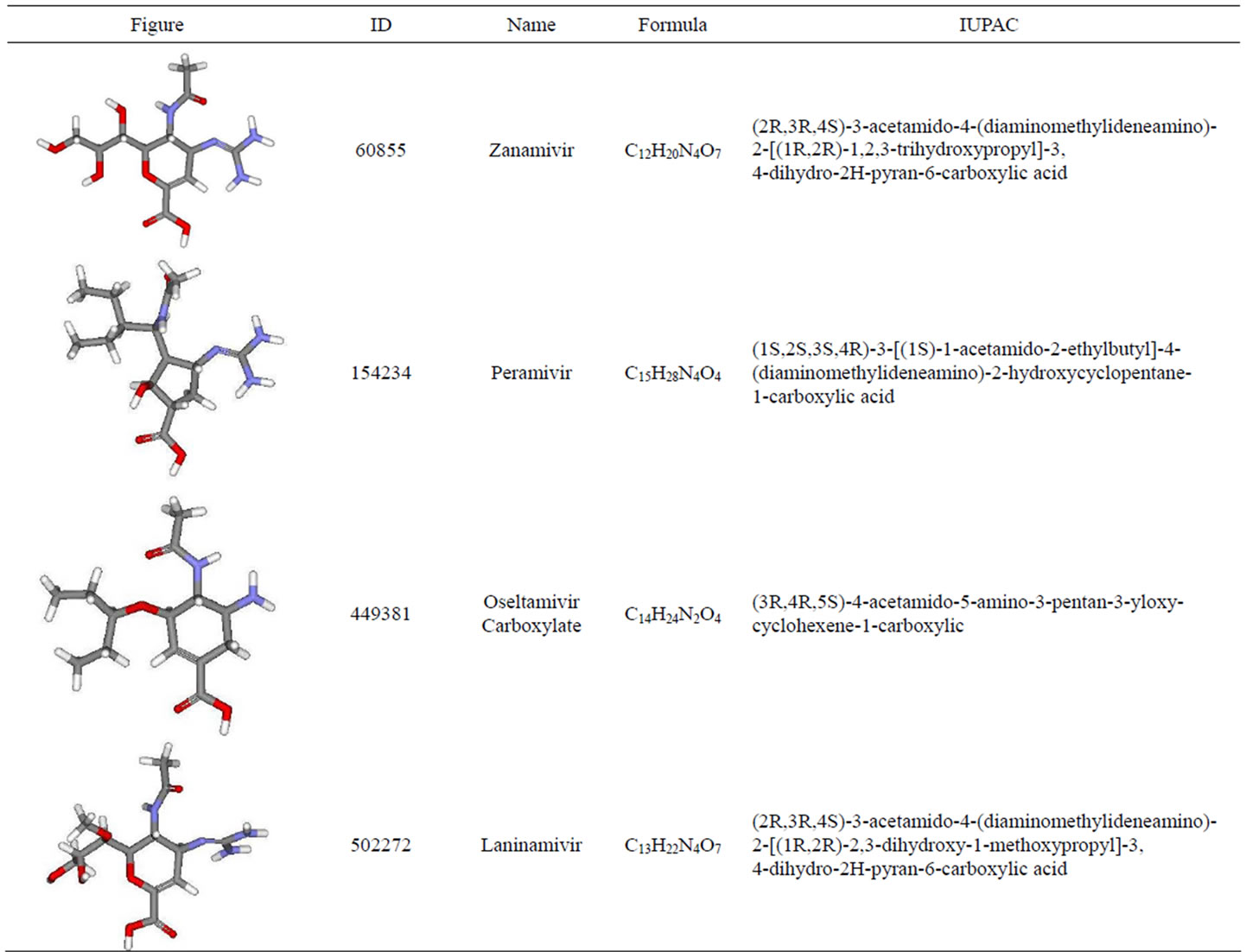
Table 1. Neuraminidase inhibitors and its properties.
2.2. Neuraminidase-Inhibitors Docking (CDOCKER)
The attachment of inhibitors into neuraminidase molecule has completed with CDOCKER [51]. The parameters set with 10 random top hits and conformations, 1000 random conformations dynamics steps and conformations dynamics target temperature, including electrostatic interactions and orientation van der Waals energy threshold, and use CHARMm for forcefield and ligand partial charge. Candidate poses were created using random rigidbody with six degrees of freedom (3 rotations/3 translations) followed by simulated annealing. Sphere generated to fill the active site to facilitate ligand matching in the sphere centre and find possible ligand orientations. Following that, a final minimization used to refine the ligand process. A pose with minimum interaction energy would be ranked in the top of 10 random top hits.
2.3. Minimization Energy
The best docked pose with minimum interaction energy still has a great RMS gradient around 13,000 kcal/ (mol × Å). A proper energy minimization is needed in order to produce an optimum geometrical structure. The energy minimization process was divided by two main steps with different algorithms, steepest descent and conjugate gradient. The steepest descent was calculated with targeted energy gradient 0.5 kcal/ (mol × Å) and 1,000,000 steps maximum. This step used rough calculation to gain the targeted energy gradient. The second step was executed with targeted energy gradient 0.1 kcal/ (mol × Å) and 1,000,000 steps maximum. This algorithm used very smooth calculation to gain optimum geometrical structure with minimum energy. Both methods use Generalized Born Molecular Volume (GBMV) implicit solvent energy to mimicking the ambience, a non bond list radius of 14 Å, and a switching function was applied between 10 - 12 Å for a computational efficiency. To gain a longrange electrostatic energy contribution, it was visualized in a spherical cutoff mode.
2.4. Solvent Accessible Surface (SAS)
Solvent accessible surface was measured two times. First measurement was done before the NAI’s were docked into neuraminidases. The second was after complex molecules reach energy gradient 0.1 kcal/(mol × Å). The measurement were done using a probe radius 1.4 Å. The changes of solvent accessible surface (DSAS) were calculated by subtracting the first with the second SAS of active site residues of neuraminidase.
Small ligand binding sites are usually clefts or pockets where catalysis takes a place. The interaction in the binding site would cause active site residues were buried in complex. It could be determined by the decreasing of the surface area that solvent could access. This is known as solvent accessible surface changes. In this experiment, the active site residues of neuraminidase for SAS calculation were chosen followed the template 3CKZ structure definition and subsites were divided according Stoll et al. [52] subsites characterization with the addition E277 into S5 subsite residues (look Figure 1).
3. RESULTS
3.1. Lipinski’s Rule Analysis
The rule of 5 (also called Lipinski’s rules), made by Lipinski et al. [53], predicts poor absorption or permeation of drugs. Although there are several categories that violate this rule (such antibiotics, antifungal, vitamins and cardiac glycosides), most of drugs marketed follow this rule. Lipinski’s rule analysis are needed to observe since the common drugs is taken orally and its must be able to cross the cell membrane or barrier. This section will give complementary information about neuraminidase inhibitors profile and will not be compared to the results of the experiment.
Table 2 listed all information needed to analyze the Lipinski rule in 4 neuraminidase inhibitors involved in the experiment. Under the Lipinski’s rule column has written 0 and 1 which identify ligand whether it violated or not the Lipinski’s rule. There are 2 out of 4 inhibitors that violated the Lipinski’s rule, zanamivir and laninamivir. The Lipinski’s rule was violated through the number of hydrogen bond donors which over the limit that Lipinski created: 5. Overlimited hydrogen bond donors is happen because laninamivir and zanamivir are bulkier than other neuraminidase inhibitors used in this experiment. Out of that, other requirements are fulfilled for all neuraminidase inhibitors.
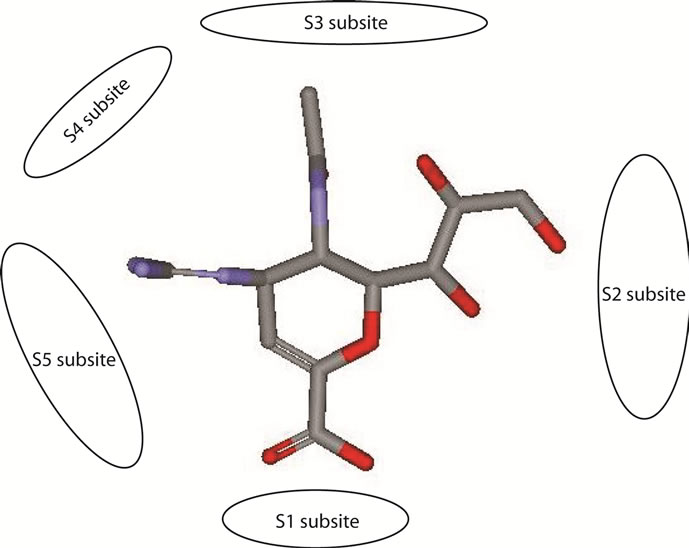
Figure 1. Zanamivir without H atoms trapped in the active site.
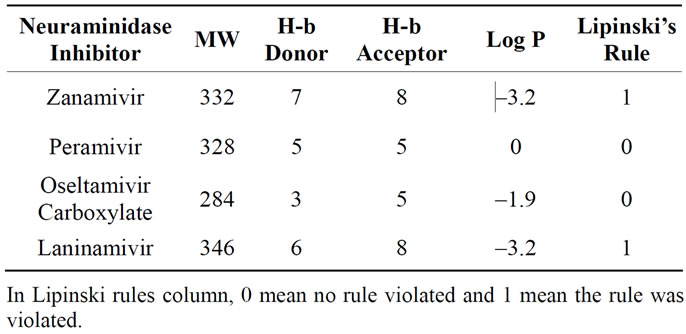
Table 2. The Lipinski rules analysis.
3.2. Structural
3.2.1. NA-Inhibitors Interaction
Table 3 provided hydrogen bonds formed between active site residues and inhibitors before and after minimization. Zanamivir shows better interaction with NA active site than the other inhibitors. The numbers of hydrogen bond formed increase from 11 to 13. Following zanamivir, laninamivir has increases numbers of hydrogen bond formed from 9 to 10. Then oseltamivir carboxylate with 6 hydrogen bonds formed before and after minimization. And peramivir which lost one of its hydrogen bond and leave only 6 hydrogen bonds formed after minimization.
Zanamivir most interaction was made by S1 subsite residues (R118, R292 and R371). It showed by the changes of the orientation of interaction. Before minimization there is no hydrogen bonds formed between zanamivir and R292 and shows four hydrogen bonds which formed by R118 and R371 with zanamivir. After minimization, the distance between R118 and zanamivir decreases and the carboxyl group of zanamivir moved closer to R292 and formed two hydrogen bonds. Total there are five hydrogen bonds in the S1 subsite after minimization.
The improvement of interaction also made in S2 subsite and S5. In the S2 subsite, the improvement could be indicated from an addition of a hydrogen bond formed between E119 and zanamivir. Other residue in S2 subsite, E227, did not have hydrogen bond with all of inhibitors experimented. In the S5 subsite, the changes of distances of E276 and E277 from zanamivir are indicated a good attraction in this subsite that moved the inhibitor more close to the binding pocket.
In this research also noted that only when docked with zanamivir hydrogen bond in R152 did not formed, even after energy minimization. For other inhibitors, the interaction could be seen as a good attraction. It was showed as an improvement in hydrogen bond distance and an addition of hydrogen bond formed in this residue. Further research is needed to answer the hesitancy whether the absence of hydrogen bond in R152 could enhance the NA-inhibitor interaction in other subsites.
The lack of interaction between neuraminidase and peramivir showed by the loss of one hydrogen bond from total hydrogen bonds formed and unstable movement which showed by the changes of interaction orientation. Peramivir have a strong interaction with D151 and R152 in the S3 subsite. It can be seen through the changes of their distances. Unfortunately, that condition did not occurred with other hydrogen bonds formed in other subsites. There are E227, R371 and Y406 which lost its hydrogen bonds with peramivir.
The interaction between neuraminidase with oseltamivir carboxylate showed the movement from the region near Y347 and R371 into R118 and R152. Two hydrogen bonds formed by Y347 before minimization has lost and changed by R118 and R152 which keep oseltamivir carboxilate has six hydrogen bonds with neuraminidase. Following that, the distance of hydrogen bond formed between R371 and inhibitor has increased. Although being unstable in R371, oseltamivir carboxylate has a good interaction with other residue in S1 subsite, R292.
Interesting fact showed that in both peramivir and oseltamivir carboxylate does not have any formed hydrogen bond with S5 subsite. The neuraminidase interaction with this two inhibitors depended mostly with S1 subsite. Even the strong interaction between those inhibitors and R292 which adjacent with S5 subsite did not helped to create any hydrogen bond in S5 subsite. In zanamivir and laninamivir, the binding in S5 subsite shows an attraction that could be seen from the hydrogen bonds distance which became shorter.
The hydrogen bonds list and its distances data of the interaction between NA with laninamivir showed that laninamivir has moved closer into the active site residues. Strong attraction could be seen in S1 and S5 subsites. The hydrogen bonds with S1 subsite were formed only by R292 and R371. With laninamivir, R292 has produced a good interaction which showed by the addition of R292-laninamivir hydrogen bonds formed.
The strong interaction in S5 subsite showed by the unbroken hydrogen bonds formed in NA active site with laninamivir. Three hydrogen bonds, formed by E276 and E277, remain stable with little movements. Most of the residues in others subsites has a good attraction with laninamivir. It showed by the changes of hydrogen bonds distance formed which became shorter after minimization.
3.2.2. ΔSAS
The changes of solvent accessible surface of specific residues could show the buried areas in the enzyme (in this case neuraminidase) after the substrate (neuraminidase inhibitors) were docked and minimized. Table 4 shows the DSAS in the subsites after docked and minimized with zanamivir, laninamivir, peramivir and oseltamivir carboxylate. Overall, the DSAS of most active site resi
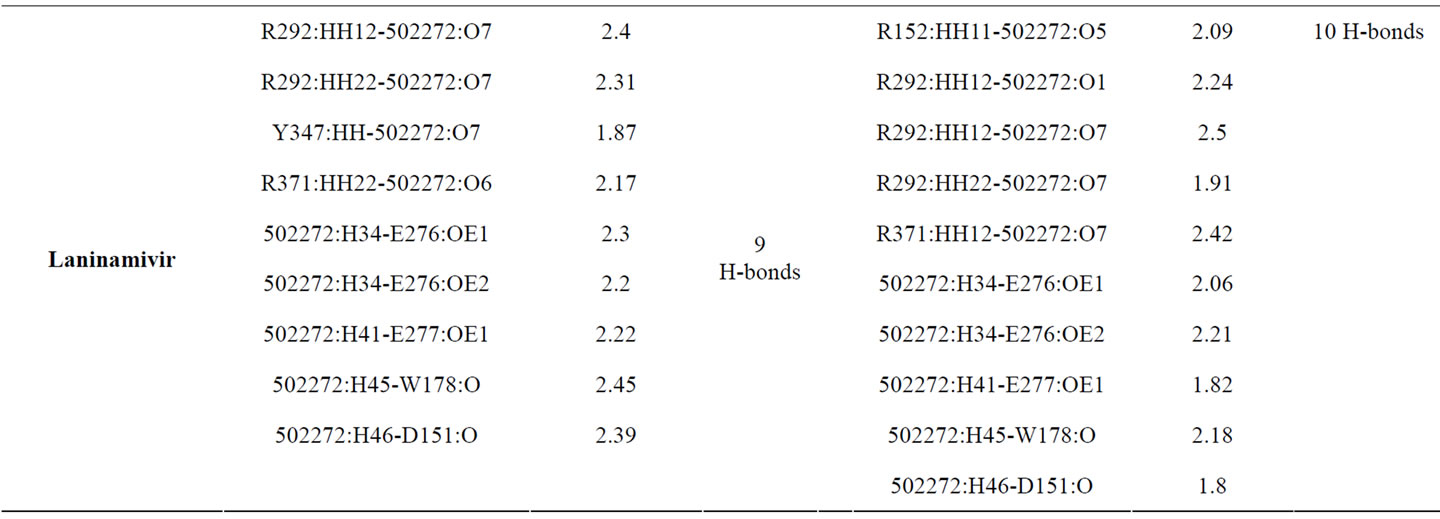
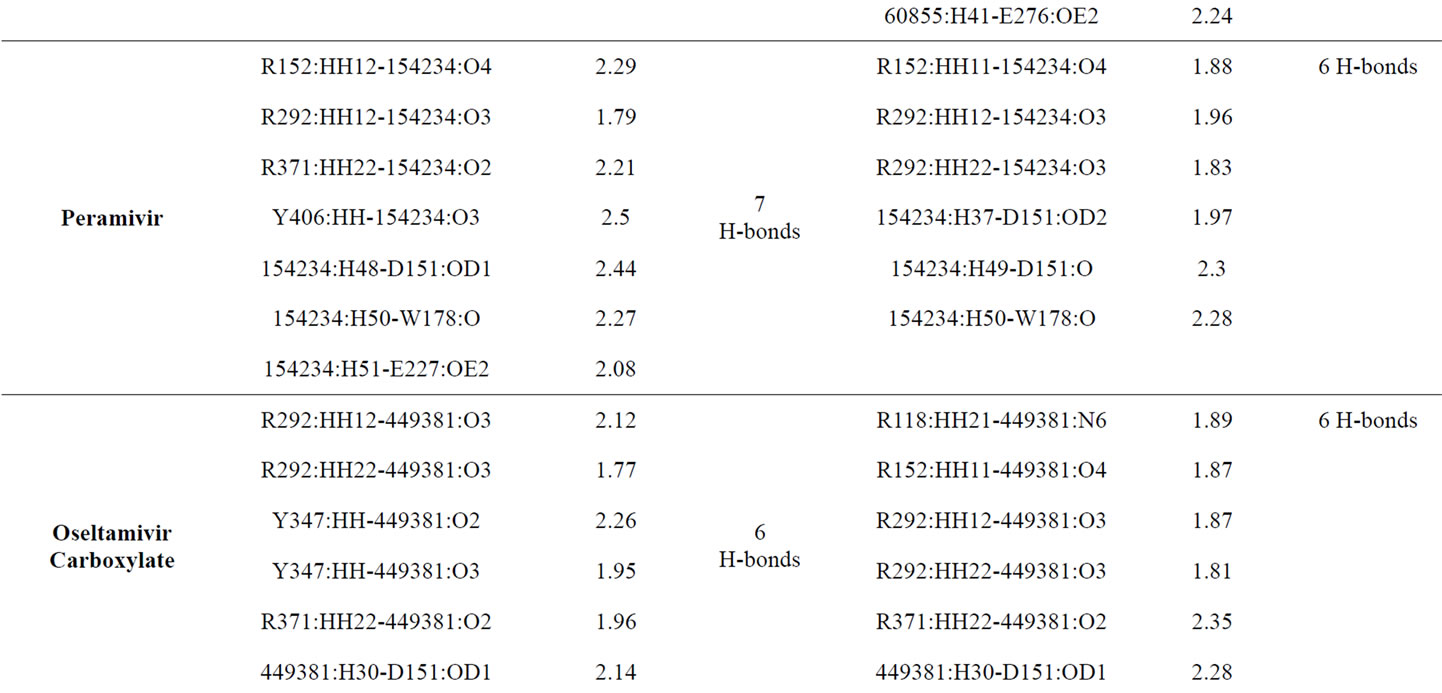
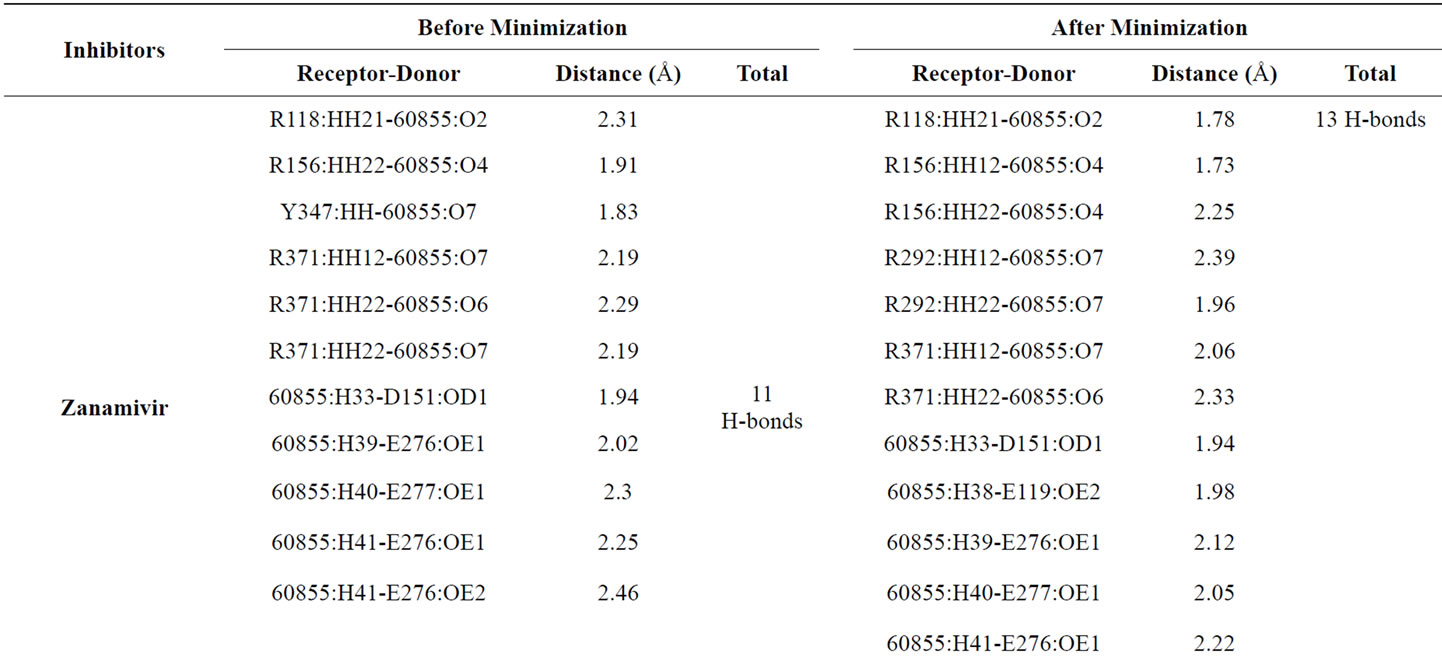
Table 3. The hydrogen bonds formed before and after minimization.
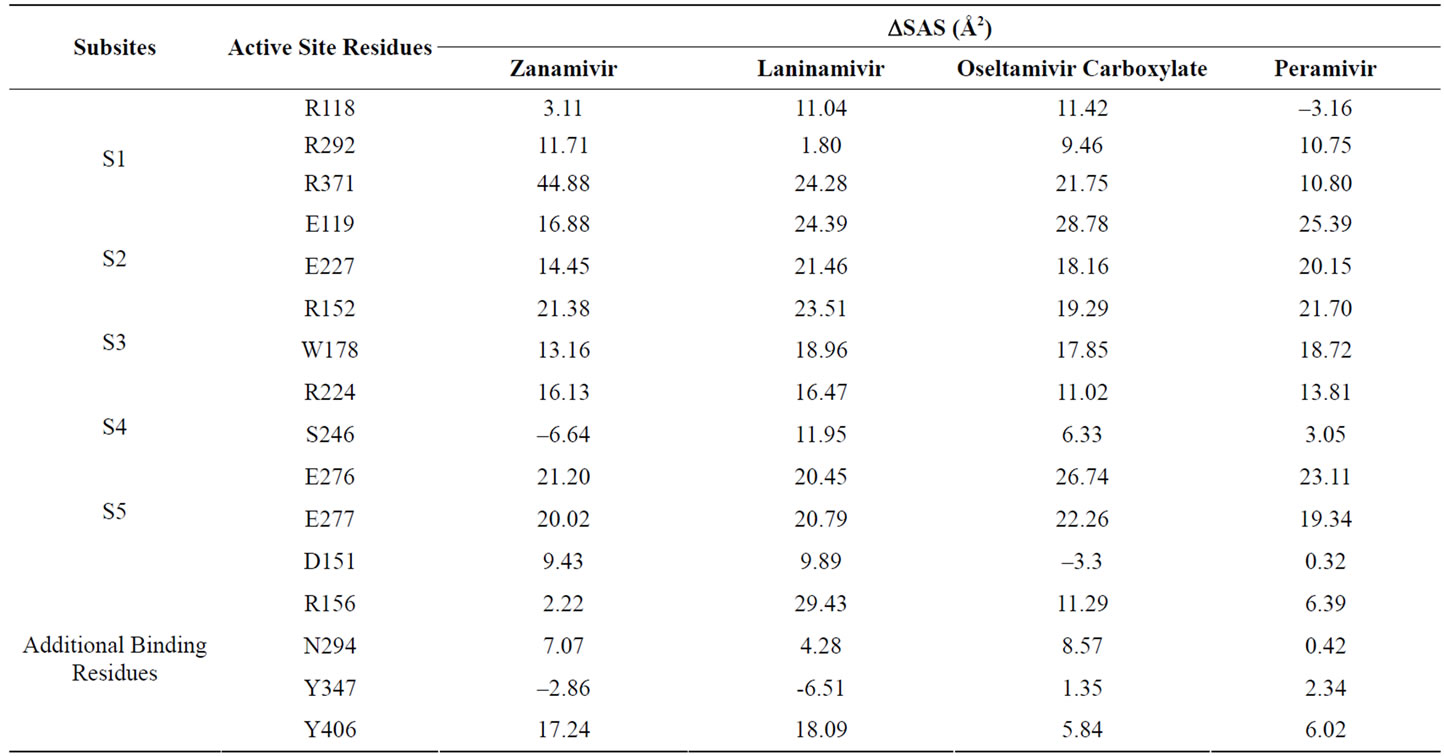
Table 4. ΔSAS of active site residues before and after minimization shows the burials residues by the inhibitors when complexed. Positive values are decrease and negative values are increase.
dues are decrease (showed by positive value). Only a few residues have an increases DSAS.
In the S1 subsite, the largest change was carried out between R371 and zanamivir interaction. This is also being the largest decrease in this experiment. Except R118, the site that also known as triad arginine, decrease its solvent accessible surfaces area when binding with zanamivir more than 7 Å2 and be the largest compared by other inhibitors. Different with S1 subsite, DSAS of S2 subsite residues when interact with zanamivir has been the lowest in decreasement compared with other inhibitors. Even though the data shows similar order, it interesting that the inhibitor which has considerable hydrogen bonds formed with neuraminidase was decreased its DSAS the less.
Another DSAS similar order happened between four inhibitors also made in S3 and S5 subsites. Although the strength of interaction, which is indicated by the hydrogen bond formed, in those subsite are different, there are no significant differences in DSAS. This is shows that almost all residues in S3 and S5 subsites are buried in the complex by inhibitor. It seems to be caused by the similar part of inhibitors facing those subsites. It has to have an impact in the similar order of DSAS.
In the S4 subsite, there is only one buried residue which has DSAS more than 7 Å2 in all inhibitors, R224. Even R224 did not form any hydrogen bond, this residue was buried in complex and may be interacted with electrostatic interaction. Other S4 subsite residue, S246, showed an increase DSAS in zanamivir (shown by negative value) and only buried over 7 Å2 in complex by laninamivir. It shows that S246 has a tendency to repel any atoms around its surface.
From an additional residues data, interesting event shows by D151, Y347 and Y406. In D151 and Y406 residues, both of them had decrease DSAS over 7 Å2 only in zanamivir and laninamivir. Those events did not appear in other inhibitors. It is really wondering since zanamivir and laninamivir was known from the table 3 as inhibitors that formed relatively numberous hydrogen bonds than other inhibitors docked in this experiment. Another interesting event showed by Y347 which has DSAS decreases below 7 Å2 in all inhibitors. It may be related with the lost hydrogen bond formed by Y347 in 3 from 4 inhibitors after minimization. Even in complex molecule with peramivir, Y347 did not produced hydrogen bond formed.
3.3. Interaction Energy Analysis
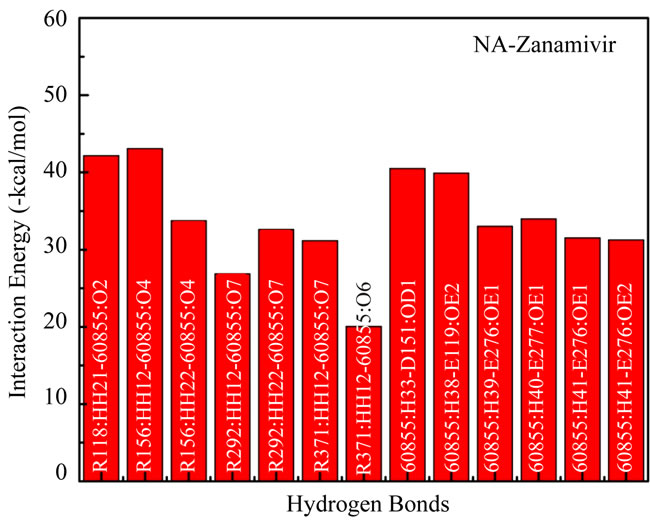
In this study were calculated interaction energy, which compound of electrostatic energy and van der Waals energy, between two sets of atoms in a specified structure. This could help identified which hydrogen bond interacted stronger and contributed much in receptor-ligand binding. The results of interaction energies of four inhibitors were plotted in Figure 2.
Figure 2(a) shows the plot of interaction energies from atoms which formed hydrogen bonds between NA and zanamivir. The strongest interaction (the lowest in
 (a)
(a)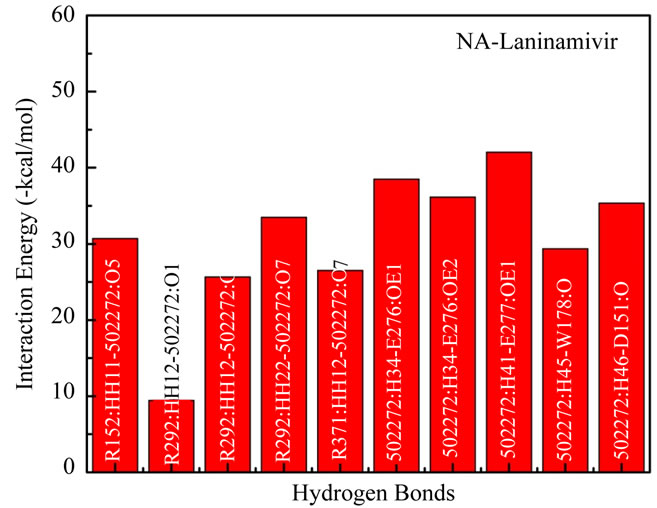 (b)
(b)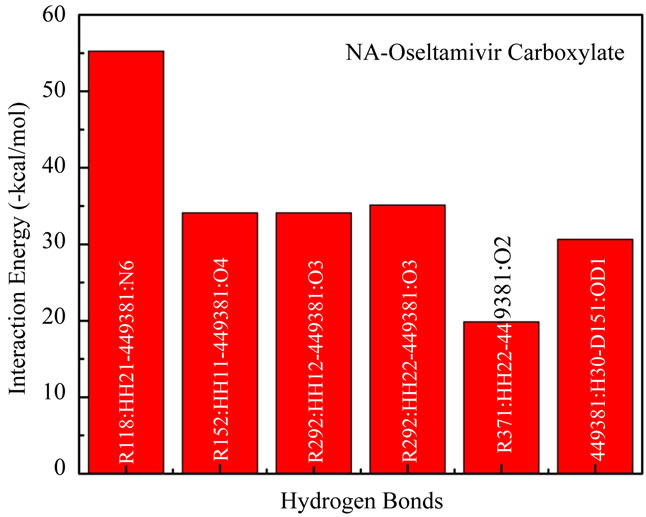 (c)
(c)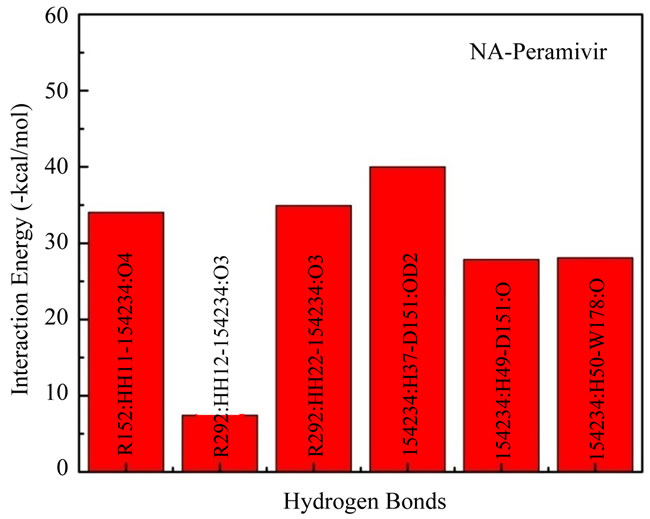 (d)
(d)
Figure 2. Interaction energy between functional residues and inhibitor which formed hydrogen bond linkage directly.
teraction energy) was made between R156: HH12- 60855: O4 with –43.08 kcal/mol. The highest interaction energy was made between R371:HH22-60855:O6 with –20.07 kcal/mol. Total energy from 13 interaction energies –439.96 kcal/mol gives a lowest total interaction energy compared with other inhibitors in this experiment.
Figure 2(b) plotted the interaction energies from hydrogen bonds formed in the NA subsite when interacted with laninamivir. The lowest interaction energy was made by 502272:H41-E277:OE1 with –42.03 kcal/mol. And the highest energy was made by R292:HH12-502272:O1 with –9.45 kcal/mol. Total interaction energy of 10 nonbonded interactions in hydrogen bonds formed atoms is –307.19 kcal/mol.
The interaction energies in NA subsite when interacted with oseltamivir carboxylate plotted in Figure 2(c). The lowest interaction energy was made by R118: HH21- 449381:N6 with –55.26 kcal/ mol and be the lowest interaction energy in this experiment. The highest was made by R371:HH22-449381:O2 with –19.84 kcal/mol.
Total interaction energy of 6 non-bonded interactions between atoms which formed hydrogen bonds is –209.11 kcal/mol which is larger than peramivir.
In Figure 2(d) was plotted the interaction energies of atoms which formed hydrogen bonds in NA subsite when interacted with peramivir. The lowest interaction energy was made by 154234:H37-D151:OD2 with –40.01 kcal/mol and the highest interaction energy was made by R292: HH12-154234:O3 with –7.45 kcal/mol. The interaction energy which made by R292:HH12-154234:O3 is also the highest interaction energy in this experiment. Total interaction energy of atoms which formed hydrogen bonds in NA-peramivir complex is –172.37 kcal/mol and be the highest compared than other inhibitors in this experiment.
4. DISCUSSION
Molecular docking investigation is able to give detail information in receptor-ligand binding affinity. In this study, main focuses pointed at structural and energetic changes that could indicate the binding affinity of NA-inhibitors. The structural changes viewed from hydrogen bonds which formed and solvent accessible surface area change. The result shows that the H274Y mutant neuraminidase N1 has the affinities to bind stronger with zanamivir and laninamivir.
The result also may have an association with the resistance of oseltamivir in the virus which its NA has mutated at 274 from histidine into tyrocine. Currently, oseltamivir is used mainly for influenza treatment, but the generation and/or circulation of oseltamivir resistant mutants influenza virus in humans has become a major concern [54-58]. In Yamashita et al. experiment, oseltamivir carboxylate shows IC50 ratios of 32 to 8,400 to the H274Y (H1N1), R292K, E119V, and N294S (H3N2) mutants, the IC50 ratios of R-125489 and zanamivir for these strains are 0.69 to 2.8 and 0.72 to 1.7, respectively [59]. Other studies carried out by Maki Kiso et al. also explain similar results that oseltamivir resistant mutants, especially H274Y, could be handled by zanamivir and laninamivir [60]. This is coherence with our experiment results which shows that only zanamivir and laninamivir has relatively good binding affinities with H274Y mutant NA active site. The experiment to seek the threshold of interaction energy which may be able to give a reference to determine the resistance of NA virus is needed for future development in this research area. It may cut the time and cost on in vivo and in vitro characterization.
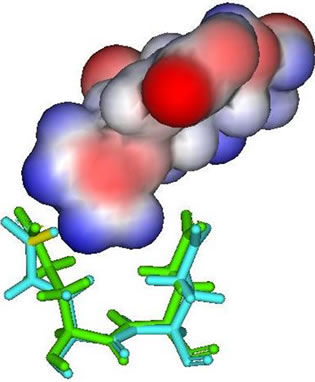
In the result data, both peramivir and oseltamivir carboxylate do not formed a hydrogen bond in S5 subsite. This is indicated that the interaction between neuraminidase with both peramivir and oseltamivir carboxylate disturbed at S5 subsite. Different partial charge of each inhibitor may have a direct effect to the interaction in this subsite (Figure 3). But also, the correlation between H274Y mutation and the weak interaction in functional residues-substrate binding has an influence. The residue substitution from histidine to tyrocine at position 274 may contribute in distracted interaction of E276 and E277 with substrate. The changes of partial charges in position 274 may have indirect affection to both E276 and E277 besides the changes in molecular volume which happen (Figure 4). More experiments for the effects of residual changes in this position with much more neuraminidase to investigate those partial charge changes deeper.
5. CONCLUSION
This research shows four inhibitors (zanamivir, laninamivir, oseltamivir carboxylate and peramivir) affinities when attached into neuraminidase and gives a binding strength comparison between all of them. The hydrogen bonds and interaction energy shows that zanamivir and laninamivir is preferable to treat the patient. Lipinski rule
 (a)
(a)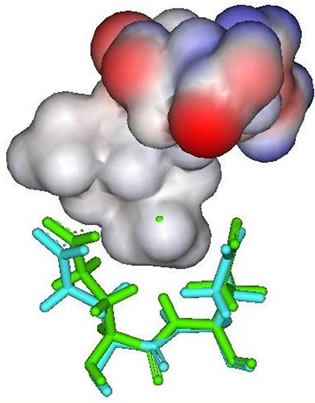 (b)
(b) (c)
(c) (d)
(d)
Figure 3. E276 and E277 (stick figures) with: (a) Zanamivir; (b) Peramivir; (c) Oseltamivir carboxylate; (d) Laninamivir.
 (a)
(a) (b)
(b)
Figure 4. Partial charge of Histidin (left) and Tyrocine (right). Negative charge shown by red and positive charge by blue.
analysis shows that zanamivir and laninamivir would be better taken non-oral (inhalation or injection). Overall, the DSAS of most active site residues are decrease (showed by positive value). Only a few residues have an increases DSAS. From the results, this research results proposed zanamivir and laninamivir as potent inhibitor to inhibit Indonesian H274Ymutant neuraminidase.
6. ACKNOWLEDGEMENTS
We would like to express gratitude towards Ding Ming Chee of Accelrys Singapore for the Accelrys Discovery Studio 2.1 trial sent to us.
REFERENCES
- Sedyaningsih, E.R., Isfandari, S., Soendoro, S. and Supari, S.F. (2008) Towards mutual trust, transparency and equity in virus sharing mechanism: The avian influenza case of Indonesia. Annals Academy of Medicine Singapore, 37, 482-488
- http://www.who.int/csr/disease/avian_influenza/avianinfluenza_factsheetJan2006/en/index.html
- Cheung, C.L., Rayner, J.M., Smith, G.J.D., Wang, P., Naipospos, T.S.P., Zhang, J., Yuen, K.Y., Webster, R.G., Peiris, J.S.M. and Chen, H. (2006) Distribution of amantadine-resistant H5N1 avian influenza variants in Asia. The Journal of Infectious Diseases, 193, 1626-1629. doi:10.1086/504723
- Tu, Q., Pinto, L.H., Luo, G., Shaughnessy, M.A., Mullaney, D., Kurtz, S., Krystal, M. and Lamb, R.A. (1996) Characterization of M2 ion channel activity by BL-1743, an inhibitor of influenza A virus. Journal of Virology, 70, 4246-4252.
- Li, K.S., Guan, Y., Wang, J., Smith, G.J., Xu, K.M., Duan, L., et al. (2004) Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature, 430, 209-213. doi:10.1038/nature02746
- Puthavathana, P., Auewarakul, P., Charoenying, P.C., Sangsiriwut, K., Pooruk, P., Boonnak, K., et al. (2005) Molecular characterization of the complete genome of human influenza H5N1 virus isolates from Thailand. Journal of General Virology, 86, 423-433. doi:10.1099/vir.0.80368-0
- Hay, A.J., et al. (1993) Options for the control of influenza virus II. Excerpta Medica, Amsterdam, 281-288.
- Hay, A.J., Wolstenholme, A.J., Skehel, J.J. and Smith, M.H. (1985) The molecular basis of the specific anti-influenza action of amantadine. The EMBO Journal, 4, 3021-3024.
- Hayden, F.G., Sperber, S.J., Belshe, R.B., Clover, R.D., Hay, A.J. and Pyke, S. (1991) Recovery of drug resistant influenza A virus during therapeutic use of rimantadine. Antimicrob Agents Chemother, 35, 1741-1747.
- Peiris, J.S., Yu, W.C., Leung, C.W., et al. (2004) Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet, 363, 617-619. doi:10.1016/S0140-6736(04)15595-5
- Le, Q.M., Kiso, M., Someya, K., et al. (2005) Avian flu: Isolation of drug-resistant H5N1 virus. Nature, 437, 1108. doi:10.1038/4371108a
- Jefferson, T., Demicheli, V., Rivetti, D., Jones, M., Di Pietrantonj, C., Rivetti, A. (2006) Antivirals for influenza in healthy adults: Systematic review. Lancet, 367, 303- 313. doi:10.1016/S0140-6736(06)67970-1
- Bright, R.A., Shay, D.K., Shu, B., et al. (2006) Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. Journal of the American Medical Association (JAMA), 295, 891-894. doi:10.1001/jama.295.8.joc60020
- Hayden, F.G., Belshe, R.B., Clover, R.D., Hay, A.J., Oakes, M.G. and Soo, W. (1989) Emergence and apparent transmission of rimantadine-resistant influenza A virus in families. New England Journal of Medicine, 321, 1696- 1702. doi:10.1056/NEJM198912213212502
- Hall, C.B., Dolin, R., Gala, C.L., et al. (1987) Children with influenza A infection: Treatment with rimantadine. Pediatrics, 80, 275-282.
- Shiraishi, K., Mitamura, K., Sakai-Tagawa, Y., Goto, H., Sugaya, N. and Kawaoka, Y. (2003) High frequency of resistant viruses harboring different mutations in amantadine-treated children with influenza. Journal of Infectious Diseases, 188, 57-61. doi:10.1086/375799
- Englund, J.A., Champlin, R.E., Wyde, P.R., et al. (1998) Common emergence of amantadineand rimantadineresistant influenza A viruses in symptomatic immunocompromised adults. Clinical Infectious Diseases, 26, 1418-1424. doi:10.1086/516358
- Hayden, F.G. (2001) Perpectives on antiviral use during pandemic influenza. Philosophical Transactions of the Royal Society B: Biological Sciences, 356, 1877-1884.
- Russell, R.J., Haire, L.F., Stevens, D.J., Collins, P.J., Lin, Y.P., Blackburn, G.M., Hay, A.J., Gamblin, S.J. and Skehel, J.J. (2006) The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature London, United Kingdom, 443, 45-49. doi:10.1038/nature05114
- Chachra, R. and Rizzo, R.C. (2008) Origins of resistance conferred by the R292K neuraminidase mutation via molecular dynamics and free energy calculations. Journal of Chemical Theory and Computation, 4, 1526-1540. doi:10.1021/ct800068v
- McKimm-Breschkin, J.L., Sahasrabudhe, A., Blick, T.J., McDonald, M., Colman, P.M., Hart, G.J., Bethell, R.C. and Varghese, J.N. (1998) Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en derivatives. Journal of Virology, 72, 2456-2462.
- Mishin, V.P., Hayden, F.G. and Gubareva, L.V. (2005) Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrobial Agents and Chemotherapy, 49, 4515-4520. doi:10.1128/AAC.49.11.4515-4520.2005
- Sheu, T.G., Deyde, V.M., Okomo-Adhiambo, M., Garten, R.J., Xu, X., Bright, R.A., Butler, E.N., Wallis, T.R., Klimov, A.I. and Gubareva, L.V. (2008) Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrobial Agents and Chemotherapy, 52, 3284- 3292.
- Wetherall, N.T., Trivedi, T., Zeller, J., Hodges-Savola, C., McKimm-Breschkin, J.L., Zambon, M. and Hayden, F.G. (2003) Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza clinical isolates to neuraminidase inhibitors: Report of the neuraminidase inhibitor susceptibility network. Journal of Clinical Microbiology, 41, 742-750. doi:10.1128/JCM.41.2.742-750.2003
- McKimm-Breschkin, J.L., Trivedi, T., Hampson, A., Hay, A., Klimov, A., Tashiro, M., Hayden, F.G. and Zambon, M. (2003) Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrobial Agents and Chemotherapy, 47, 2264-2272. doi:10.1128/AAC.47.7.2264-2272.2003
- Yen, H., Ilyushina, N.A., Salomon, R., Hoffmann, E., Webster, R.G. and Govorkova, E.A. (2007) Neuraminidase inhibitor-resistant recombinant A/Vietnam/1203/04 (H5N1) influenza viruses retain their replication efficiency and pathogenicity in vitro and in vivo. Journal of Virology, 81, 12418-12426. doi:10.1128/JVI.01067-07
- Meijer, A., Lackenby, A., Hungnes, O., Lina, B., van der Werf, S., Schweiger, B., Opp, M., Paget, J., van de Kassteele, J., Hay, J. and Zambon, M. (2009) Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 season. Emerging Infectious Diseases, 15, 552- 560. doi:10.3201/eid1504.081280
- Monto, A.S., McKimm-Breschkin, J.L., Macken, C., Hampson, A.W., Hay, A., Klimov, A., Tashiro, M., Webster, R.G., Aymard, M., Hayden, F.G. and Zambon, M. (2006) Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrobial Agents and Chemotherapy, 50, 2395-2402. doi:10.1128/AAC.01339-05
- Tamura, D., Mitamura, K., Yamazaki, M., Fujino, M., Nirasawa, M., Kimura, K., Kiso, M., Shimizu, H., Kawakami, C., Hiroi, S., Takahashi, S., Hata, M., Minagawa, H., Kimura, Y., Kaneda, S., Sugita, S., Horimoto, T., Sugaya, N. and Kawaoka, Y. (2009) Oseltamivir-resistant influenza A viruses circulating in Japan. Journal of Clinical Microbiology, 47, 1424-1427. doi:10.1128/JCM.02396-08
- McNicholl, I.R. and McNicholl, J.J. (2001) Neuraminidase inhibitors: Zanamivir and oseltamivir. Annals of Pharmacotherapy, 35, 57-70. doi:10.1345/aph.10118
- Russell, R.J., Haire, L.F., Stevens, D.J., Collins, P.J., Lin, Y.P., Blackburn, G.M., Hay, A.J., Gamblin, S.J. and Skehel, J.J. (2006) The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design, Nature, 443, 45-49. doi:10.1038/nature05114
- Collins, P.J., Haire, L.F., Lin, Y.P., Liu, J., Russell, R.J., Walker, P.A., Skehel, J.J., Martin, S.R., Hay, A.J. and Gamblin, S.J. (2008) Crystal structures of oseltamivirresistant influenza virus neuraminidase mutants, Nature, 453, 1258-1261. doi:10.1038/nature06956
- http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi?go=database
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W. and Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389-3402. doi:10.1093/nar/25.17.3389
- Zdobnov, E.M. and Apweiler, R. (2001) InterProScan— An integration platform for the signature-recognition methods in InterPro. Bioinformatics, 17, 847-848.
- Arnold, K., Bordoli, L., Kopp, J. and Schwede, T. (2006). The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics, 22, 195-201. doi:10.1093/bioinformatics/bti770
- Kiefer, F., Arnold, K., Künzli, M., Bordoli, L. and Schwede, T. (2009). The SWISS-MODEL repository and associated resources. Nucleic Acids Research, 37, D387- D392. doi:10.1093/nar/gkn750
- Schwede, T., Kopp, J., Guex, N. and Peitsch, M.C. (2003) SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Research, 31, 3381-3385. doi:10.1093/nar/gkg520
- Guex, N. and Peitsch, M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modelling. Electrophoresis, 18, 2714-2723. doi:10.1002/elps.1150181505
- Peitsch, M.C. (1995) Protein modeling by E-mail. Bio/ Technology, 13, 658-660.
- http://www.pdb.org/pdb/home/home.do
- Sali, A., Pottertone, L., Yuan, F., van Vlijmen, H. and Karplus, M. (1995) Evaluation of comparative protein modeling by MODELLER. Proteins, 23, 318-326. doi:10.1002/prot.340230306
- Fiser, R.K. and Sali, A. (2000) Modeling of loops in protein structures. Protein Science, 9, 1753-1773. doi:10.1110/ps.9.9.1753
- Laskowski, R.A., MacArthur, M.W., Moss, D.S. and Thornton, J.M. (1993) PROCHECK—A program to check the stereochemical quality of protein structures. Journal of Applied Crystallography, 26, 283-291. doi:10.1107/S0021889892009944
- Morris, A.L., MacArthur, M.W., Hutchinson, E.G. and Thornton, J.M. (1992) Stereochemical quality of protein structure coordinates. Proteins: Structure, Function, and Bioinformatics, 12, 345-364. doi:10.1002/prot.340120407
- Shen, M.-Y. and Sali, A. (2006) Statistical potential for assessment and prediction of protein structures. Protein Science, 15, 2507-2524. doi:10.1110/ps.062416606
- Lüthy, R., Bowie, J.U. and Eisenberg, D. (1992) Assessment of protein models with three-dimensional profiles. Nature, 356, 83-85. doi:10.1038/356083a0
- Ramachandran, G.N., Ramakrishnan, C. and Sasisekharan, V. (1963) Stereochemistry of polypeptide chain configurations. Journal of Molecular Biology, 7, 95-99. doi:10.1016/S0022-2836(63)80023-6
- Kabsch, W. and Sander, C. (1983) Biopolymers dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577-2637. doi:10.1002/bip.360221211
- http://pubchem.ncbi.nlm.nih.gov/
- Wu, G., Robertson, D.H., Brooks, C.L.III and Vieth, M. (2003) Detailed analysis of grid-based molecular docking: A case study of CDOCKER—A CHARMm-based MD docking algorithm. Journal of Computational Chemistry, 24, 1549. doi:10.1002/jcc.10306
- Stoll, V., Stewart, K.D., Maring, C.J., Muchmore, S., Giranda, V., Gu, Y.Y., Wang, G., Chen, Y., Sun, M., Zhao, C., Kennedy, A.L., Madigan, D.L., Xu, Y., Saldivar, A., Kati, W., Laver, G., Sowin, T., Sham, H.L., Greer, J. and Kempf, D. (2003) Influenza neuraminidase inhibitors: Structure-based design of a novel inhibitor series. Biochemistry, 42, 718-727. doi:10.1021/bi0205449
- Lipinski, C.A., Lombardo, F., Dominy, B.W. and Feeney, P.J. (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews, 23, 3-25. doi:10.1016/S0169-409X(96)00423-1
- Hatakeyama, S., Sugaya, N., Ito, M., Yamazaki, M., Ichikawa, M., Kimura, K., Kiso, M., Shimizu, H., Kawakami, C., Koike, K., Mitamura, K. and Kawaoka, Y. (2007) Emergence of influenza B viruses with reduced sensitivity to neuraminidase inhibitors. Journal of the American Medical Association (JAMA), 297, 1435-1442. doi:10.1001/jama.297.13.1435
- Hayden, F.G. (2006) Antiviral resistance in influenza viruses—Implications for management and pandemic response. New England Journal of Medicine, 354, 785-788. doi:10.1056/NEJMp068030
- Kiso, M., Mitamura, K., Sakai-Tagawa, Y., Shiraishi, K., Kawakami, C., Kimura, K., Hayden, F. G., Sugaya, N. and Kawaoka, Y. (2004) Resistant influenza A viruses in children treated with oseltamivir: Descriptive study. Lancet, 364, 759-765. doi:10.1016/S0140-6736(04)16934-1
- Lackenby, A., Hungnes, O., Dudman, S.G., Meijer, A., Paget, W.J., Hay, A.J. and Zambon, M.C. (2008) Emergence of resistance to oseltamivir among influenza A (H1N1) viruses in Europe. Eurosurveillance, 13, 1-2.
- Le, Q.M., Kiso, M., Someya, K., Sakai, Y.T., Nguyen, T.H., Nguyen, K.H.L., Pham, N.D., Ngyen, H.H., Yamada, S., Muramoto, Y., Horimoto, T., Takada, A., Goto, H., Suzuki, T., Suzuki, Y. and Kawaoka, Y. (2005) Avian flu: Isolation of drug-resistant H5N1 virus. Nature (London), 437, 1108. doi:10.1038/4371108a
- Yamashita, M., Tomozawa, T., Kakuta, M., Tokumitsu, A., Nasu, H. and Kubo, S. (2009) CS-8958, a Prodrug of the new neuraminidase inhibitor R-125489, shows long-acting anti-influenza virus activity. Antimicrobial Agents and Chemotherapy, 53, 186-192. doi:10.1128/AAC.00333-08
- Kiso, M., Kubo, S., Ozawa, M., Le, Q.M., Nidom, C.A., et al. (2001) Efficacy of the new neuraminidase inhibitor CS-8958 against H5N1 influenza viruses. PLoS Pathogens, 6, 1-10.
NOTES
*Corresponding author.

