Paper Menu >>
Journal Menu >>
 J. Biomedical Science and Engineering, 2008, 1 85-90 Published Online August 2008 in SciRes. http://www.srpublishing.org/journal/jbise JBiSE Retrospective analysis of chronic hepatitis C in un- treated patients with nonlinear mixed effects mod- el Jian Huang1, Kathleen O’Sullivan1, John Levis2, Elizabeth Kenny-Walsh3, Orla Crosbie3 & Liam Jo- seph Fanning2 1 Statistical Consultancy Unit, University College Cork, Ireland. 2 Molecular Virology, Department of Medicine, Hospital Cork University, Ireland. 3 Depart- ment of Gastroenterology and Hepatology, University Hospital Cork,Ireland. Correspondence should be addressed to Jian Huang (j.huang@ucc.ie). ABSTRACT It is well known that viral load of the hepatitis C virus (HCV) is related to the efficacy of interferon therapy. The complex biological parameters that impact on viral load are essentially unknown. The current knowledge of the hepatitis C virus does not provide a mathematical model for viral load dynamics w ithin untreated patients. We car- ried out an empirical modelling to investigate whether different fluctuation patterns exist and how these patterns (if exist) are related to host- specific factors. Data was prospectively col- lected from 147 untreated patients chronically infected with hepatitis C, each contributing be- tween 2 to 10 years of measurements. We pro- pose to use a three parameter logistic model to describe the overall pattern of viral load fluctua- tion based on an exploratory analysis of the data. To incorporate the correlation feature of longitu- dinal data and patient to patient variation, we introduced random effects components into the model. On the basis of this nonlinear mixed ef- fects modelling, we investigated effects of host- specific factors on viral load fluctuation by in- corporating covariates into the model. The pro- posed model provided a good fit for describing fluctuations of viral load measured with varying frequency over different time intervals. The aver- age viral load growth time was significantly dif- ferent between infection sources. There was a large patient to patient variation in viral load as- ymptote. Keywords: Logistic model, Viral load, Viral genotype, Mixed effects modelling 1. INTRODUCTION Approximately 3% of the world population is infected by the hepatitis C virus (HCV). This virus is a single stranded positive sense RNA virus and does not exist as a single clonotype. It is found as a complex mixture of similar but non-identical isolates, hence, quasispecies. There are seven different genotypes of HCV each with a unique population of subtypes. The nomenclature used to describe these genotypes is numerical, while subtypes are described alphabetically. The amount of virus present in serum at any one time is referred to as the viral load, which can be measured in serum by RT-PCR (a method based on amplification of genomic RNA [1]). A wide range of viral load fluctuation over time was observed within some untreated patients [2]. Treatment efficacy is reduced when viraemia is greater than 5.7-6.0 log10 IU/mL [3]. Knowledge of viral load fluctuations could lead to a more optimised treatment initiation time point. To date several studies have attempted to elucidate viral load fluctuation within untreated patients. Halfon et al. [2] showed that viral load fluctuation within untreated pa- tients was significant. Arase et al. [4] illustrated the ratio of the maximum viral load to the minimum viral load was related to acute exacerbation. Pontisso et al. [5] demon- strated that the mean difference between the maximum viral load and minimum viral load was significantly dif- ferent between normal transaminases and fluctuating transaminases. Our previous studies [6, 7] have showed that viral load does change over time in some patients and exhibits periods of apparent stability in others. The complex biological parameters that impact on HCV viral load are essentially unknown. However, what is known is that the magnitude of the viral load at any one time represents the output of the equilibrium between viral production and host mediated viral clearance. The phenomenon of replicative homeostasis may explain viral load fluctuation over time within an untreated patient [8]. Replicative homeostasis consists of a series of autoregula- tory feedback epicycles that link RNA polymerase func- tion, RNA replication and viral production and presents a model which may rationalize why viraemia modulates over time. Replicative homeostasis results dynamic equi- librium controlled by the specificity of the interactions between mutant or wild type envelope proteins and the replicas, the RNA dependant RNA polymerase (RDRP). In other words, a highly progressive RDRP exhibits a low SciRes Copyright © 2008  86 J. Huang et al. / J. Biomedical Science and Engineering 1 (2008) 85-90 SciRes Copyright © 2008 JBiSE fidelity of replication yielding a high intracellular concen- tration of mutant type envelope proteins. This mutant population competes with wild type forms, resulting in a progressive increase in fidelity and a shift in the dynamic equilibrium which results in the generation of a dominant viral quasispecies and which may be reflected in a change in the absolute magnitude of the viral load. The rate of oscillation between high and low fidelity is likely to be influenced, in a unique way within each viral-host pairing by the visibility of the antigenic quasispecies to immune enhanced viral clearance. Low quasispecies complex is associated with increased antigen concentration, if the threshold of immune activation is passed, immune po- tency is increased and active clearance of particular viral isolates can take place. The removal of the dominant qua- sispecies may be followed by a parallel temporally mis- matched decrease in viral load. Based on interaction of HIV with cells of the immune system, various mathematical models have been devel- oped to fit HIV viral load data [9, 10]. These models were developed to describe viral dynamics during antiviral treatment. Thus, they cannot be used for long term inves- tigations of viral load progression in untreated patients. The current knowledge of HCV does not provide a mathematical model for describing long term HCV dy- namics within an untreated patient. We carried out an empirical modelling analysis to investigate whether dif- ferent fluctuation patterns exist and how these patterns (if exist) are related to host-specific factors. The analysis comprised a two-step approach: first, a statistical model was developed for describing overall viral load fluctua- tion as a function of time, taking individualization into account. Then, we examined effects of host-specific fac- tors on viral load fluctuation by incorporating covariates into the model. A mixed effects logistic model for viral load fluctua- tion is developed in Section 2. By sequential modelling, the effects of host-specific factors (covariates) on viral load fluctuation are investigated in Section 3. The results are discussed in Section 4. 2. MODELLING VIRAL LOAD FLUCTUA- TION 2.1. Data The data used consisted of 147 untreated patients chroni- cally infected with hepatitis C, each contributing between 2 to 10 years of measurements. The study population was divided according to likely source of infection. Group A consisted of 85 individuals whose sole risk factor for ac- quisition of hepatitis C was iatrogenic infection throughreceipt of HCV genotype 1 subtype b (HCV 1) contaminated Anti-D immunoglobulin [11]. Group B consisted of 62 individuals whose risk factors for acquisi- tion of hepatitis C infection likely to be one or more of the followings: intravenous drug use, receipt of Time (year) Viral load (log10IU/mL) 3 4 5 6 7 02468100246810024681002468100246810 3 4 5 6 7 3 4 5 6 73 4 5 6 7 3 4 5 6 73 4 5 6 7 3 4 5 6 73 4 5 6 7 3 4 5 6 73 4 5 6 7 3 4 5 6 73 4 5 6 7 3 4 5 6 73 4 5 6 7 3 4 5 6 7 024681002468100246810 Figure 1. For each patient viral load is plotted against time from the first viral load sampling. contaminated blood, or blood products other than Anti-D immunoglobulin or infection was of undefined aetiology (sexual). The genotype composition of Group B is 1 and 3. 2.2. Models for viral load fluctuation Plotting viral load against time post infection is a natural way for visualising viral load fluctuation. However, due to the asymptomatic nature of HCV, the exact time of when infection occurred is not available for patients in group B. Figure 1 shows the profiles of the viral load of the 147 available patients against time from the first viral load sampling. The study duration time ranged from 24 to 120 months (25% percentile, median, 75% percentile: 60, 84, 108 months). The mean intervals of blood sampling from each patient varied from 3 to 23 months (25% per- centile, median, 75% percentile: 9, 10, 13 months). Figure 1 shows a wide range of viral load fluctuation in  J. Huang et al. / J. Biomedical Science and Engineering 1 (2008) 85-90 87 SciRes Copyright © 2008 JBiSE 0246810 34567 Tim e log10.IU.per.mL Lowess logistic quadratic Figure 2. Scatter plot of viral load overlaid with the LOWESS, logistic and quadratic fitted curves. some untreated patients. In addition, the viral load pro- files vary between patients. To identify an appropriate model to describe the overall fluctuation of viral load as a function of time, the locally-weighted polynomial regres- sion smoothing (the R function LOWESS) was performed on the data (Figure 2). The pattern obtained from the LOWESS shows that viral load increase over some time period, followed by a period of the more stabilized viral load. Such patterns can be described by logistic models [12] ,))/)81ln()(exp(1/( ijijij ty ε γ β α + − −+= (1) where ij y is the logarithm (base 10) of the measurement of viral load for the ith patient at the jth measurement time (from the first viral load sampling) ij t and the model er- ror ij ε is assumed to be i.i.d. N (0,2 σ ). The parameter α is the asymptote (the limit of viral load growth), β is the midpoint, the time when the most rapid viral load growth occurs. The scale parameter γ is the growth time, time interval during which growth progresses from 10% to 90% of the asymptote [12]. )81ln( is introduced into model (1) to facilitate interpretation of γ . As a comparison we fed the quadratic polynomial model to data as well. The fitted curves corresponding to the quadratic polynomial model (AIC=2896.37) and lo- gistic model (AIC=2896.01) are also shown in Figure 2. As can be seen from Figure 2 both models captured the basic structure of the LOWESS smooth with the logistic one performing better at the tail. Having a smaller AIC value the logistic model was selected to describe the overall pattern of viral load fluctuation. It is also evident from Figure 1 that there exists a large patient to patient variation in viral load fluctuation. To account for this patient to patient variation random com- ponents were introduced into model (1) yielding the fol- lowing mixed effects model ,)))/(ln(81) ))(((/(1)( iji iijiij r btexpay εγ βα ++× +−−++= (2) where ),,( iii rba are assumed to be the i.i.d random vec- tor, independent of the model errors and follow the nor- mal distribution N(0,Σ). α is replaced by i a + α to ac- count for patient to patient variation in the viral load as- ymptote. α is called the fixed effect and represents the mean level of the viral load asymptote for the population. i a is called the random effect and represents the individ- ual patient departure from the mean level. Similarly, the fixed effects β and γ represent the mean levels of the midpoint and growth time for the population, respectively. The random effects i band i rare the individual patient departures from the mean levels of the midpoint and growth time, respectively. The R package nlme [13] was used to fit nonlinear mixed effects models. In model (2), all three parameters consisted of a fixed effect term and a random effect term. Such models might be over parameterized. In these cases, the variance-covariance matrix of random effects become seriously ill-conditioned, making convergence difficult or impossible. We adopted model building strategies sug- gested in [13] to determine an adequate but parsimonious model. We began with fitting model (2) to data. Convergence was achieved. However, convergence was sensitive to changes in the initial values and the algorithm failed to converge using values slightly different from the fitted values. Checking the fitted model we found that the esti- mated covariance matrix Σ ) has off diagonal terms of zero. Hence, we refitted model (2) with a diagonal co- variance matrix Σ . Then the possibility of eliminating one or more random effects from the model was investigated. First, we compared models generated by eliminating one of the three random effects from model (2) and found that the model with random components in α and β had the largest likelihood value, termed as model A. Then we considered models generated by removing two of the three random effects from model (2) and found that the model with a random effect in α had the largest likeli- hood value, termed as model B. To establish the signifi- cance of random effects, we followed the procedure de- scribed by Verbeke and Molenberghs [14], who provide an outline for testing the need for random effects by com- paring the log-likelihood between the nested models with and without random effects. The asymptotic null distribu- tion of the test statistics is a mixture of Chi-squares. The results are summarised in Table 1. As can be seen from the table there are significant random effects in α (P<0.0001) and β (P<0.0001). Model B was preferred. Hence we re-parameterized model (2) as follows ij iijiij bty εγ βαα +× +−−++= ))/)81ln( ))((exp(1/()( (3) When fitting model (3) to data, we estimate the stan- dard deviations of the random effects as 0.63 (log10IU/mL) and 1.32 (year) for the asymptote and midpoint of growth time, respectively. The time variable used in the model is the time from the first viral load sampling. It may not be related to the time since onset of infection. A patient can enter the study at any time since onset of infection. For example, some  88 J. Huang et al. / J. Biomedical Science and Engineering 1 (2008) 85-90 SciRes Copyright © 2008 JBiSE Table 1. Comparisons of nested models to establish significance of random effects. Random effects LR test P-Value a Model (1) vs Model B <0.001 b Model A vs Model A <0.001 c Model A vs Model (2) Nonsignificant may present two months after the time of infection; others may present one year later. Such time differences can be accounted for in the model by random effects in β . Ran- dom effects in β allow the logistic growth curve to fit viral load profiles measured at different time spans from onset of infection. Significant patient to patient variation in β (P<0.001) may reflect that patients may have en- tered the study at different time from the times of infec- tion. The significant random effects in α (P<0.001) indi- cates that there exist large patient to patient variation in the asymptote. However, from a clinical perspective our current understanding of hepatitis C disease is such that this variation is difficult to interpret with certainty. 3. ANALYSIS OF COVARIATES In this section, we examined the effects of covariates on the viral load fluctuation by sequentially incorporating them into model (3). A similar approach has been used in [15]. The covariates considered are viral genotype, gender, infection source (Group A and Group B) and age (< 45 years verses ≥ 45 years). In model (3) there are three fixed effects terms, α β and . γ Any given covariate may have a significant effect on at least one of the terms. For example, gender had significant a effect on γ (P<0.05) but not on α and . β When a covariate was incorporated into the model only were significant terms retained. Inter- actions were considered only if a fixed term was signifi- cantly affected by at least two covariates. Initially, one covariate was incorporated into model (3). Incorporating age into model (3) failed to produce any significant term. Hence, three one-covariate models were produced. The one-covariate model with the maximum likelihood was selected as the optimum one-covariate model. Subse- quently, one of the remaining covariates was incorporated into this model and yielded no significant term. The selec- tion process was stopped. The process is summarized in Table 2. The final model selected to fit the viral load data is model (4) with γ influenced by infection source and can be expressed as follows ,)))/()81ln( ))((exp(1/()( ijr iijiij btay εγγ βα =+× +−−++= (4) where r γ represents infection source effect on the growth time γ . The significance of this term was assessed using the F-tests (P<0.0001). As an alternative approach we incorporated covariates into model (3) using the backward elimination approach. The selection process began with incorporating all the Table 2. Sequentially incorporating covariates into model (3) to determine important covariates. Significant term is indicated in the bracket. Models P-value Log- likelihood Time (model (3)) -946.40 Time + gender ( γ ) <0.001 -931.34 Time + infection source ( γ ) <0.001 -925.63 Time + genotype ( γ ) <0.001 -946.01 covariates into model (3). Then non significant terms were removed from the model recursively. Using a significance level of 0.05, this approach also selected model (4) as the final model. A likelihood ratio test was used to test the difference be- tween the fixed effects represented by model (3) and(4). The result favors model (4) with P<0.0001. Plotting the standardized residuals against the fitted values of model (4) and the Normal plot of the residuals (not shown) did not illustrate any concerns about the model assumptions. The individual fitted curves (Figure 3) indicate good fits to the data, keeping in mind that the data were irregularly and sparsely sampled. The fitting of model (4) to the data is summarised in Table 3. The results show that the mean viral load as- ymptote for the population was 6.44 (log10IU/mL). The most rapid viral load growth occurred, on average, 3.29 years before patients entered the study. The mean growth time for the population was 11.04 years. The patients in Group A experienced, on average, significantly longer growth time (delta = 3.80 year) compared to those in Group B. Since all patients in Group A are female and have vi- ral genotype 1, to separate the gender and genotype ef- fects from the infection source effect, the previous ap- proach to evaluating the effects of covariates by sequen- tially incorporating them into model (3) was applied to Group B. No significant one-covariate model was pro- duced. This finding is very different from the results de- scribed earlier. Using the full data we found that all co- variates except age had a significant effect on viral load growth pattern (see Table 2). One possible explanation is that the gender and genotype significance found in the previous analysis may be due to the significance of infec- tion source. Another possibility is that number of patient in Group B was small and there may have not been suffi- cient power to identify gender and genotype significance. 4. CONCLUSIONS AND DISCUSSIONS We have developed a mixed effects logistic model for describing the viral load fluctuation within untreated pa- tients with chronically infected HCV. The model’s diag- nostic analysis indicated that the proposed model pro- vided a good fit to the data. Due to the asymptomatic na- ture of HCV, it is impossible to determine the exact time when a patient was infected. Hence the time variable used in the model is the time since an individual patient pre- 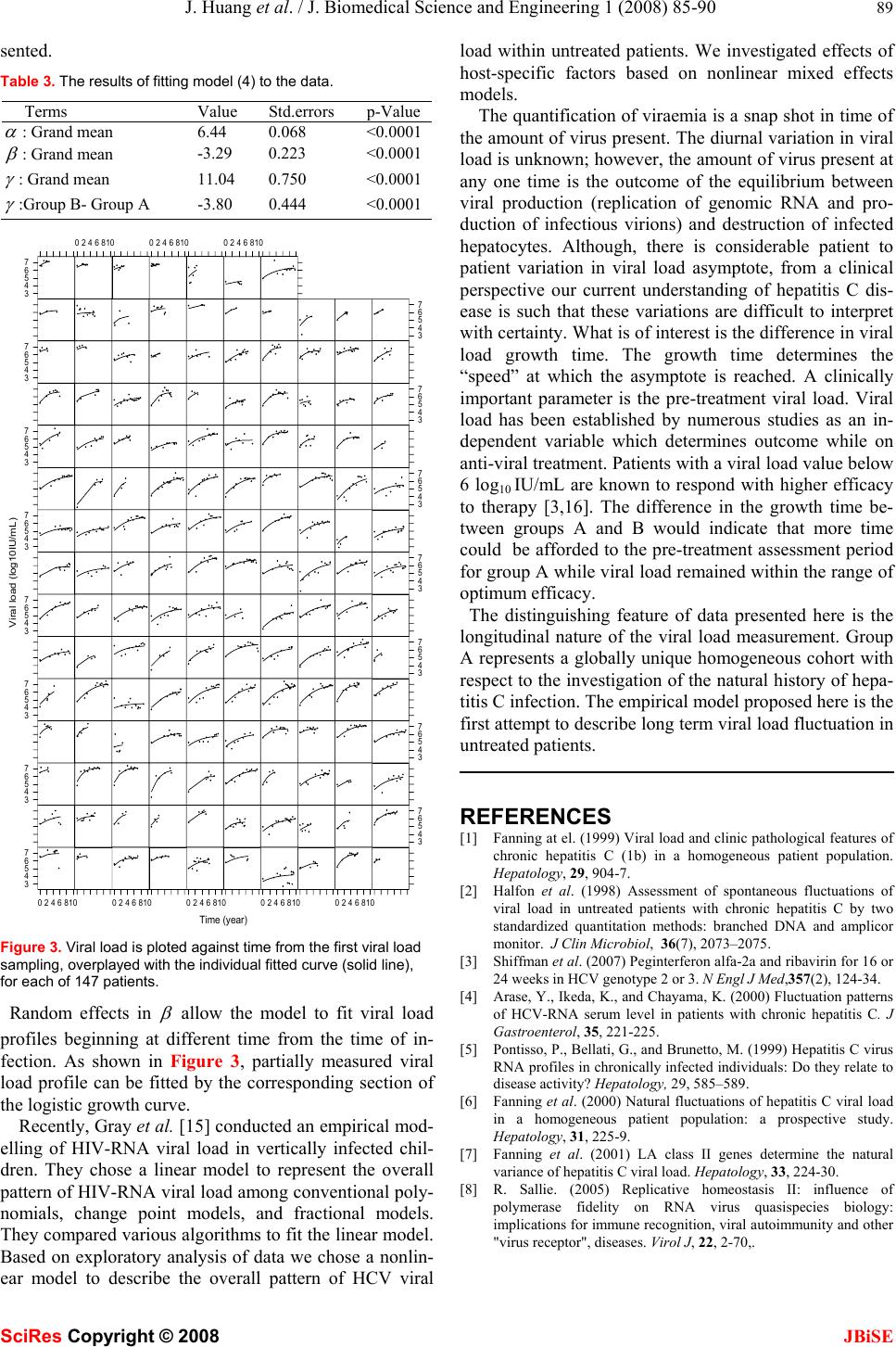 J. Huang et al. / J. Biomedical Science and Engineering 1 (2008) 85-90 89 SciRes Copyright © 2008 JBiSE sented. Table 3. The results of fitting model (4) to the data. Terms Value Std.errors p-Value α : Grand mean 6.44 0.068 <0.0001 β : Grand mean -3.290.223 <0.0001 γ : Grand mean 11.040.750 <0.0001 γ :Group B- Group A -3.800.444 <0.0001 Time (year) Viral load (log10IU/mL) 3 4 5 6 7 02468100246810 024681002468100246810 3 4 5 6 7 3 4 5 6 73 4 5 6 7 3 4 5 6 73 4 5 6 7 3 4 5 6 73 4 5 6 7 3 4 5 6 73 4 5 6 7 3 4 5 6 73 4 5 6 7 3 4 5 6 73 4 5 6 7 3 4 5 6 7 02468100246810 0246810 Figure 3. Viral load is ploted against time from the first viral load sampling, overplayed with the individual fitted curve (solid line), for each of 147 patients. Random effects in β allow the model to fit viral load profiles beginning at different time from the time of in- fection. As shown in Figure 3, partially measured viral load profile can be fitted by the corresponding section of the logistic growth curve. Recently, Gray et al. [15] conducted an empirical mod- elling of HIV-RNA viral load in vertically infected chil- dren. They chose a linear model to represent the overall pattern of HIV-RNA viral load among conventional poly- nomials, change point models, and fractional models. They compared various algorithms to fit the linear model. Based on exploratory analysis of data we chose a nonlin- ear model to describe the overall pattern of HCV viral load within untreated patients. We investigated effects of host-specific factors based on nonlinear mixed effects models. The quantification of viraemia is a snap shot in time of the amount of virus present. The diurnal variation in viral load is unknown; however, the amount of virus present at any one time is the outcome of the equilibrium between viral production (replication of genomic RNA and pro- duction of infectious virions) and destruction of infected hepatocytes. Although, there is considerable patient to patient variation in viral load asymptote, from a clinical perspective our current understanding of hepatitis C dis- ease is such that these variations are difficult to interpret with certainty. What is of interest is the difference in viral load growth time. The growth time determines the “speed” at which the asymptote is reached. A clinically important parameter is the pre-treatment viral load. Viral load has been established by numerous studies as an in- dependent variable which determines outcome while on anti-viral treatment. Patients with a viral load value below 6 log10 IU/mL are known to respond with higher efficacy to therapy [3,16]. The difference in the growth time be- tween groups A and B would indicate that more time could be afforded to the pre-treatment assessment period for group A while viral load remained within the range of optimum efficacy. The distinguishing feature of data presented here is the longitudinal nature of the viral load measurement. Group A represents a globally unique homogeneous cohort with respect to the investigation of the natural history of hepa- titis C infection. The empirical model proposed here is the first attempt to describe long term viral load fluctuation in untreated patients. REFERENCES [1] Fanning at el. (1999) Viral load and clinic pathological features of chronic hepatitis C (1b) in a homogeneous patient population. Hepatology, 29, 904-7. [2] Halfon et al. (1998) Assessment of spontaneous fluctuations of viral load in untreated patients with chronic hepatitis C by two standardized quantitation methods: branched DNA and amplicor monitor. J Clin Microbiol, 36(7), 2073–2075. [3] Shiffman et al. (2007) Peginterferon alfa-2a and ribavirin for 16 or 24 weeks in HCV genotype 2 or 3. N Engl J Med,357(2), 124-34. [4] Arase, Y., Ikeda, K., and Chayama, K. (2000) Fluctuation patterns of HCV-RNA serum level in patients with chronic hepatitis C. J Gastroenterol, 35, 221-225. [5] Pontisso, P., Bellati, G., and Brunetto, M. (1999) Hepatitis C virus RNA profiles in chronically infected individuals: Do they relate to disease activity? Hepatology, 29, 585–589. [6] Fanning et al. (2000) Natural fluctuations of hepatitis C viral load in a homogeneous patient population: a prospective study. Hepatology, 31, 225-9. [7] Fanning et al. (2001) LA class II genes determine the natural variance of hepatitis C viral load. Hepatology, 33, 224-30. [8] R. Sallie. (2005) Replicative homeostasis II: influence of polymerase fidelity on RNA virus quasispecies biology: implications for immune recognition, viral autoimmunity and other "virus receptor", diseases. Virol J, 22, 2-70,.  90 J. Huang et al. / J. Biomedical Science and Engineering 1 (2008) 85-90 SciRes Copyright © 2008 JBiSE [9] Wu, H. (2005) Statistical methods for HIV dynamic studies in AIDS clinical trials. Statistical Methods in Medical Research, 14, 171-192. [10] Donnelly, C.A. and Cox, D.R. (2001) Mathematical biology and medical statistics: contributions to the understanding of AIDS epidemiology. Stat Methods Mes Res, 10(2), 141-54. [11] Kenny-Walsh E. (1999) Clinical outcomes after hepatitis C infection from contaminated anti-D immune globulin. Irish Hepatology Research Group. N Engl J Med, 16, 1228-33. [12] Meyer, P.S., Yung, J.W., and Ausubel, J.H. (1999) A Primer on Logistic Growth and Substitution: The Mathematics of the Loglet Lab Software. Technological Forecasting and Social Change, 61, 247-271. [13] Pinheiro, J.C., and Bates, D.M. (2000) Mixed-Effects Models in S and S-PLUS, Springer. [14] Verbeke, G., Molenberghs, G. (2000) Linear Mixed Models for Longitudinal Data. Springer, New York. [15] L. Gray, M. Cortina-Borja and ML Newell. (2004) Modelling HIV-RNA viral load in vertically infected children. Statist. Med, 23, 769-781. [16] Zeuzem et al. (2006) Efficacy of 24 weeks treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C infected with genotype 1 and low pretreatment viremia. J Hepatol, 44, 97-103. |

