 American Journal of Plant Sciences, 2011, 2, 180-189 doi:10.4236/ajps.2011.22020 Published Online June 2011 (http://www.SciRP.org/journal/ajps) Copyright © 2011 SciRes. AJPS Molecular Markers for Tm-2 Alleles of Tomato Mosaic Virus Resistance in Tomato Ainong Shi1,2*, Richard Vierling2,6, Richa rd Gra zzini3, Pengyin Chen4, Homer Caton1, Dilip Panthee5 1Syngenta Seeds, Slater, USA; 2Indiana Crop Improvement Association and Department of Agronomy, Purdue University, West Lafayette, USA; 3GardenGenetics LLC, Bellefonte, USA; 4Department of Crop, Soil, and Environmental Sciences, University of Arkansas, Fayetteville, USA; 5Department of Horticultural Science, North Carolina State University, Mountain Horticultural Crops Research and Extension Center, Raleigh, USA; National Corn Growers Association, Cepi Drive Chesterfield, USA; 6National Corn Growers Association, Chesterfield, USA. Email: *ainong.shi@syngenta.com, ainong_shi@hotmail.com Received January 23rd, 2011; revised March 26th, 2011; accepted March 31st, 2011. ABSTRACT Tomato mosaic virus (To MV) is one of the most infectious virus diseases in tomato (Solanum lycopersicum L). The practical and effective method o f controllin g this disease is through gene tic control by u sing major resistance genes. So far, three genes Tm-1, Tm-2 and Tm-22 conferring resistance to ToMV have been reported and utilized in toma to culti- var development. Marker assisted selection (MAS) has become very important and useful tool in selection of ToMV re- sistant tomato lines or hybrids. The objective of this research was to identify allele-specific PCR-bas ed, cleaved ampli- fied polymorphic sequence (CAPS), and allele-derived single nucleotide polymorphism (SNP) markers for Tm-2 loci. Four allele-specific PCR-based markers were iden tified: one for Tm-2, one for Tm-22, and two for th e susceptib le allele tm-2. Three allele-derived CAPS markers were identified, which can identify and distinguish three alleles, tm-2, Tm-2 and Tm-22 in tomato germplasm. Three SNP markers were developed specific for Tm-2 locus. These markers will pro- vide breeders with a tool in selection of Tm-2 and Tm-22 resistance genes i n tomato breeding pr ogr a m. Keywords: Cleaved Amplified PolymorphicSequence,Marker-Assisted Selection, Single Nucleotide Polymorphism, Solanum Lycopersicum, Tomato, Tomato Mosaic Virus (ToMV), Tm -2 1. Introduction Tomato mosaic virus (ToMV) is one of the most infec- tious virus diseases in tomato (Solanum lycopersicum L. formerly, Lycopersicon esculentum Mill). The practical and effective method of controlling this disease is through introducing major resistance gene(s). So far, three genes, Tm-1, Tm-2 and Tm-22 of ToMV resistance have been reported and used in tomato cultivar develop- ment [1-5]. The Tm-1 gene was introgressed into S. ly- copersicum from the wild tomato species S. habrochaites and mapped to chromosome 5 and it conferred resistance to ToMV strains 0 and 2 [1,4,6,7]. The Tm-1 gene has been cloned and the sequences were stored in GenBank with five accessions DJ344478 to DJ344481, and DJ344505. The other two genes Tm-2 and Tm-22 were introgressed from S. peruvianum and were located closely to the centromere of chromosome 9 and are considered to be allelic and Tm-2 conferred resistance to ToMV strains 0 and 1, whereas allele Tm-22 conferred resistance to ToMV strains 0, 1 and 2 [2,3,5,8-10]. Molecular markers have been widely used in genetic mapping and marker-assisted selection (MAS) for dis- ease resistance in tomato [6]. For Tm-2 genes of ToMV resistance, Ohmori et al. [11] reported 13 random ampli- fied polymorphic DNA (RAPD) markers linked to the Tm-2 locus. From the 13 RAPD markers, Motoyoshi et al. [11] found two markers, OPE16 (900) and OPN31 (1000), nearest to the Tm-2 locus. Six out of the 13 RAPD mark- ers distributed within 0.7 cM on chromosome 9 were cloned and sequenced to be converted into sequence characterized amplified region (SCAR) markers [11]. Sobir et al. [12] and Dax et al. [13] identified co-dominant SCAR markers tightly linked to Tm-22. Three alleles, Tm-2, Tm-22, and tm-2 at Tm - 2 locus have been cloned and sequenced (GenBank Accessions: AF536199, AF536200, and AF536201) and three cleaved amplified polymorphic sequences (CAPS) markers were developed to distinguish the three alleles in tomato [14]. Based on the two CAPS markers reported by Lanfemeijer et al.  Molecular Markers for Tm-2 Alleles of Tomato Mosaic Virus Resistance in Tomato181 [14], Arens et al. [15] developed a co-dominant assay combining tetra primers designed from the SNP regions of the two CAPS markers using a method called ARMS-PCR SNP and this Tetra-primer ARMS-PCR assays can amplify PCR products with different DNA fragment sizes from tomato lines carrying Tm-2 or Tm-22, and susceptible allele tm-2. Single nucleotide polymorphism (SNP) is becoming to be the most useful as molecular marker in genome map- ping, association studies, diversity analysis, and tagging genes for economically important traits in crop plants because of their abundance and automated highthroughput genotyping [16-18]. SNPs have been discovered and verified in tomato [19-22] and successfully used in se- lecting resistance to bacterial speck and bacterial spot in tomato [22,23]. The objective of this research was to identify Tm-2 allele-specific PCR-based markers, CAPS markers, and SNP markers for MAS in tomato breeding. 2. Materials and Methods 2.1. Plant Materials Twenty-three tomato genotypes including released or commercial cultivars and accessions were used in this research (Table 1). Seeds of 15 tomato accessions (LA series) were obtained from the C.M. Rick Tomato Genetics Resource Center, Dept. of Plant Sciences, University of California, Davis, CA 95616 (http://tgrc.ucdavis.edu), six cultivars were purchased from commercial sources, and two lines NY07-461 and NY07-464 were obtained from Cornell University; refined selections from a cross be- tween “Brandywine” and “Rose de Berne” (received as NY07-461 (Brandyrose #1) and NY07-464 (Brandyrose #2)). Among the 23 genotypes, three cultivars, Royal Red (LA2088), CLN2264F (LA4285), and LA3433 are homozygous for Tm-2 (Tm-2/Tm-2); Mogeor (LA3471), Sophya, and VFNT Cherry (LA1221) are homozygous for Tm-22 (Tm-22/Tm-22); Swt Cluster, Bush Celebrity, and Golden Girl are heterozygous for Tm-22 (Tm-22/tm-2); and the other 14 genotypes contained the susceptible allele tm-2 (Table 1). 2.2. DNA Extraction, PCR Amplification and DNA Sequencing Genomic DNA was extracted from fresh leaves of greenhouse-grown plants using the CTAB (Cetyl- trimethylammonium Bromide) method [24,25]. The gene- specific primers for Tm-2, Tm-22, or tm-2 at Tm-2 locus were designed from the sequences of the GenBank accessions AF536199, AF536200, and AF536201 using the primer design tool—Primer-BLAST (http://www.ncbi. nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=Bl astHome). AF536199 is the accession in GenBank that contains the sequence of the tm-2 susceptible allele; AF536200 is the sequence Tm-2 resistance allele; and AF536201 is the sequence of Tm-22 resistance allele. The size of the AF536199 sequence of the allele tm-2 is 2820 bp; AF536200 is 2819 bp; and AF536201 is 9837 bp [3]. Primers Tm2RS-f2, Tm2RS-r2, Tm2RS-f3, and Tm2RS- r3 were designed for the three sequences of AF536199, AF536200, and AF536201; Tm2S-f1, Tm2S-r1, Tm2S-f2, and Tm2S-r2 for AF536199 only; Tm2R-f1c and Tm2R- r4 for both AF536200 and AF536201; Tm2R-r3 for AF536200 only; and Tm2aR-r3 for AF536201 only (Ta- ble 2). PCR amplification was performed in an eppendorf thermal cycler (Eppendorf, Westbury, NY) following standard PCR procedures with minor modifications. Briefly, each 50 µl PCR reaction mixture consisted of 29.8 µl sterilized ddH2O, 10 µl 5x Mango Taq reaction buffer (Bioline, London, UK), 3 µl MgCl2 (25 mM), 1.5 µl dNTP (2.5 mM each), 1.5 µl each primer (5 µM), 0.2 µl Mango Taq DNA polymerase (5 U/µl) (Bioline, Lon- don, UK), and 2.5 µl template DNA (30 ng/µl). For the primer pairs, Tm2RS-f2/r2 and Tm2RS-f3/r3, PCR pro- cedure consisted of an initial denaturation step of 94˚C for 4 min followed by 36 to 38 cycles of 15 to 30 second at 94˚C for denaturation, 15 to 30 s at 51 to 56˚C for an- nealing, and 40 to 75 s at 72˚C for extension depending on primer pairs with a final extension step at 72˚C for 5 min (Table 3). The PCR fragments were separated by gel electrophoresis with 1.5% agarose gel in 0.5 X TAE buffer, stained with ethidium bromide, and visualized with UV light. The PCR fragments were sequenced in the Purdue Genomics Core Facility, Purdue University, West Lafay- ette, IN 47907. Before sequencing of PCR products, PCR products were purified using QIA quick PCR Purification Kit (Qiagen Inc, Valencia, CA) following the manufac- turer’s instructions of the protocol. The sequences ampli- fied from the two primer pairs Tm2RS-f2/-r2 and Tm2RS- f3/-r3 in tomato were submitted to GenBank using a DNA sequence submission and update tool—Sequin (http://www.ncbi.nlm.nih.gov/Sequin/). 2.3. SNP Identification and Genotyping SNP discovery was postulated from available Tm-2 se- quences of AF536199, AF536200, and AF536201. The multiple sequence alignment showed that 68 SNPs and one InDel were observed among the three sequences. One of the approaches to SNP validation is through re-sequencing of the same gene among various genotypes. Two Tm-2 gene-specific primer pairs Tm-2RS-f2/r2 and Tm-2RS-f3/r3 were designed from the sequences of AF536199, AF536200, and AF536201 using Primer- LAST (Table 2) and used to run PCR among tomato B Copyright © 2011 SciRes. AJPS  Molecular Markers for Tm-2 Alleles of Tomato Mosaic Virus Resistance in Tomato Copyright © 2011 SciRes. AJPS 182 Table 1. Allele-specific PCR-based, allele-derived CAPS, and allele-derived SNP markers for Tm-2 loci in 23 tomato geno- types. Allele-specific PCRa Allele-derived CAPS SNP genotyping by Sequenom b Cultivar/ accession Genotype at Tm-2 locus Tm2S-f1/ Tn2S-r1 Tm2S-f2/ Tm2S-r2 Tm2R-f1 c/ Tm2R-r3 Tm2R-f1c/ Tm2aR-r3 Tm2R-fi c/Tm2-r 4 Tm2rs- f3/r3 PshAIHpaI BsiHKAI Tm2- snp1 Tm2- snp46 Tm2- snp53 NY07-461 tm-2/tm-2 + + − − − 703 bp538 bp + 165 bp703 bp 703 bp T T C CG G NY07-464 tm-2/tm-2 + + − − − 703 bp538 bp + 165 bp703 bp 703 bp T T C CG G Riesentraube tm-2/tm-2 + + − − − 703 bp538 bp + 165 bp703 bp 703 bp T T C CG G LA0656 tm-2/tm-2 + + − − − 703 bp538 bp + 165 bp703 bp 703 bp T T C CG G LA1792 tm-2/tm-2 + + − − − 703 bp538 bp + 165 bp703 bp 703 bp T T C CG G LA1802 tm-2/tm-2 + + − − − 703 bp538 bp + 165 bp703 bp 703 bp T T C CG G LA3386 tm-2/tm-2 + + − − − 703 bp538 bp + 165 bp703 bp 703 bp T T C CG G Anahu (LA0655) tm-2/tm-2 + + − − − 703 bp 538 bp + 165 bp703 bp 703 bp T T C CG G Motelle (LA2823) tm-2/tm-2 + + − − − 703 bp538 bp + 165 bp703 bp 703 bp T T C CG G Mobox (LA2821) tm-2/tm-2 + + − − − 703 bp538 bp + 165 bp703 bp 703 bp T T C CG G Ontario 7710 (LA2396) tm-2/tm-2 + + − − − 703 bp538 bp + 165 bp703 bp 703 bp T T C CG G Peto 95-43 (LA3528) tm-2/tm-2 + + − − − 703 bp538 bp + 165 bp703 bp 703 bp T T C CG G Rehovot 13 (LA3129) tm-2/tm-2 + + − − − 703 bp538 bp + 165 bp703 bp 703 bp T T C CG G UC-204C (LA3130) tm-2/tm-2 + + − − − 703 bp 538 bp + 165 bp703 bp 703 bp T T C CG G Royal Red Cherry (LA2088) Tm-2/Tm-2 − − + − + 703 bp703 bp458 bp + 245 bp 358 bp + 353 bp C C T TA A CLN2264F (LA4285) Tm-2/Tm-2 − − + − + 703 bp703 bp458 bp + 245 bp 358 bp + 353 bp C C T TA A LA3433 Tm-2/Tm-2 − − + − + 703 bp703 bp458 bp + 245 bp 358 bp + 353 bp C C T TA A Mogeor (LA3471) Tm-22/ Tm-22 − − − + + 703 bp703 bp458 bp + 245 bp 703 bp C C T TA G Sophya Tm-22/ Tm-22 − − − + + 703 bp703 bp458 bp + 245 bp 703 bp C C T TA A VFNT Cherry (LA1221) Tm-22/ Tm-22 − − − + + 703 bp703 bp458 bp + 245 bp 703 bp C C T TA A Swt Cluster Tm-22/tm-2 + + − + + 703 bp 703bp + 538 bp + 165 bp 703 bp + 458 bp + 245 bp 703 bp C T C TA G Bush Celebrity Tm-22/tm-2 + + − + + 703 bp 703 bp + 538 bp + 165 bp 703 bp + 458 bp + 245 bp 703 bp C T C TA G Golden Girl Tm-22/tm-2 + + − + + 703 bp 703 bp + 538 bp + 165 bp 703 bp + 458 bp + 245 bp 703 bp C T C TA G a ‘+’ signifies the PCR fragment (band) present and ‘−’ signifies the PCR fragment (band) absent; b The SNP type such as [A T] signifies a heterogeneous SNP type and [A A] signifies a homogenous SNP type. 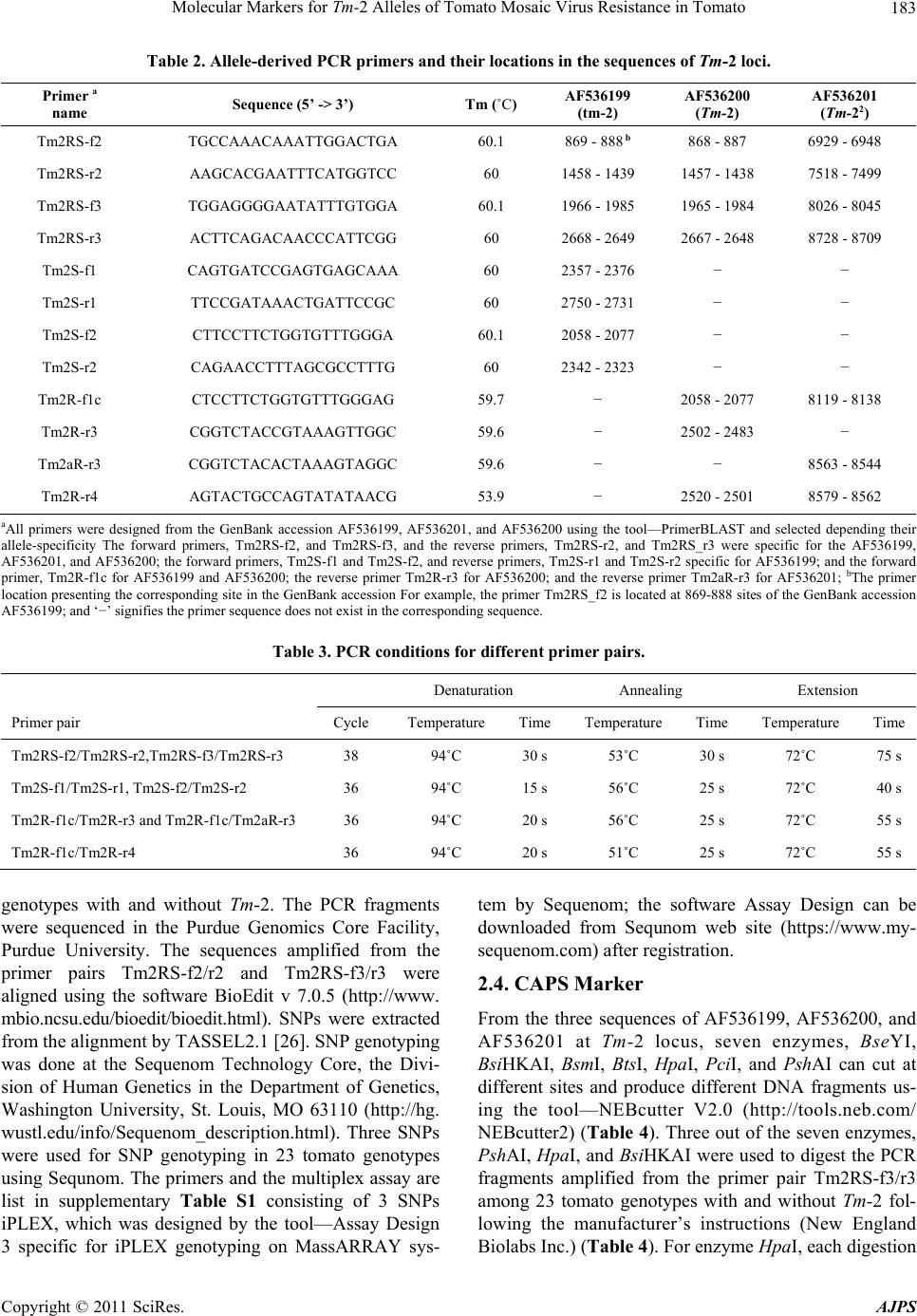 Molecular Markers for Tm-2 Alleles of Tomato Mosaic Virus Resistance in Tomato183 Table 2. Allele-derived PCR primers and their locations in the sequences of Tm-2 loci. Primer a name Sequence (5’ -> 3’) Tm (˚C) AF536199 (tm-2) AF536200 (Tm-2) AF536201 (Tm-22) Tm2RS-f2 TGCCAAACAAATTGGACTGA 60.1 869 - 888 b 868 - 887 6929 - 6948 Tm2RS-r2 AAGCACGAATTTCATGGTCC 60 1458 - 1439 1457 - 1438 7518 - 7499 Tm2RS-f3 TGGAGGGGAATATTTGTGGA 60.1 1966 - 1985 1965 - 1984 8026 - 8045 Tm2RS-r3 ACTTCAGACAACCCATTCGG 60 2668 - 2649 2667 - 2648 8728 - 8709 Tm2S-f1 CAGTGATCCGAGTGAGCAAA 60 2357 - 2376 − − Tm2S-r1 TTCCGATAAACTGATTCCGC 60 2750 - 2731 − − Tm2S-f2 CTTCCTTCTGGTGTTTGGGA 60.1 2058 - 2077 − − Tm2S-r2 CAGAACCTTTAGCGCCTTTG 60 2342 - 2323 − − Tm2R-f1c CTCCTTCTGGTGTTTGGGAG 59.7 − 2058 - 2077 8119 - 8138 Tm2R-r3 CGGTCTACCGTAAAGTTGGC 59.6 − 2502 - 2483 − Tm2aR-r3 CGGTCTACACTAAAGTAGGC 59.6 − − 8563 - 8544 Tm2R-r4 AGTACTGCCAGTATATAACG 53.9 − 2520 - 2501 8579 - 8562 aAll primers were designed from the GenBank accession AF536199, AF536201, and AF536200 using the tool—PrimerBLAST and selected depending their allele-specificity The forward primers, Tm2RS-f2, and Tm2RS-f3, and the reverse primers, Tm2RS-r2, and Tm2RS_r3 were specific for the AF536199, AF536201, and AF536200; the forward primers, Tm2S-f1 and Tm2S-f2, and reverse primers, Tm2S-r1 and Tm2S-r2 specific for AF536199; and the forward primer, Tm2R-f1c for AF536199 and AF536200; the reverse primer Tm2R-r3 for AF536200; and the reverse primer Tm2aR-r3 for AF536201; bThe primer location presenting the corresponding site in the GenBank accession For example, the primer Tm2RS_f2 is located at 869-888 sites of the GenBank accession AF536199; and ‘−’ signifies the primer sequence does not exist in the corresponding sequence. Table 3. PCR conditions for different primer pairs. Denaturation Annealing Extension Primer pair Cycle Temperature Time Temperature Time Temperature Time Tm2RS-f2/Tm2RS-r2,Tm2RS-f3/Tm2RS-r3 38 94˚C 30 s 53˚C 30 s 72˚C 75 s Tm2S-f1/Tm2S-r1, Tm2S-f2/Tm2S-r2 36 94˚C 15 s 56˚C 25 s 72˚C 40 s Tm2R-f1c/Tm2R-r3 and Tm2R-f1c/Tm2aR-r3 36 94˚C 20 s56˚C 25 s 72˚C 55 s Tm2R-f1c/Tm2R-r4 36 94˚C 20 s51˚C 25 s 72˚C 55 s genotypes with and without Tm-2. The PCR fragments were sequenced in the Purdue Genomics Core Facility, Purdue University. The sequences amplified from the primer pairs Tm2RS-f2/r2 and Tm2RS-f3/r3 were aligned using the software BioEdit v 7.0.5 (http://www. mbio.ncsu.edu/bioedit/bioedit.html). SNPs were extracted from the alignment by TASSEL2.1 [26]. SNP genotyping was done at the Sequenom Technology Core, the Divi- sion of Human Genetics in the Department of Genetics, Washington University, St. Louis, MO 63110 (http://hg. wustl.edu/info/Sequenom_description.html). Three SNPs were used for SNP genotyping in 23 tomato genotypes using Sequnom. The primers and the multiplex assay are list in supplementary Table S1 consisting of 3 SNPs iPLEX, which was designed by the tool—Assay Design 3 specific for iPLEX genotyping on MassARRAY sys- tem by Sequenom; the software Assay Design can be downloaded from Sequnom web site (https://www.my- sequenom.com) after registration. 2.4. CAPS Marker From the three sequences of AF536199, AF536200, and AF536201 at Tm-2 locus, seven enzymes, BseYI, BsiHKAI, BsmI, BtsI, HpaI, PciI, and PshAI can cut at different sites and produce different DNA fragments us- ing the tool—NEBcutter V2.0 (http://tools.neb.com/ NEBcutter2) (Table 4). Three out of the seven enzymes, PshAI, HpaI, and BsiHKAI were used to digest the PCR fragments amplified from the primer pair Tm2RS-f3/r3 among 23 tomato genotypes with and without Tm-2 fol- lowing the manufacturer’s instructions (New England iolabs Inc.) (Table 4). For enzyme HpaI, each digestion B Copyright © 2011 SciRes. AJPS 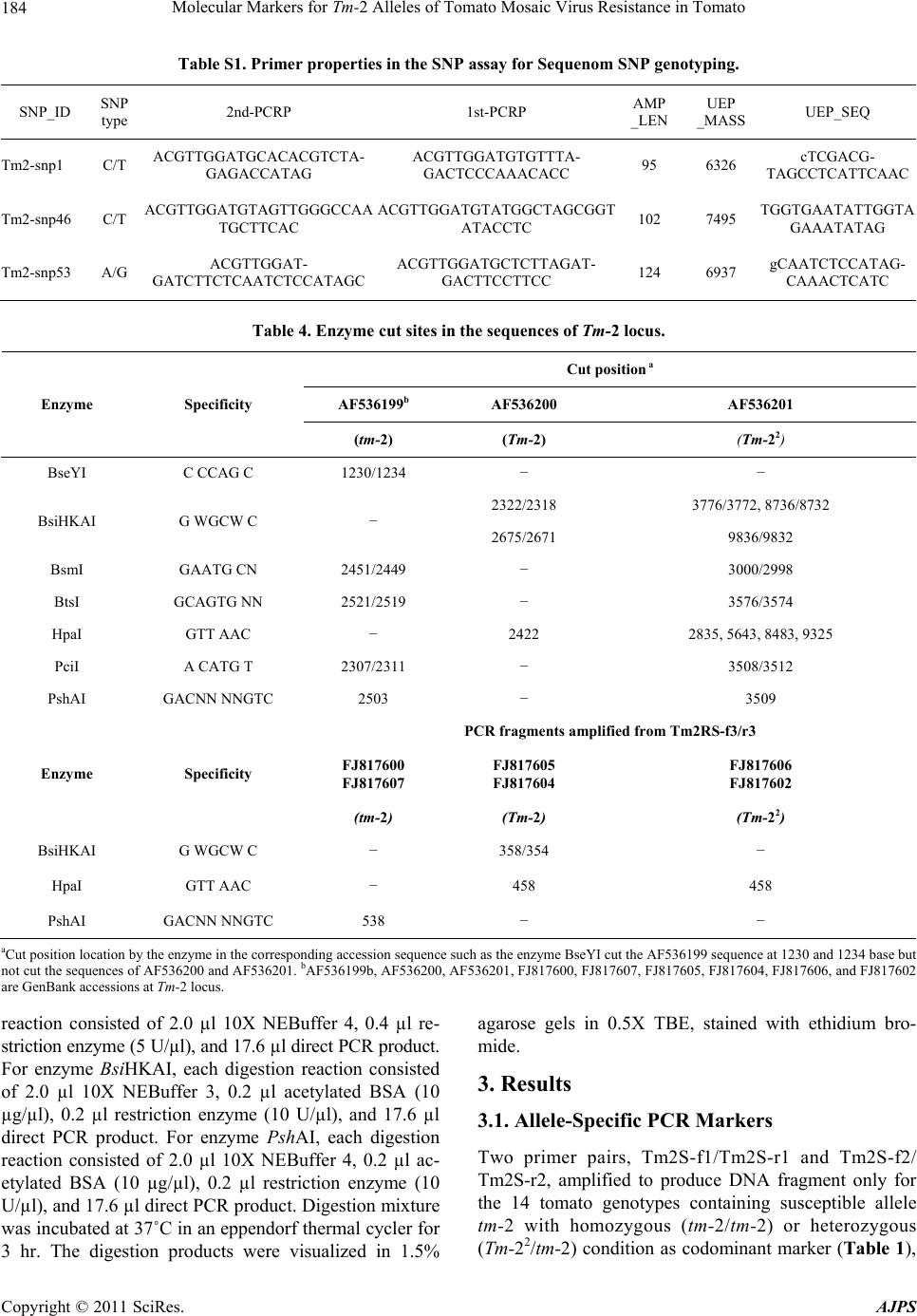 Molecular Markers for Tm-2 Alleles of Tomato Mosaic Virus Resistance in Tomato 184 Table S1. Primer properties in the SNP assay for Seque nom SNP ge notyping. SNP_ID SNP type 2nd-PCRP 1st-PCRP AMP _LEN UEP _MASS UEP_SEQ Tm2-snp1 C/T ACGTTGGATGCACACGTCTA- GAGACCATAG ACGTTGGATGTGTTTA- GACTCCCAAACACC 95 6326 cTCGACG- TAGCCTCATTCAAC Tm2-snp46 C/T ACGTTGGATGTAGTTGGGCCAA TGCTTCAC ACGTTGGATGTATGGCTAGCGGT ATACCTC 102 7495 TGGTGAATATTGGTA GAAATATAG Tm2-snp53 A/G ACGTTGGAT- GATCTTCTCAATCTCCATAGC ACGTTGGATGCTCTTAGAT- GACTTCCTTCC 124 6937 gCAATCTCCATAG- CAAACTCATC Table 4. Enzyme cut sites in the sequences of Tm-2 locus. Cut position a AF536199b AF536200 AF536201 Enzyme Specificity (tm-2) (Tm-2) (Tm-22) BseYI C CCAG C 1230/1234 − − 2322/2318 3776/3772, 8736/8732 BsiHKAI G WGCW C − 2675/2671 9836/9832 BsmI GAATG CN 2451/2449 − 3000/2998 BtsI GCAGTG NN 2521/2519 − 3576/3574 HpaI GTT AAC − 2422 2835, 5643, 8483, 9325 PciI A CATG T 2307/2311 − 3508/3512 PshAI GACNN NNGTC 2503 − 3509 PCR fragments amplified from Tm2RS-f3/r3 FJ817600 FJ817607 FJ817605 FJ817604 FJ817606 FJ817602 Enzyme Specificity (tm-2) (Tm-2) (Tm-22) BsiHKAI G WGCW C − 358/354 − HpaI GTT AAC − 458 458 PshAI GACNN NNGTC 538 − − aCut position location by the enzyme in the corresponding accession sequence such as the enzyme BseYI cut the AF536199 sequence at 1230 and 1234 base but not cut the sequences of AF536200 and AF536201. bAF536199b, AF536200, AF536201, FJ817600, FJ817607, FJ817605, FJ817604, FJ817606, and FJ817602 are GenBank accessions at Tm-2 locus. reaction consisted of 2.0 µl 10X NEBuffer 4, 0.4 µl re- striction enzyme (5 U/µl), and 17.6 µl direct PCR product. For enzyme BsiHKAI, each digestion reaction consisted of 2.0 µl 10X NEBuffer 3, 0.2 µl acetylated BSA (10 µg/µl), 0.2 µl restriction enzyme (10 U/µl), and 17.6 µl direct PCR product. For enzyme PshAI, each digestion reaction consisted of 2.0 µl 10X NEBuffer 4, 0.2 µl ac- etylated BSA (10 µg/µl), 0.2 µl restriction enzyme (10 U/µl), and 17.6 µl direct PCR product. Digestion mixture was incubated at 37˚C in an eppendorf thermal cycler for 3 hr. The digestion products were visualized in 1.5% agarose gels in 0.5X TBE, stained with ethidium bro- mide. 3. Results 3.1. Allele-Specific PCR Markers Two primer pairs, Tm2S-f1/Tm2S-r1 and Tm2S-f2/ Tm2S-r2, amplified to produce DNA fragment only for the 14 tomato genotypes containing susceptible allele tm-2 with homozygous (tm-2/tm-2) or heterozygous (Tm-22/tm-2) condition as codominant marker (Table 1), Copyright © 2011 SciRes. AJPS  Molecular Markers for Tm-2 Alleles of Tomato Mosaic Virus Resistance in Tomato185 indicating the two primers can be used as PCR-based markers to select the susceptible allele tm-2 in tomato. The primer pair Tm2R-f1c/Tm2R-r3 only amplified the tomato genotypes containing resistance allele Tm-2. However, the primer pair Tm2R-f1c/Tm2aR-r3 amplified the tomato genotypes with Tm-22 with homozygous or heterozygous condition. The primer pair Tm2R-f1c/ Tm2R-r4 amplified the DNA from tomato genotypes containing either Tm-2 or Tm-22 with homozygous or heterozygous condition, indicating the primer pair can be used to select either Tm-2 or Tm-22 gene (Table 1). 3.2. Allele-Derived CAPS Markers The enzyme PshAI cut the PCR fragment amplified with the primer pair Tm2RS-f3/r3 at the site 537 base into two fragments with the size of 538 bp and 165 bp for the to- matoes containing susceptible allele tm-2; The enzyme HapI cut the PCR fragment into 458 bp and 245 bp among those tomatoe lines carrying either Tm-2 or Tm22 and; BsiHKAI cut the fragment into 358 bp and 353 bp in those containing Tm-2 (Table 1). This indicated that combination of the three markers can be used to identify and distinguish the three alleles at Tm-2 locus in tomato lines. 3.3. Gene-Derived SNP Markers Both SNP Tm2-snp1 and Tm2-snp46 had three types of SNP [C C], [T T], and [C T] showing co-dominant pat- tern (Table 1). The type [T T] in Tm2-snp1 and [C C] in Tm2-snp46 were specific for those tomato genotypes containing susceptible homozygous alleles tm-2/tm-2. On the other hand, the type [C C] in Tm2-snp1 and [T T] in Tm2-snp46 were specific to those tomato genotypes car- rying resistance homozygous alleles Tm-2/Tm-2 or Tm22/Tm22. [C T] in both SNPs was for the three tomato genotypes containing heterozygous Tm22/tm-2. The SNP Tm2-snp53 had three types [A A], [A G], and [G G]. The tomato lines containing homozygous or heterozygous alleles for resistance allele Tm-2 or Tm22 had the SNP Tm2-snp53 type [A A] or [A G], but those carrying ho- mozygous susceptible allele tm-2 had [G G] type in Tm2- snp53 (Table 1). Results indicated that these SNPs can differentiate resistance and susceptible allele at Tm-2 locus. 3.4. Sequence Analysis and Potential SNP Identification The primer pairs Tm2RS-f2/r2 and Tm2RS-f3/r3 pro- duced DNA fragments with the size of 490 bp, and 703 bp, respectively in all 23 tomato genotypes (Table 1). The 13 DNA fragment sequences amplified from the two primer pair were stored at GenBank with the accessions no. from FJ17595 to FJ17607. From the multiple sequence alignment among the eight PCR fragments (FJ17600 to FJ17607) amplified from the primer pair Tm2RS-f3/r3 with the corresponding DNA segments of the AF536201, AF536200, AY742887, and AF536199, 40 SNPs were found (Supplementary Table S2). Among them, 36 SNPs were capable to discriminate the tomato genotypes with the susceptible allele tm-2 and those with the resistance alleles either Tm-2 or Tm-22. Four SNPs, SNP9, SNP30, SNP32, and SNP33 were further capable to differentiate the genotypes carrying Tm-2 and Tm-22. Two SNPs, SNP9 and SNP32 had triple alleles. SNP1 [C/T] was selected as one of the multiplex assay for SNP genotyping in 23 tomato genotypes by Sequenom. From the multiple sequence alignment among the five PCR fragments (FJ17595 to FJ17599) amplified from the primer pair Tm2RS-f2/r2 with the corresponding DNA segments of the AF536201, AF536200, AY742887, and AF536199, eight SNPs were observed (Supplementary Table S3). Among them, six SNPs showed the differ- ences between the tomato lines with susceptible allele tm-2 and resistance alleles including Tm-2 or Tm-22. Two SNPs, SNP42 and SNP45 showed the differences be- tween those carrying Tm-2 and Tm-22. SNP46 [C/T] was also selected as one of the multiplex assay for SNP genotyping in 23 tomato genotypes by Sequenom. Besides the 48 SNPs verified from the re-sequencing of the PCR products amplified the two primer pairs, ten SNPs and one InDel were also postulated among the three Tm-2 sequences AF536201, AF536200, and AF536199 (Supplementary Table S4). Among them, nine SNPs and one InDel showed the difference between the tomato genotypes with susceptible allele tm-2 and resistance alleles including Tm-2 or Tm-22. The SNP SNP53 showed the difference between Tm-2 and Tm-22 and was selected as one of the multiplex assay for SNP genotyping in 23 tomato genotypes by Sequenom. 4. Discussion Three types of molecular marker development for Tm-2 loci were developed in this research. From known Tm-2 gene cloning sequences AF536199 (tm-2), AF536200 (Tm-2), and AF536201 (Tm-22), two gene-specific primer pairs, Tm2RS-f2/r2 and Tm2RS-f3/r3 were de- signed. The PCR products amplified from tomato geno- types contained either resistance allele Tm-2, Tm-22 or susceptible allele tm-2 using the two primer pairs were sequenced. After multiple sequence alignment, 40 SNPs were found and validated from the sequences amplified from Tm2RS-f3/r3 (Table S2) and eight SNPs and one InDel from Tm2RS-f2/r2 (Table S3). Two SNPs, snp1 and snp46 selected from sequences amplified from the two primer pairs plus the SNP snp53 picked up from other location of the sequene (Table S4) were selected c Copyright © 2011 SciRes. AJPS 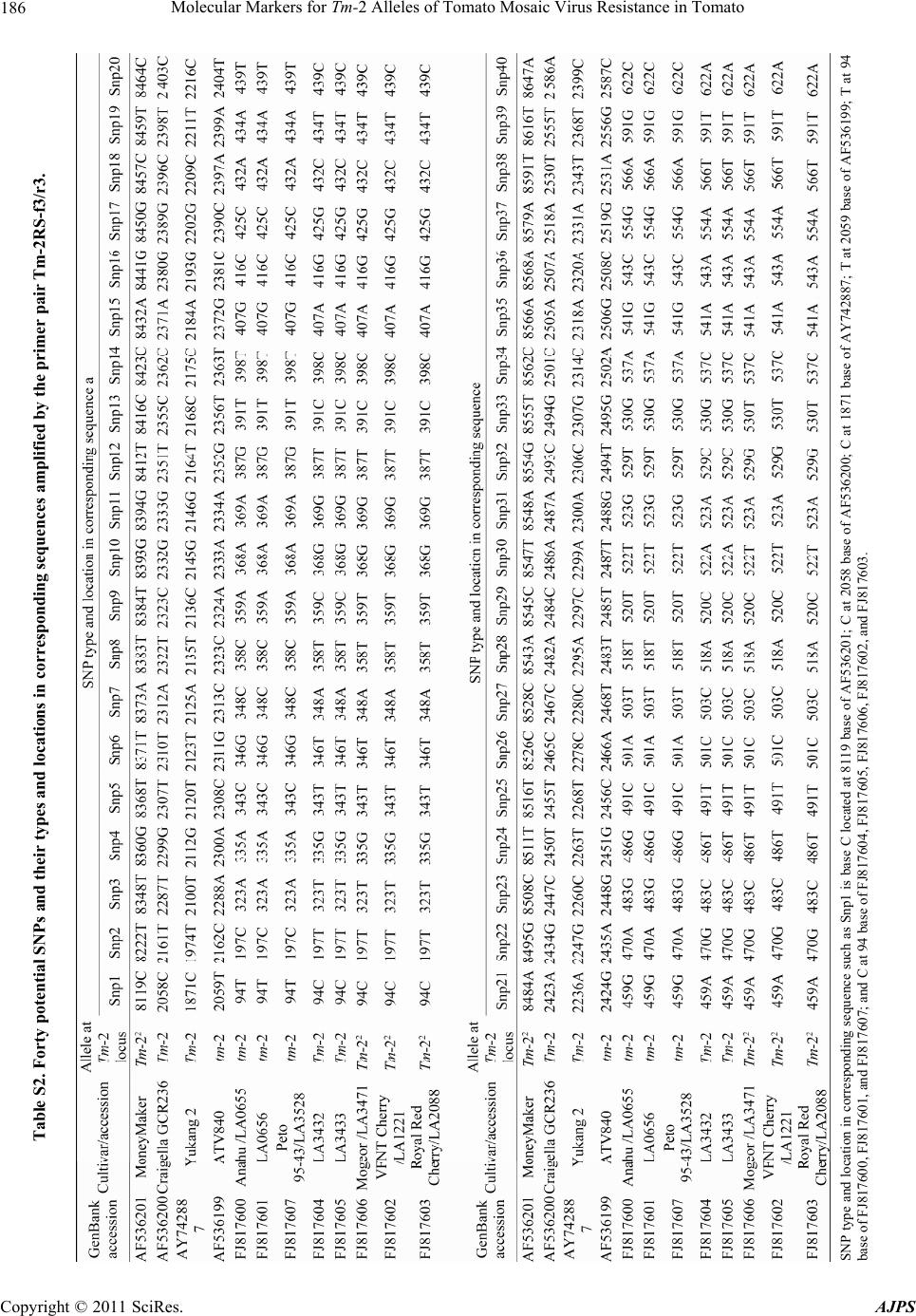 Molecular Markers for Tm-2 Alleles of Tomato Mosaic Virus Resistance in Tomato Copyright © 2011 SciRes. AJPS 186 SNP type and location in corresponding sequence such as Snp1 is base C located at 8119 base of AF536201; C at 2058 base of AF536200; C at 1871 base of AY742887; T at 2059 base of AF536199; T at 94 base of FJ817600, FJ817601, and FJ817607; and C at 94 base of FJ817604, FJ817605, FJ817606, FJ817602, and FJ817603. Table S2. Forty potential SNPs and their types and locations in corresponding sequenc e s amplifie d by the primer pair Tm-2RS-f3/r3.  Molecular Markers for Tm-2 Alleles of Tomato Mosaic Virus Resistance in Tomato187 Table S3. Eight potential SNPs and their types and locations in corresponding sequences amplified by the primer pair Tm-2RS-f2/r2. SNP type and location in corresponding sequence a GenBank accession Cultivar/ accession Allele at Tm-2 locus SNP41 SNP42 SNP43 SNP44SNP45 SNP46 SNP47 SNP48 AF536201 MoneyMaker Tm-22 6962G 7017A 7043G 7085T 7106G 7291T 7454T 7485G AF536200 Craigella GCR236 Tm2 901G 956T 982G 1024T1045T 1230T 1393T 1424G AF536199 ATV840 tm-2 902T 957A 983T 1025C1046G 1231C 1394G1425A AY742887 Yukang 2 Tm2 714G 769T 795G 837T 858T 1043T 1206T 1237G FJ817595 Anahu/LA0655 tm2 34T 89A 115T 157C 178G 363C 526G 557A FJ817596 LA0656 tm2 34T 89A 115T 157C 178G 363C 526G 557A FJ817599 Peto 95-43/LA3528 tm2 34T 89A 115T 157C 178G 363C 526G 557A FJ817597 VFNT Cherry/LA1221 Tm-22 34G 89A 115G 157T 178G 363T 526T 557G FJ817598 LA3433 Tm2 34G 89T 115G 157T 178T 363T 526T 557G aSNP type and location in corresponding sequence such as Snp41 is base G located at 6962 base of AF536201; G at 901 base of AF536200; T at 902 base of AF536199; G at 714 base of AY742887; T at 34 base of FJ817595, FJ817596, and FJ817599; and G at 34 base of FJ817597 and FJ817598. Table S4. Ten potential SNPs and one InDel and their types and locations in corresponding sequences among Tm-2 alleles except above 48 SNPs in Table S1 and S2. SNP type and location in corresponding sequence a GenBank accession Cultivar Allele at Tm-2 locus SNP49 SNP50SNP51SNP52SNP53SNP54SNP55 SNP56 SNP57 SNP58InDel1 AF536199 ATV840 tm-2 73A 109A457G487C500G 612T 704C849G 2672A 2733G89A AF536200 Craigella GCR236 Tm-2 73C 108T456A486G499G611C703T848A 2671G 2732C88-89d AF536201 MoneyMaker Tm-2 2 6134C 6169T 6517A6547G6560A6672C6764T6909A 8732G 8793C88-89d aSNP type and location in corresponding sequence such as Snp49 is base A located at 73 base of AF536199; C at 73 base of AF536200; and C at 6134 base of AF536201. to use SNP genotyping for identification of Tm-2 alleles and verified that they were useful to identify difference alleles, tm-2, Tm-2, and Tm-22 in 23 tomato genotypes by Sequnom technology (Table 1). Meanwhile, allele-spe- cific PCR primers were designed for each allele at Tm-2 locus and five allele-specific PCR-based markers were identified: one for Tm-2, one for Tm-22, one for both Tm-2 and Tm-22, and two for the susceptible allele tm-2 in 23 tomato genotypes (Table 1). The allele-derived CAPS markers were designed from the sequences ampli- fied from primer pair Tm2RS-f3/r3 digested by the three enzymes, BsiHKAI, HpaI, and PshAI, which can identify and distinguish the three alleles, tm-2, Tm- 2, and Tm-22 among 23 tomato genotypes (Table 1). The five allele-specific PCR markers were dominant markers, which can’t distinguish the homozygous and heterozygous genotypes at Tm-2 locus individually, but the combination of these markers can do (Tabl e 1). The three CAPS markers performed the same way as the al- lele-specific PCR-based markers did (Table 1). The two SNP markers, snp1 and snp46 were co-dominant ones and they can distinguish the homozygous and heterozy- gous genotypes at Tm-2 locus individually. However, we did not identify a SNP marker that can distinguish Tm-22 from Tm-2. The SNP53 should distinguish Tm-22 from Tm-2 because AF53200 (Tm-2) has a “G” at 499 base of the sequence but AF536201 (Tm-22) has an “A” base at the corresponding site of the sequence (Table S4). But our SNP genotyping data did not support Tm-2-carrying tomato lines carried a “G” base in the corresponding site, such as Royal Red Cherry (LA2088), CLN2264F (LA4285), and LA3433 contained Tm-2 allele but they had [A A] type for snp53 (Table 1). Besides the snp53, SNP9, SNP30, SNP32, SNP33, SNP42, and SNP45 also showed difference in their SNP types for Tm-22 from Tm-2 (Tables S2 and S3) and maybe they can be used in SNP genotyping for distinguishing Tm-22 from Tm-2. But, they need to be validated in further research. How- ever, the snp9 was validated to be used to distinguish Tm-22 from Tm-2 by the CAPS marker through digestion Copyright © 2011 SciRes. AJPS  Molecular Markers for Tm-2 Alleles of Tomato Mosaic Virus Resistance in Tomato 188 the PCR products amplified from Tm-2RS-f3/r3 by en- zyme EsiHKAI (Tables 1 and 4). Only those tomato lines contained Tm-2 allele were digested by EsiHKAI for the DNA segments amplified from T2RS-f3/r3 be- cause Tm-2 carrying tomato genotypes had a “C” base at 359 base of the sequences but others had an “A” base at 359. The SNP32 in supplementary Table S2 and SNP41 in Table S3 were used as a tetra-primer ARMS-PCR assays by Arens et al. [15] to produce different PCR fragment sizes for detection of three alleles tm-2, Tm-2, and Tm-22 in tomato genotypes. Three types of molecular markers, allele-specific PCR, allele-derived CAPS, and allele-derived SNP were iden- tified for the Tm-2 locus for ToMV resistance (Table 1). Lanfermeijer et al. in 2005 [14] reported three CAPS markers which can distinguish the three alleles, tm-2, Tm-2, and Tm-22 in tomato genotypes and the enzyme HapI was also reported that cut the same site of the se- quences from resistant cultivars, Craigella GCR236 (Tm-2), Craigella GCR267 (Tm22), and ATV847 (Tm22), but not in the susceptible cultivars, GCR26 (tm-2) and ATV840 (tm-2). However, in their research, the primer pair, PrRuG151 (GAGTTCTTCCGTTCAAATCCTAA- GCTTGAGAAG)/PrRuG086 (CTACTACACTCACGT- TGCTGTGATGCAC) was used to amplify DNA frag- ments among the five cultivars. The primer PrRuG151 was located at 7824 to 7856 base and PrRuG086 at 8908 to 8881 base of the AF536201 sequence and the primer pair produced a 1085 bp DNA fragment. The enzyme HpaI cut the DNA fragments into two with 660 bp and 424 bp fregments for Craigella GCR236 (Tm-2), Craig- ella GCR267 (Tm-22), and ATV847 (Tm-22), but not for GCR26 (tm-2) and ATV840 (tm-2). In our research, the primer pair Tm-2RS-f3/r3 was used and produced a 703 bp DNA fragment of PCR products. The fragment was digested with HpaI into two fragments with 538 bp and 165 bp for tomato genotypes carrying Tm-2 and Tm-22 but not for tm-2 (Table 1). HpaI can cut the sequence “GTT AAC” in DNA fragments amplified from prmier pair of either PrRuG151/PrRuG086 or Tm-2RS-f3/r3. Because the resistant genotypes contained Tm-2 and Tm-22 had the sequence “GTT AAC”, but the susceptible genotypes carrying tm-2 had the “GTT GAC” in corre- sponding site of the sequence. The SNP snp21 in Table S2 was the same site of HpaI cutting site. These results indicated that the CAPS marker digested by HpaI and the SNP marker SNP21 [A/G] can be used as a markers to identify and distinguish the susceptible allele tm-2 from the resistance allele Tm-2 or Tm-22. After all, these al- lele-specific PCR-based, CAPS, and gene-derived SNP markers for Tm-2 locus will provide breeders to select the allele tm-2, Tm - 2, and Tm-22 of ToMV resistance in advancing the MAS for tomato breeding. 5. Acknowledgements We thank G. Moriarty, M. Glos and M. Jahn (Cornell University) and C. R. Lawn (Restoring our Seed) for providing tomato line NY07-461 and NY07-464 that was developed with support from USDA-SARE proposal LNE04-204 and LNE02-160. We also thank Roger Chetelat and Sheh May Tam at Tomato Genetics Resource Center, Department of Plant Sciences, University of Cali- fornia for providing 16 tomato accessions. We are grate- ful to Xianming Chen at USDA-ARS and Washington State University for review of the manuscript. REFERENCES [1] M. R. Foolad, “Genome Mapping and Molecular Breed- ing of Tomato,” International Journal of Plant Genomics, Vol. 2007, 2007, Article ID 64358. doi:10.1155/2007/64358 [2] T. J. Hall, “Resistance at the Tm-2 Locus in the Tomato to Tomato Mosaic Virus,” Euphytica, Vol. 29, No. 1, 1989, pp. 189-197. doi:10.1007/BF00037266 [3] F. C. Lanfermeijer, J. Dijkhuis, M. J. G. Sturre, P. de Haan and J. Hille, “Cloning and Characterization of the Durable Tomato Mosaic Virus Resistance Gene Tm-2 from Lycopersicon Esculentum,” Plant Molecular Biol- ogy, Vol. 52, No. 5, 2003, pp. 1037-1049. 2 doi:10.1023/A:1025434519282 [4] H. Levesque, F. Vedel, C. Mathieu and A. G. L. de Courcel, “Identification of a Short Rdna Spacer Sequence Highly Specific of a Tomato Line Containing Tm-1 Gene Introgressed from Lycopersicon Hirsutum,” Theoretical and Applied Genetics, Vol. 80, No. 5, 1990, pp. 602-608. doi:10.1007/BF00224218 [5] J. Pelham, “Resistance in Tomato to Tobacco Mosaic Virus,” Euphytica, Vol. 15, 1966, pp. 258-267. doi:10.1007/BF00022331 [6] M. R. Foolad and A. Sharma, “Molecular Markers as Selection Tools in Tomato Breeding,” ISHS Acta Hor- ticulturae, Vol. 695, 2005, pp. 225-240. [7] T. Ohmori, M. Murata and F. Motoyoshi, “Molecular Characterization of RAPD and SCAR Markers Linked to the Tm-1 Locus in Tomato,” Theoretical and Applied Genetics, Vol. 92, 1996, pp. 151-156. [8] S. D. Tanksley, M. W. Ganal, J. P. Prince, M. C. de Vicente, M. W. Bonierbale, P. Broun, T. M. Fulton, J. J. Giovannoni, S. Grandillo and G. B. Martin, “High Density Molecular Linkage Maps of the Tomato and Potato Ge- nomes,” Genetics, Vol. 132, No. 4, 1992, pp. 1141-1160. [9] D. J. Vakalounakis, H. Laterrot, A. Moretti, E. K. Ligoxi- gakis and K. Smardas, “Linkage between Fr1 (Fusar ium Oxysporum f sp Radicis-Lycope rsic i Resistance) and Tm-2 (Tobacco Mosaic Virus Resistance-2) Loci in Tomato (Ly- copersicon Esculentum),” Annals of Applied Biology, Vol. 130, 1997, pp. 319-323. doi:10.1111/j.1744-7348.1997.tb06835.x [10] T. Ohmori, M. Murata and F. Motoyoshi, “Identification Copyright © 2011 SciRes. AJPS  Molecular Markers for Tm-2 Alleles of Tomato Mosaic Virus Resistance in Tomato Copyright © 2011 SciRes. AJPS 189 of RAPD Markers Linked to the Tm-2 Locus in Tomato,” Theoretical and Applied Genetics, Vol. 90, 1995, pp. 307-311. doi:10.1007/BF00221969 [11] F. Motoyoshi, T. Ohmori and M. Murata, “Molecular Characterization of Heterochromatic Regions around the Tm-2 Locus in Chromosome 9 of Tomato,” Symposium of the Society for Experimental Biology, Vol. 50, 1996, pp. 65-70. [12] Sobir, T. Ohmori, M. Murata and F. Motoyoshi, “Molecu- lar Characterization of the SCAR Markers Tightly Linked to the Tm-2 Locus of the Genus Lycope rsi con,” Theoretical and Applied Genetics, Vol. 101, No. 1-2, 2000, pp. 64-69. doi:10.1007/s001220051450 [13] E. Dax, O. Livneh, E. Aliskevicius, N. Kedar, N. Gavish, J. Milo, F. Geffen, A. Blumenthal, H. D. Rabinowich and I. Sela, “A SCAR Marker Linked to the Tomv Resistance Gene, Tm2, in Tomato,” Euphytica, Vol. 101, No. 1, May 1998, pp. 73-77. 2 doi:10.1023/A:1018307326636 [14] F. C. Lanfermeijer, J. Warmink and J. Hille, “The Prod- ucts of the Broken Tm-2 and the Durable Tm-2 Resis- tance Genes from Tomato Differ in Four Amino Acids,” Journal of Experimental Botany, Vol. 56, No. 421, No- vember 2005, pp. 2925-2933. 2 doi:10.1093/jxb/eri288 [15] P. Arens, C. Mansilla, D. Deinum, L. Cavellini, A. Moretti, S. Rolland, H. van der Schoot, D. Calvache, F. Ponz, C. Collonnier, R. Mathis, D. Smilde, C. Caranta and B. Vosman, “Development and Evaluation of Robust Molecular Markers Linked to Disease Resistance in Tomato for Distinctness, Uniformity and Stability Testing,” Theoretical and Applied Genetics, Vol. 120, No. 3, 2010, pp. 655-664. doi:10.1007/s00122-009-1183-2 [16] I. Y. Choi, D. L. Hyten, L. K. Matukumalli, Q. Song, J. M. Chaky, C. V. Quigley, K. Chase, K. G. Lark, R. S. Reiter, M. Yoon, E. Hwang, S. Yi, N. D. Young, R. C. Shoemaker, C. P. van Tassell, J. E. Specht and P. B. Cre- gan, “A Soybean Transcript Map: Gene Distribution, Haplotype and Single Nucleotide Polymorphism Analy- sis,” Genetics, Vol. 176, No. 1, May 2007, pp. 685-696. doi:10.1534/genetics.107.070821 [17] S. Giancola, H. I. McKhann, A. Berard, C. Camilleri, S. Durand, P. Libeau, F. Roux, X. Rebound, I. G. Gut and D. Brunel, “Utilization of Three High-Throughput SNP Genotyping Methods, the GOOD Assay, Amplifluor and Taqman, in Diploid and Polyploidy Plants,” Theoretical and Applied Genetics, Vol. 112, No. 6, 2006, pp. 1115- 1124. doi:10.1007/s00122-006-0213-6 [18] J. A. Labate and A. M. Baldo, “Tomato SNP Discovery by EST Mining and Resequencing,” Molecular Breeding, Vol. 16, No. 4, November 2005, pp. 343-349. doi:10.1007/s11032-005-1911-5 [19] J. M. Jimenez-Gomez and J. N. Maloof, “Sequence Di- versity in Three Tomato Species: Snps, Markers, and Molecular Evolution,” BMC Plant Biology, Vol. 9, July 2009, p. 85. doi:10.1186/1471-2229-9-85 [20] J. A. Rafalski, “Application of Single Nucleotide Poly- morphisms in Crop Genetics,” Current Opinion in Plant Biology, Vol. 5, No. 2, April 2002, pp. 94-100. doi:10.1016/S1369-5266(02)00240-6 [21] A. Shi, R. Vierling, R. Grazzini, P. Chen, H. Caton and Y. Weng, “Development of Single Nucleotide Polymor- phism Markers for Selection of Ve Gene of Tomato Ver- ticillium Wilt Resistance,” International Research Jour- nal of Plant Science, Vol. 1, No. 2, August 2010, pp. 034-042. [22] W. Yang, S. A. Miller, J. W. Scott, J. B. Jones and D. M. Francis, “Ming Tomato Genome Sequence Databases for Molecular Markers: Application to Bacterial Resistance and Marker Assisted Selection,” Acta Hort (ISHS), Vol. 695, 2005, pp. 241-250. [23] W. Yang, X. Bai, E. Kabelka, C. Eaton, S. Kamoun, E. van-der-Knaap and D. Francis, “Discovery of Single Nu- cleotide Polymorphisms in Lycopersicon Esculentum by Computer Aided Analysis of Expressed Sequence Tags,” Molecular Breeding, Vol. 14, No. 1, 2004, pp. 21-34. doi:10.1023/B:MOLB.0000037992.03731.a5 [24] N. Acciarri, G. L. Rotino, G. Tamietti, D. Valentino, S. Voltattorni and E. Sabatini, “Molecular Markers for Ve1 and Ve 2 Verticillium Resistance Genes from Italian To- mato Germplasm,” Plant Breeding, Vol. 126, No. 6, De- cember 2007, pp. 617-621. doi:10.1111/j.1439-0523.2007.01398.x [25] J. J. Doyle and J. L. Doyle, “Isolation of Plant DNA from Fresh Tissue,” Focus, Vol. 12, No. 1, 1990, pp. 13-15. [26] P. J. Bradbury, A. Zhang, D. E. Kroon, T. C. Casstevens, Y. Ram-Doss and E. S. Buckler, “TASSEL Software for Association Mapping of Complex Traits in Diverse Sam- ples,” Bioinformatics, Vol. 23, No. 19, June 2007, pp. 2633-2635. doi:10.1093/bioinformatics/btm308
|