 American Journal of Plant Sciences, 2011, 2, 111-119 doi:10.4236/ajps.2011.22013 Published Online June 2011 (http://www.SciRP.org/journal/ajps) Copyright © 2011 SciRes. AJPS 111 Difference of Curcumin Content in Curcuma longa L. (Zingiberaceae) Caused by Hybridization with Other Curcuma Species Hiroshi Hayakawa1,2, Yukio Minaniya1, Katsura Ito1, Yoshinori Yamamoto1, Tatsuya Fukuda1 1Faculty of Agriculture, Kochi University, Nankoku, Japan; 2United Graduate School of Agricultural Sciences, Ehime University, Nankoku, Japan. Email: tfukuda@kochi-u.ac.jp Received November 29th, 2010; revised January 24th, 2011; accepted January 30th, 2011. ABSTRACT Curcumin, which is traditionally known to have effects on various types of diseases in humans, is found in Curcuma longa L. Previous reports have indica ted that the curcumin co ntent varies b etween the different lines of this sp ecies. To clarify the differences in the amounts of curcumin between the lines, we investigated the outcomes of cultivation ex- periments with the hybridization or introgression between C. longa and other Curcuma species using the matK gene of chloroplast DNA (cpDNA) and the external transcribed spacer (ETS) of nuclear DNA (nrDNA). The results show that there is heterogeneity of the ETS and incongruence between the matK and the ETS phylogenetic trees, suggesting that hybridization and introgression had taken place in the diversification of the various lines of C. longa. Moreover, al- though all of the lines had the same cpDNA haplotype of C. longa, the lines of homogeneous C. longa had a high con- tent of curcumin, whereas the lines created by hybridization and introgression with other Curcuma species had a me- dium or low level. These results suggest that the difference of cu rcumin conten t among the various lines of C. longa was caused by hybridization and introgression with other Curcuma species. Keywords: Curcuma longa, Curcumin, Hybridization, Introgression, Molecular Analysis, Nuclear DNA 1. Introduction Many chemicals in plants are potential drugs for humans and natural products from plants are found in many therapeutic formulations. Moreover, conscious efforts to search for desirable traits in plants have been underway for the past century, and in recent decades species with desirable traits have come to be regarded as important biological resources in need of conservation [1]. Curcuma longa L., which belongs to the ginger family, Zingiberaceae, is a perennial widely used as a spice, a colorant and also as a major ingredient of curry powder [2]. This species has a long history of use as a traditional medicine in China and India [3], reflecting it’s diverse and beneficial health effects. In addition, the curcuma species contains phenolic compounds found in the plant’s rhizomes. Traditionally, curcumin is well-known to have therapeutic effects on a variety of human diseases, and the cancer preventive activity of curcumin is being inten- sively studied all over the world. Experiments in animal models indicate that it is a preventive agent against vari- ous types of cancer [4]. Specifically, curcumin inhibits the cell growth of various cancer cell lines, induces apoptosis of cancer cells [5-7], and was effective on the cell-cycle regulation of cancer cells [8]. According to previous study there is a difference in the curcumin content among individuals of C. longa [9,10], however, it remains unclear why the curcumin content is different. To identify the lines with high curcumin con- tent, Hayakawa et al. [10,11] developed a molecular marker. Recent molecular phylogenetic study using chloroplast DNA (cpDNA) sequences indicated that C. longa has some closely related species. Also, a large number of previous studies of species within the genus Drosophila had illuminated the relationship between genetic distance and reproductive isolation [12,13]. One possibility is that the difference in curcumin content within C. longa was caused by including hybrids be- tween C. longa and other Curcuma species. It is very difficult to detect polymorphisms of morphology of the rhizomes and cpDNA sequences within this species. Al- 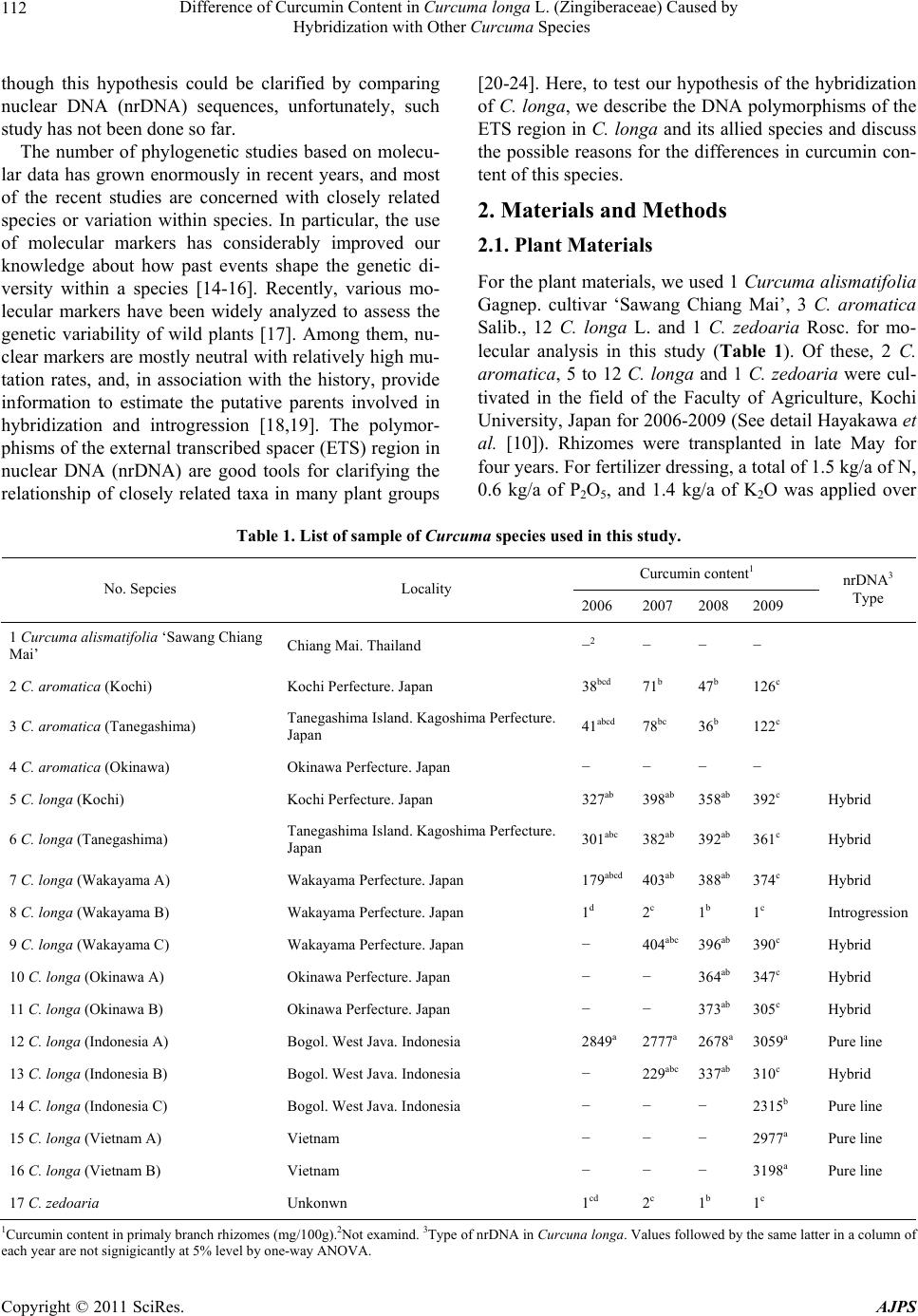 Difference of Curcumin Content in Curcuma longa L. (Zingiberaceae) Caused by 112 Hybridization with Other Curcuma Species though this hypothesis could be clarified by comparing nuclear DNA (nrDNA) sequences, unfortunately, such study has not been done so far. The number of phylogenetic studies based on molecu- lar data has grown enormously in recent years, and most of the recent studies are concerned with closely related species or variation within species. In particular, the use of molecular markers has considerably improved our knowledge about how past events shape the genetic di- versity within a species [14-16]. Recently, various mo- lecular markers have been widely analyzed to assess the genetic variability of wild plants [17]. Among them, nu- clear markers are mostly neutral with relatively high mu- tation rates, and, in association with the history, provide information to estimate the putative parents involved in hybridization and introgression [18,19]. The polymor- phisms of the external transcribed spacer (ETS) region in nuclear DNA (nrDNA) are good tools for clarifying the relationship of closely related taxa in many plant groups [20-24]. Here, to test our hypothesis of the hybridization of C. longa, we describe the DNA polymorphisms of the ETS region in C. longa and its allied species and discuss the possible reasons for the differences in curcumin con- tent of this species. 2. Materials and Methods 2.1. Plant Materials For the plant materials, we used 1 Curcuma alismatifo lia Gagnep. cultivar ‘Sawang Chiang Mai’, 3 C. aromatica Salib., 12 C. longa L. and 1 C. zedoaria Rosc. for mo- lecular analysis in this study (Table 1). Of these, 2 C. aromatica, 5 to 12 C. longa and 1 C. zedoaria were cul- tivated in the field of the Faculty of Agriculture, Kochi University, Japan for 2006-2009 (See detail Hayakawa et al. [10]). Rhizomes were transplanted in late May for four years. For fertilizer dressing, a total of 1.5 kg/a of N, 0.6 kg/a of P2O5, and 1.4 kg/a of K2O was applied over Table 1. List of sample of Curcuma species used in this study. Curcumin content1 No. Sepcies Locality 2006 2007 2008 2009 nrDNA3 Type 1 Curcuma alismatifolia ‘Sawang Chiang Mai’ Chiang Mai. Thailand −2 − − − 2 C. aromatica (Kochi) Kochi Perfecture. Japan 38bcd 71b 47b 126c 3 C. aromatica (Tanegashima) Tanegashima Island. Kagoshima Perfecture. Japan 41abcd 78bc 36b 122c 4 C. aromatica (Okinawa) Okinawa Perfecture. Japan − − − − 5 C. longa (Kochi) Kochi Perfecture. Japan 327ab 398ab 358ab 392c Hybrid 6 C. longa (Tanegashima) Tanegashima Island. Kagoshima Perfecture. Japan 301abc 382ab 392ab 361c Hybrid 7 C. longa (Wakayama A) Wakayama Perfecture. Japan 179abcd 403ab 388ab 374c Hybrid 8 C. longa (Wakayama B) Wakayama Perfecture. Japan 1d 2 c 1 b 1 c Introgression 9 C. longa (Wakayama C) Wakayama Perfecture. Japan − 404abc 396ab 390c Hybrid 10 C. longa (Okinawa A) Okinawa Perfecture. Japan − − 364ab 347c Hybrid 11 C. longa (Okinawa B) Okinawa Perfecture. Japan − − 373ab 305c Hybrid 12 C. longa (Indonesia A) Bogol. West Java. Indonesia 2849a 2777a2678a 3059a Pure line 13 C. longa (Indonesia B) Bogol. West Java. Indonesia − 229abc 337ab 310c Hybrid 14 C. longa (Indonesia C) Bogol. West Java. Indonesia − − − 2315b Pure line 15 C. longa (Vietnam A) Vietnam − − − 2977a Pure line 16 C. longa (Vietnam B) Vietnam − − − 3198a Pure line 17 C. zedoaria Unkonwn 1cd 2 c 1 b 1 c 1Curcumin content in primaly branch rhizomes (mg/100g).2Not examind. 3Type of nrDNA in Curcuna longa. Values followed by the same latter in a column of ach year are not signigicantly at 5% level by one-way ANOVA. e Copyright © 2011 SciRes. AJPS  Difference of Curcumin Content in Curcuma longa L. (Zingiberaceae) Caused by 113 Hybridization with Other Curcuma Species four years. In addition, 200 kg/a of compost fertilizer, 15 kg/a of magnesia lime, and 30 kg/a of chicken droppings were applied. The experimental plots were arranged in a randomized complete design with two replicates, which formed three rows. Due to a lack of seed rhizomes, some lines of C. longa were examined with/without replicate using two or three rows. Samples were harvested in early December for four years. The total curcumin content (mg/100g) of primary branch rhizomes was measured by using High Performance Liquid Chromatography (HPLC), according to the method described by Sato et al. [25]. 2.2. Molecular Analyses For the molecular analyses, total DNA was isolated from fresh root using a Plant Genomic DNA Mini Kit (VIO- GENE, Sunnyvale, USA), according to the manufac- turer’s protocol. We amplified the maturase K (matK) gene from cpDNA and the external transcribed spacer (ETS) region of 18S-26S rDNA from nrDNA with prim- ers designed by Johnson and Soltis [26] and Starr et al. [27], respectively. The isolated DNA was amplified by PCR in a 50 µl reaction solution containing approxi- mately 50 ng total DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM of each dNTP, 1.25 units Taq DNA polymerase (TaKaRa) and 0.5 µM of each primer pair. We used the following thermal cycle profile for amplification by the PCR Thermal Cycler Dice (TaKaRa): 1 min at 94˚C, 2 min at 48˚C, and 2 min at 72˚C for 45 cycles, followed by 15 min of final exten- sion at 72˚C. After amplification, the PCR products of the matK and ETS region were subjected to electropho- resis in 1% low-melting-temperature agarose gels to re- move by-products and purify amplified products. We sequenced the purified PCR products using a BigDye Terminator ver. 3.1 (Applied BioSystems) and ABI Prism 3100 Genetic Analyzer (Applied BioSystems) according to the manufacturer’s instructions. For sequencing, we used the same primers as those used for amplification. To construct a phylogenetic tree based on the matK sequences of Curcuma and its allied species and the ETS sequences of Curcuma species, the amplified regions were aligned using ClustalW [28] and were improved manually using MEGA 4 [29]. Phylogenetic relationships were analyzed using the neighbor-joining (NJ) method with PAUP* 4.08b [30]. The NJ analyses were per- formed using MEGA 4 with Kimura’s two-parameter model. For the NJ analyses, bootstrapping with 1000 pseudo-replicates was chosen to examine the robustness of the clades and their phylogenetic relationships. The matK sequences were collected from DDBJ/EMBL/ GenBank International DNA databases (Table 2). For the ETS region, because C. longa with a medium curcumin content could not determine its sequence caused by putative heterozygosity, we carried out PCR- RFLP analysis after checking the sequencing results and alignments. The result of the alignments indicated that an autapomorphic character of the nrDNA was the restric- tion site Hinf I. After designating the restriction sites, the amplified products were digested by Hinf I at 37˚C for more than an hour. The digested DNAs were separated on 1.0% agarose gel and the size of each band was de- termined. 3. Results 3.1. Curcumin Content The gradient of curcumin content between species de- creased as follows; South Asian C. longa > domestic C. longa and C. longa (Indonesia B) > C. aromatica > C. zedoaria and C. longa (Wakayama B) (Table 1) [10] and the level of curcumin content was divided into three groups; high, medium and low. 3.2. Molecular Analyses To construct the molecular phylogenetic tree of Curcuma and its allied species, we determined the sequences of the matK gene of Curcuma cpDNA and seven outgroup taxa (Table 2). The lengths of the matK gene of the Curcuma species varied from 1539 bp (C. alismatifolia ‘Sawang Chiang Mai’ and C. long a (Wakayama B)) to 1554 bp (C. thorelii). The result of the phylogenetic analysis of matK indicated that C. longa had a conserved sequence in this species and was closely related to C. aromatica and C. zedoaria, whereas C. alismatifolia ‘Sawang Chiang Mai’ was located in the basal position of the phylogenetic tree and the sister to C. thorelii with a high boot strap value (Figure 1). In addition, we sequenced the ETS region of nrDNA to detect polymorphisms among Curcuma species. The lengths of the ETSs of Curc uma species were 514 bp (C. alismatifolia) to 517 bp (C. aromatica). The sequences have been deposited into the DDBJ/EMBL/GenBank International DNA databases (C. alismatifo lia , AB588183; C. aromatica: AB588181, C. longa: AB588182, AB588185 and AB588186, C. zedoaria: AB588184). The results of the phylogenetic analyses of the ETSs indi- cated that C. longa and closely related species were di- vided into two monophyletic groups: clade 1 and clade 2 (Figure 2). Clade 1 consisted of all individuals of C. longa and clade 2 consisted of C. longa, C. aromatica and C. zedoaria. Although all homogeneous C. longa with its high curcumin content appeared in clade 1, C. longa (Wakayama B) with its low curcumin content was loc ted in clade 2 (Figure 2, Table 1). This suggested a Copyright © 2011 SciRes. AJPS 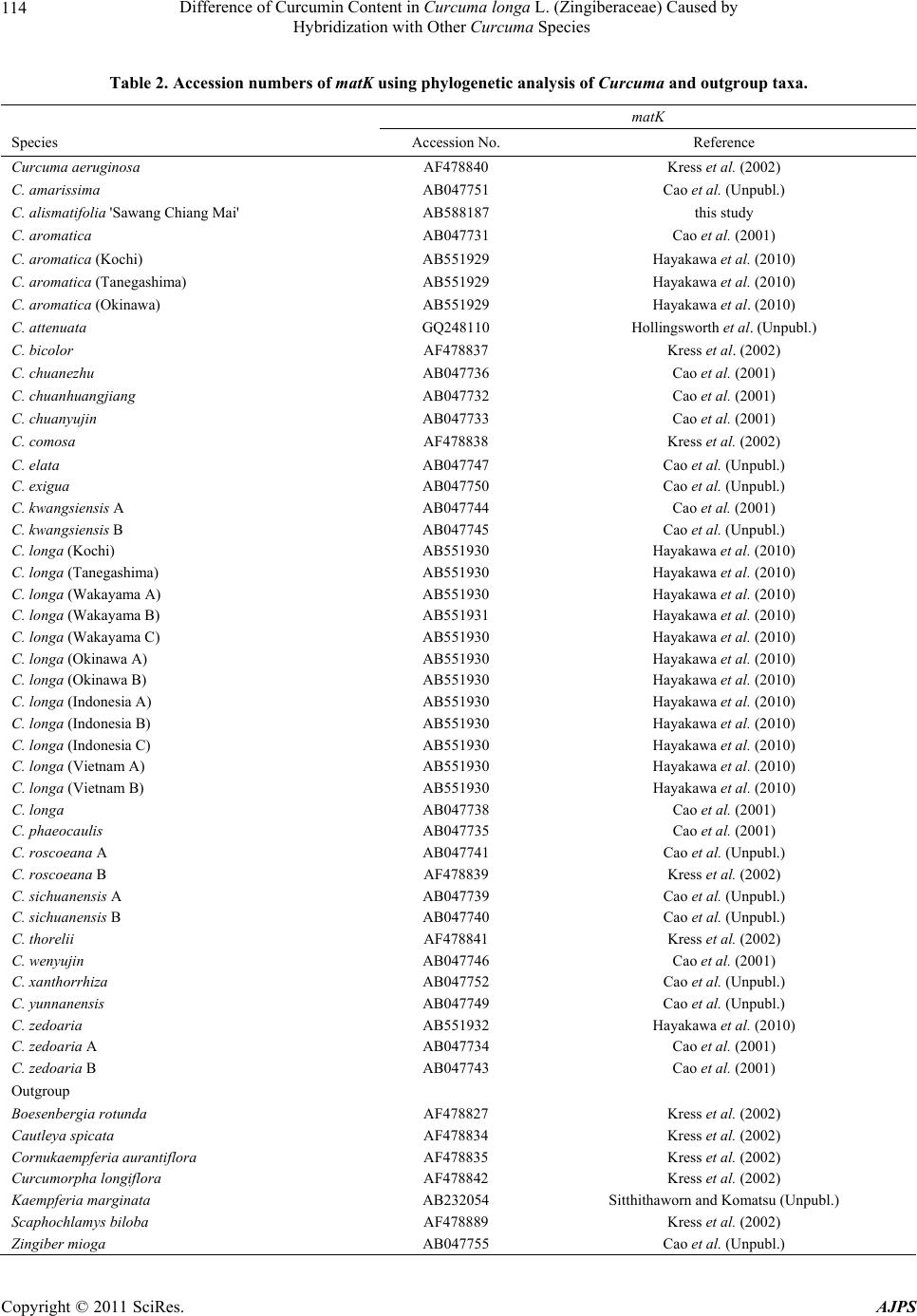 Difference of Curcumin Content in Curcuma longa L. (Zingiberaceae) Caused by 114 Hybridization with Other Curcuma Species Table 2. Accession numbers of matK using phylogenetic analysis of Curcuma and outgroup taxa. matK Species Accession No. Reference Curcuma aeruginosa AF478840 Kress et al. (2002) C. amarissima AB047751 Cao et al. (Unpubl.) C. alismatifolia 'Sawang Chiang Mai' AB588187 this study C. aromatica AB047731 Cao et al. (2001) C. aromatica (Kochi) AB551929 Hayakawa et al. (2010) C. aromatica (Tanegashima) AB551929 Hayakawa et al. (2010) C. aromatica (Okinawa) AB551929 Hayakawa et al. (2010) C. attenuata GQ248110 Hollingsworth et al. (Unpubl.) C. bicolor AF478837 Kress et al. (2002) C. chuanezhu AB047736 Cao et al. (2001) C. chuanhuangjiang AB047732 Cao et al. (2001) C. chuanyujin AB047733 Cao et al. (2001) C. comosa AF478838 Kress et al. (2002) C. elata AB047747 Cao et al. (Unpubl.) C. exigua AB047750 Cao et al. (Unpubl.) C. kwangsiensis A AB047744 Cao et al. (2001) C. kwangsiensis B AB047745 Cao et al. (Unpubl.) C. longa (Kochi) AB551930 Hayakawa et al. (2010) C. longa (Tanegashima) AB551930 Hayakawa et al. (2010) C. longa (Wakayama A) AB551930 Hayakawa et al. (2010) C. longa (Wakayama B) AB551931 Hayakawa et al. (2010) C. longa (Wakayama C) AB551930 Hayakawa et al. (2010) C. longa (Okinawa A) AB551930 Hayakawa et al. (2010) C. longa (Okinawa B) AB551930 Hayakawa et al. (2010) C. longa (Indonesia A) AB551930 Hayakawa et al. (2010) C. longa (Indonesia B) AB551930 Hayakawa et al. (2010) C. longa (Indonesia C) AB551930 Hayakawa et al. (2010) C. longa (Vietnam A) AB551930 Hayakawa et al. (2010) C. longa (Vietnam B) AB551930 Hayakawa et al. (2010) C. longa AB047738 Cao et al. (2001) C. phaeocaulis AB047735 Cao et al. (2001) C. roscoeana A AB047741 Cao et al. (Unpubl.) C. roscoeana B AF478839 Kress et al. (2002) C. sichuanensis A AB047739 Cao et al. (Unpubl.) C. sichuanensis B AB047740 Cao et al. (Unpubl.) C. thorelii AF478841 Kress et al. (2002) C. wenyujin AB047746 Cao et al. (2001) C. xanthorrhiza AB047752 Cao et al. (Unpubl.) C. yunnanensis AB047749 Cao et al. (Unpubl.) C. zedoaria AB551932 Hayakawa et al. (2010) C. zedoaria A AB047734 Cao et al. (2001) C. zedoaria B AB047743 Cao et al. (2001) Outgroup Boesenbergia rotunda AF478827 Kress et al. (2002) Cautleya spicata AF478834 Kress et al. (2002) Cornukaempferia auran tiflora AF478835 Kress et al. (2002) Curcumorpha longiflora AF478842 Kress et al. (2002) Kaempferia marginata AB232054 Sitthithaworn and Komatsu (Unpubl.) Scaphochlamys biloba AF478889 Kress et al. (2002) Zingiber mioga AB047755 Cao et al. (Unpubl.) Copyright © 2011 SciRes. AJPS 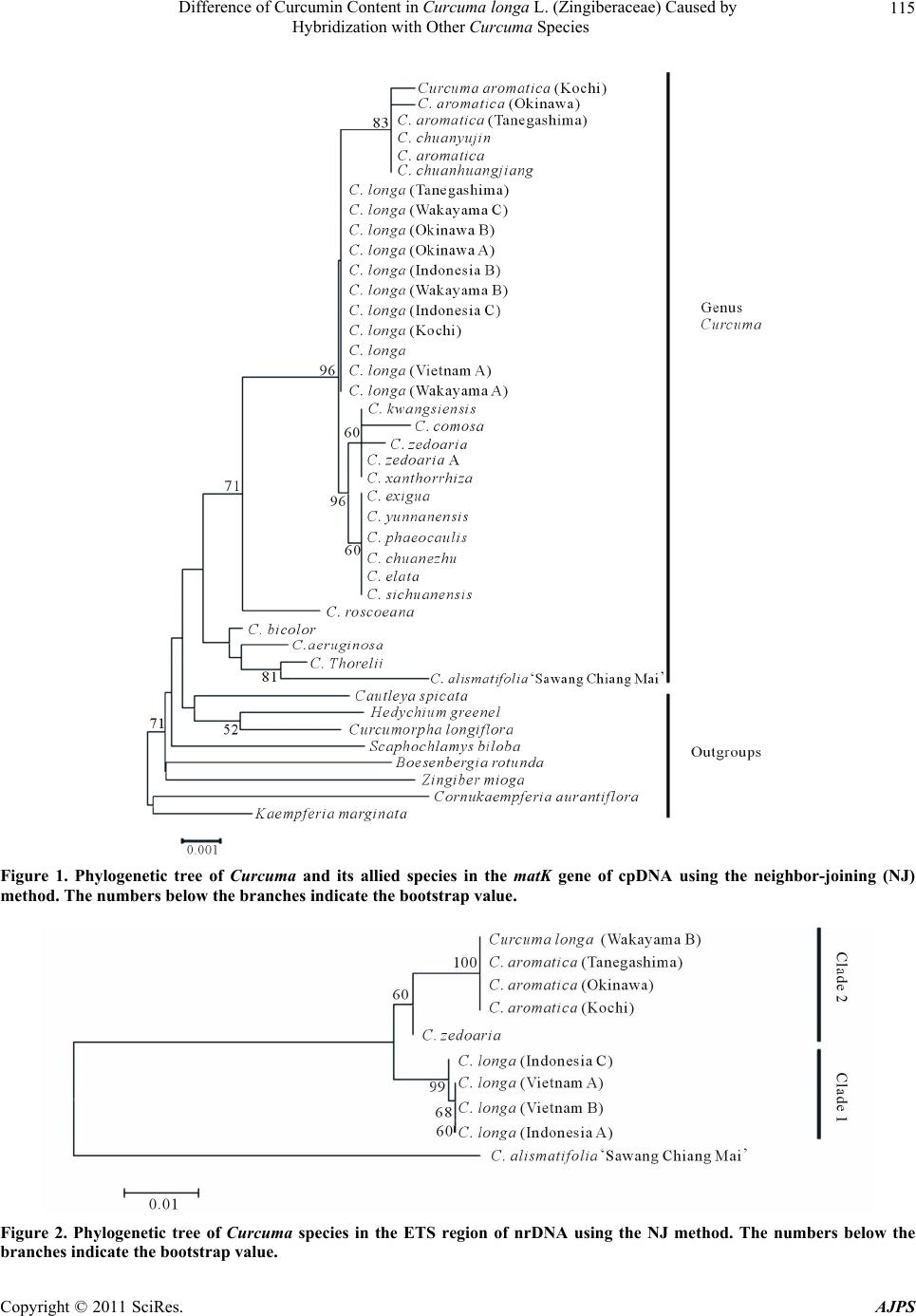 Difference of Curcumin Content in Curcuma longa L. (Zingiberaceae) Caused by 115 Hybridization with Other Curcuma Species Figure 1. Phylogenetic tree of Curcuma and its allied species in the matK gene of cpDNA using the neighbor-joining (NJ) method. The numbers below the branc he s indic a te the bootstrap value . Figure 2. Phylogenetic tree of Curcuma species in the ETS region of nrDNA using the NJ method. The numbers below the branches indicate the bootstrap value. Copyright © 2011 SciRes. AJPS  Difference of Curcumin Content in Curcuma longa L. (Zingiberaceae) Caused by Hybridization with Other Curcuma Species Copyright © 2011 SciRes. AJPS 116 4. Discussion that C. longa (Wakayama B) was introgressive with clade 2 because it had the cpDNA haplotype of C. longa. However, some individuals could not be sequenced in the ETS region because of the putative heterogeneity of C. longa and other Curcuma species. To detect their het- erogeneity, we conducted a PCR-RFLP analysis because restriction of the site of Hinf I to distinguish C. longa with other Curcuma species was in the ETS region (Fig- ure 3). The result was that the digestion pattern of all samples of homogeneous C. longa and C. aromatica showed the expected patterns, and heterogeneous C. longa showed the combined patterns of homogeneous C. longa and C. aromatica (Figure 4). Moreover, C. longa (Wakayama B) showed same band pattern as C. aro- matica. We therefore confirmed that the medium and low curcumin contents of C. longa were hybrid and intro- gressive between C. longa and other Curcuma species on clade 2 (Figure 4, Table 1). In general, hybrids typically display a mosaic of parental and intermediate morphological characters, although extreme and novel characters appear quite often. A spe- cies with morphological characteristics intermediate be- tween two recognized species has always been consid- ered to be a hybrid [31]. However, morphological char- acteristics alone, such as the rhizomes of the Curcuma species, are of limited value when identifying natural hybrids, but molecular studies have greatly enhanced our knowledge in this field [32]. Interspecific hybrids are most commonly identified by the heterogeneity of nrDNA and the incongruences between cpDNA and nrDNA phy- logenies that may indicate different parental contributions to the hybrid genome [33,34]. In particular, incongruence between cpDNA and nrDNA phylogenies is very likely the result of interspecific gene flow and subsequent C. longa C. aromatica C. longa C. aromatica C. longa C. aromatica C. longa C. aromatica C. longa C. aromatica C. longa C. aromatica C. longa C. aromatica C. longa C. aromatica C. longa C. aromatica C. longa C. aromatica Figure 3. Expected restriction sites of Hinf I for molecular characteristics of ETS regions by PCR-RFLP. H: restriction site.  Difference of Curcumin Content in Curcuma longa L. (Zingiberaceae) Caused by 117 Hybridization with Other Curcuma Species Figure 4. PCR-RFLP profile of various lines of C. longa and C. aromatica. Arrows indicate expected fragments of both C. longa and C. aromatica. M: size marker. A: C. aromatica. L: C. longa. Plant number corresponds to the numbers in Table 1. chloroplast capture. In fact, some studies indicate that introgression and asymmetric capture of the cpDNA are common phenomena in hybridized species [19,35]. Our results indicated that hybrid and introgressive individuals with other Curcuma species were included in C. longa although hybrid and introgressive have same haplotype of C. longa based on matK sequences of cpDNA (Figure 1). Moreover, it is very interesting that homogeneous C. longa has a high curcumin content, and that a heteroge- neous hybrid of the Curcuma genome has a medium amount of curcumin. Additionally, an introgressive sam- ple with incongruent haplotypes between cpDNA and nrDNA has a low content of curcumin (Table 1, Figure 4). Therefore, the pattern of decreased curcumin content was congruent with the hybridization or introgression between C. longa and other Curcuma species, such as C. aromatica, which have low curcumin content. These re- sults indicated that hybridization or introgression with other Curcuma species could affect the content of cur- cumin of C. longa. In this study, C. longa proved to be the seed parent of the hybrid and introgression samples because all of the haplotypes on the matK of cpDNA matched this species. In the future, an analysis of the curcumin content of hybrid and introgression samples in which C. longa is the pollen parent needs to be con- ducted. In addition, the recent resurgence in plant devel- opment study has been accelerated, in part, by success in elucidating the molecular genetic basis of plant devel- opmental processes, including the isolation and charac- terization of genes that synthesize curcumin in C. longa [36-38]. As a result of these studies, it is considered very important to isolate and analyze the homologous genes of C. longa apart from other Curcuma species. As for hy- brids, Allard [39] claimed that interspecific hybrids are very useful in introducing genetic divergence, and, in fact, hybrids have been used for many crops and orna- mental plants. The way it was stated now appears that it is negating the well established knowledge of hybrid vigor and our results could not support this claim because interspecific hybrids contribute to the decrease in the curcumin content of C. longa. Genetic variation is one of the fundamental underpin- nings of biological diversity. The genetic structure and history of a given species is an important research focus, because this knowledge is needed to plan species con- servation and to understand the evolutionary processes leading to diversity. Our study results suggest that re- productive isolation mechanisms were not acting in the case of the small phylogenetic distances among the Cur- cuma species (Figure 1). The evolution of reproductive isolation is one of the defining characteristics of speci- ation [40], and reproductive isolation contributes to the diversification of species by creating genetically inde- pendent lineages and phylogenetic tree branches [41]. Each branching of the tree is a speciation event; however, reproductive isolation alone does not create a new branch. Each branch by itself cannot produce the phenotypic di- vergence represented by the angular departure of a branch from the ancestral form [41]. Therefore, the di- versification in the Curcuma species may have other factors involved rather than just reproductive isolation. In the future, it will be necessary to consider the phyloge- netic implications in order to understand the detailed evolutionary history of the Curcuma species. In summary, we have provided a hypothesis for the differences of curcumin by analyzing cpDNA and nrDNA Copyright © 2011 SciRes. AJPS  Difference of Curcumin Content in Curcuma longa L. (Zingiberaceae) Caused by 118 Hybridization with Other Curcuma Species data. Our results, using a molecular approach, were highly effective in revealing the histories of hybridization and introgression of C. longa. However, our data was less effective in definitively answering the question concern- ing the differences of curcumin content. Further studies will be needed to determine whether more comprehensive samplings and additional genetic evidence support the working hypothesis we have developed here. 5. Acknowledgements We wish to thank T. Kobayashi, A. Miyazaki, R. Ara- kawa and J. Yokoyama. T. Yoshida, A. Matsuzawa, A. Hirata, Y. Muramatsu, M. Saito, R. Ueda, K. Ohga, N. Yokoyama, M. Muroi, and K. Matsuyama for providing much help. This study was partly supported by a grant- in-aid for scientific research from the Ministry of Educa- tion, Science, and Culture of Japan (T.F.). REFERENCES [1] S. D. Tanksley and S. R. McCouch, “Seed Banks and Molecular Maps: Unlocking Genetic Potential from the Wild,” Science, Vol. 277, No. 5329, 1997, pp. 1063-1066. doi:10.1126/science.277.5329.1063 [2] A. Asai and T. Miyazawa, “Dietary Curcuminoids Pre- vent High-Fat Diet-Induced Lipid Accumulation in Rat Liver and Epididymal Adipose Tissue,” Journal of Nutri- tion, Vol. 131, No. 11, 2001, pp. 2932-2935. [3] A. C. Beynen, J. J. Visser and J. A. Schouten, “Inhibitory Effect on Lithogenesis by Ingestion of a Curcuma Mix- ture (Temoe Lawak Singer),” Journal of Food Science Technology, Vol. 24, 1987, pp. 253-256. [4] P. Anand, C. Sundaram, S. Jhurani, A. B. Kunnumakkara and B. B. Aggarwal, “Curcumin and Cancer: An ‘Old-Age’ Disease with an ‘Age-Old’ Solution,” Cancer Letters, Vol. 267, No. 1, 2008, pp. 133-164. doi:10.1016/j.canlet.2008.03.025 [5] S. Aggarwal, Y. Takada, S. Singh, J. N. Myers and B. B. Aggarwal, “Inhibition of Growth and Survival of Human Head and Neck Squamous Cell Carcinoma Cells by Cur- cumin via Modulation of Nuclear Factor-Κb Signaling,” International Journal of Cancer, Vol. 111, No. 5, 2004, pp. 679-692. doi:10.1002/ijc.20333 [6] Z. Wang, Y. Zhang, S. Banerjee, Y. Li and F. H. Sarkar, “Inhibition of Nuclear Factor Kappab Activity by Gen- istein is Mediated via Notch-1 Signaling Pathway in Pan- creatic Cancer Cells,” International Journal of Cancer, Vol. 118, No. 8, 2006, pp. 1930-1936. doi:10.1002/ijc.21589 [7] N. M. Weir, K. Selvendiran, V. K. Kutala, L. Tong, S. Vishwanath, M. Rajaram, S. Tridandapani, S. Anant and P. Kuppusamy, “Curcumin Induces G2/M Arrest and Apoptosis in Cisplatin-Resistant Human Ovarian Cancer Cells by Modulating Akt and p38 MAPK,” Cancer Biol- ogy and Therapy, Vol. 6, No. 2, 2007, pp. 178-184. doi:10.4161/cbt.6.2.3577 [8] E. Liu, J. Wu, W. Cao, J. Zhang, W. Liu, X. Jiang and X. Zhang, “Curcumin Induces G2/M Cell Cycle Arrest in a p53-Dependent Manner and Upregulates Ing4 Expression in Human Glioma,” Journal of Neuro-Oncology, Vol. 85, No. 3, 2007, pp. 263-270. doi:10.1007/s11060-007-9421-4 [9] K. Aoi, K. Kaburagi, T. Seki, T. Tobata, M. Sarak and M. Kuroyanagi, “Studies on the Cultivation of Turmeric (Curcuma longa L.) I: Varietal Differences in Rhizome Yield and Curcuminoid Content,” Bulletin of National In- stitute of Hygiene Science, Vol. 104, 1986, pp. 124-128. [10] H. Hayakawa, T. Kobayashi, Y. Minamiya, K. Ito, A. Miyazaki, T. Fukuda and Y. Yamamoto, “Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content,” American Journal of Plant Siences, Vol. 2, No. 1, 2011, pp. 15-26. doi:10.4236/ajps.2011.21002 [11] H. Hayakawa, T. Kobayashi, Y. Minamiya, K. Ito, A. Miyazaki, T. Fukuda and Y. Yamamoto, “Molecular Identification of Turmeric (Curcuma longa, Zingiberceae) with a High Curcumin Content,” Journal of Japanese Botany, Vol. 85, No. 5, 2010, pp. 263-269. [12] J. A. Coyne and H. A. Orr, “Patterns of Speciation in Drosophila,” Evolution, Vol. 43, No. 2, 1989, pp. 362- 381. doi:10.2307/2409213 [13] J. A. Coyne and H. A. Orr, “Patterns of Speciation in Drosophila Revisited,” Evolution, Vol. 51, No. 1, 1997, pp. 295-333. doi:10.2307/2410984 [14] J. C. Avise, J. Arnold, R. M. Ball, E. Bermingham, T. Lamb, J. E. Neigel, C. A. Reeb and N. C. Saunders, “In- traspecific Phylogeography: The Mitochondrial DNA Bridge between Population Genetics and Systematics,” Annual Review of Ecolgy, Evolution, and Systematics, Vol. 18, No. 1, 1987, pp. 489-552. [15] J. C. Avise, “Molecular Markers, Natural History, and Evolution,” Chapman & Hall, New York, 1994. [16] J. C. Avise, “Phylogeography: The History and Forma- tion of Species,” Harvard University Press, Cambridge, 2000. [17] J. A. Doyle, “Phylogeny of Vascular Plants,” Annual Review of Ecolgy, Evolution, and Systematics, Vol. 29, No. 1, 1998, pp. 567-599. doi:10.1146/annurev.ecolsys.29.1.567 [18] L. H. Rieseberg, J. Whitton and C. R. Linder, “Molecular Marker Incongruence in Plant Hybrid Zones and Phy- logenetic Trees,” Acta Botanica Neerlandica, Vol. 45, 1996, pp. 243-262. [19] J. F. Wendel and J. J. Doyle, “Phylogenetic Incongruence: Window into Genome History and Molecular Evolution,” In: P. S. Soltis, D. E. Soltis and J. J. Doyle, Eds., Mo- lecular Systematics of Plants II, Kluwer, Dordrecht, 1998, pp. 265-296. [20] B. G. Baldwin and S. Markos, “Phylogenetic Utility of the External Transcribed Spacer (ETS) of 18S-26S rDNA: Congruence of ETS and ITS Trees of Calycadenia (Com- Copyright © 2011 SciRes. AJPS  Difference of Curcumin Content in Curcuma longa L. (Zingiberaceae) Caused by Hybridization with Other Curcuma Species Copyright © 2011 SciRes. AJPS 119 positae),” Molecular Phylogenetics and Evoltion, Vol. 10, No. 3, 1998, pp. 449-463. doi:10.1006/mpev.1998.0545 [21] C. R. Linder, L. R. Goertzen, B. V. Heuvel, J. Fran- cisco-Ortega and R. K. Jansen, “The External Transcribed Spacer of the Rdna Repeat: A New Nuclear Region for Low-Level Taxonomic Analysis of the Asteraceae and Closely Allied Families,” Molecular Phylogenetics and Evoltion, Vol. 14, 2000, pp. 285-303. doi:10.1006/mpev.1999.0706 [22] S. Markos and B. G. Baldwin, “Higher-Level Relation- ships and Major Lineages of Lessingia (Compositae, As- tereae) Based on Nuclear Rdna Internal and External Transcribed Spacers (ITS and ETS) Sequences,” System- atic Botany, Vol. 26, No. 1, 2001, pp. 168-183. [23] L. E. Urbatsch, R. P. Roberts and V. Karaman, “Phy- logenetic Evaluation of Xylothamia, Gundlachia, and Re- lated Genera (Asteraceae, Astereae) Based on Ets and Its Nrdna Sequence Data,” American Journal of Botany, Vol. 90, No. 4, 2003, pp. 634-649. doi:10.3732/ajb.90.4.634 [24] O. Hidalgo, N. Garcia-Jacas, T. Garnatje and A. Susanna, “Phylogeny of Rhaponticum (Asteraceae, Cardueae- Centaureinae) and Related Genera Inferred from Nuclear and Chloroplast DNA Sequence Data: Taxonomic and Biogeographic Implications,” Annals of Botany, Vol. 97, No. 5, 2006, pp. 704-714. doi:10.1093/aob/mcl029 [25] M. Sato, K. Shimura and K. Hashizume, “Quality Valua- tion of Turmeric Powders on Market,” Annual Report of Mie Prefecture Health and Enviromant Research Institute, Vol. 6, 2004, pp. 52-54. [26] L. A. Jonson and D. E. Soltis, “matK DNA Sequences and Phylogenetic Reconstruction in Saxifragaceae s.str.,” Systematic Botany, Vol. 19, No. 1, 1994, pp. 143-156. doi:10.2307/2419718 [27] J. R. Starr, S. A. Harris and D. A. Simpson, “Potential of the 5' and 3' Ends of the Intergenic Spacer (IGS) of Rdna in the Cyperaceae: New Sequences for Lower-Level Phylogenies in Sedges with an Example from Uncinia Pers,” Internat Journal of Plant Sciencese, Vol. 164, 2003, pp. 213-227. doi:10.1086/346168 [28] J. D. Thompson, D. G. Higgins and T. J. Gibson, “CLUS- TAL W: Improving the Sensitivity of Progressive Multi- ple Sequence Alignment through Sequence Weighting, Positions-Specific Gap Penalties and Weight Matrix Choice,” Nucleic Acids Research, Vol. 22, No. 22, 1994, pp. 4673-4680. doi:10.1093/nar/22.22.4673 [29] K. Tamura, J. Dudley, M. Nei and S. Kumar, “MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software. Version 4.0,” Molecular Biology and Evolution, Vol. 24, No. 8, 2007, pp. 1596-1599. doi:10.1093/molbev/msm092 [30] D. L. Swofford, “paup*. Phylogenetic Analysis Using Parsimony (*and Other Methods),” 4th Edition, Sinauer Associates, Sunderland, 2001. [31] V. Grant, “Plant Speciation,” Columbia University Press, New York, 1981. [32] K. Marhold, J. Lihova, M. Perny, R. Grupeand and B. Neuffer, “Natural Hybridization in Cardamine (Brassica- ceae) in the Pyrenees: Evidence from Morphological and Molecular Data,” Botanical Journal of Linnean Society, Vol. 139, No. 3, 2002, pp. 275-294. doi:10.1046/j.1095-8339.2002.00066.x [33] J. Yokoyama, T. Fukuda, A. Yokoyama and M. Maki, “The Intersectional Hybrid between Weigela hortensis and W. maximowiczii (Caprifoliaceae),” Botanical Jour- nal of the Linnean Society, Vol. 138, No. 3, 2002, pp. 369-380. doi:10.1046/j.1095-8339.2002.00033.x [34] H. Hayakawa, H. Hamachi, Y. Muramatsu, A. Hirata, Y. Minamiya, K. Matsuyama, K. Ito, J. Yokoyama and T. Fukuda, “Interspesific Hybridization between Arisaema sikokianum Franch. Et Savat. and A. tosaense Makino (Araceae) Revealed from Nuclear and Chloroplast DNA Comparisons,” Acta Phytotaxonomica et Geobotanica, Vol. 61, No. 2, 2010, pp. 57-63. [35] J. C. Avise, “Molecular Markers, Natural History, and Evolution,” 2nd Edition, Sinauer Associates, Sunderland, 2004. [36] Y. Katsuyama, M. Matsuzawa, N. Funa and S. Horiuchi, “In Vitro Synthesis of Curcumoids by Type III Polyketide Synthase from Oryza sativa,” Journal of Biological Chemistry, Vol. 282, No. 52, 2007, pp. 37702-37709. doi:10.1074/jbc.M707569200 [37] Y. Katsuyama, T. Kita and S. Horiuchi, “Identification and Characterization of Multiple Curcumin Synthases from the Herb Curcuma longa,” FEBS Letter, Vol. 583, No. 17, 2009, pp. 2799-2803. doi:10.1016/j.febslet.2009.07.029 [38] Y. Katsuyama, T. Kita, N. Funa and S. Horiuchi, “Cur- cumoid Biosynthesis by Two Type III Polyketide Syn- thases in the Herb Curcuma longa,” Journal of Biological Chemistry, Vol. 284, No. 17, 2009, pp. 11160-11170. doi:10.1074/jbc.M900070200 [39] R. W. Allard, “Principle of Plant Breeding,” Jhon Wiley and Sons, New York, 1960. [40] M. King, “Species Evolution: The Role of Chromosome Change,” Cambridge University Press, Melbourne, 1993. [41] R. Dowkins, “Mechanism of Evolution,” In: N. A. Camp- bell, J. B. Reece and L. G. Mitchell, Eds., Biology, 5th Edition, Addison Wesley Longman, Boston, 1999, pp. 412-487.
|