Paper Menu >>
Journal Menu >>
 J. Biomedical Science and Engineering, 2011, 4, 419-425 JBiSE doi:10.4236/jbise.2011.46053 Published Online June 2011 (http://www.SciRP.org/journal/jbise/). Published Online June 2011 in SciRes. http://www.scirp.org/journal/JBiSE Modeling bovine lameness with limb movement variables Yu kun Wu1, N a ga r a j Nee rcha l2, Robert Dyer3, Uri Tasch4, Parimal Rajkondawar5 1Center for Vaccine Development, University of Maryland School of Medicine, Baltimore, USA; 2Department of Mathematics & Statistics, University of Maryland Baltimore County, Baltimore, USA; 3Department of Animal & Food Sciences, University of Delaware, Newark, USA; 4Department of Mechanical Engineering, University of Maryland Baltimore County, Baltimore, USA; 5Bou-Matic, LLC., Madison, USA. Email: wu@medicine.umaryland.edu Received 13 April 2011; revised 25 April 2011; accepted 2 May 2011. ABSTRACT Lameness detection is a world-wide challenge to farm- ers and veterinarians. Traditionally, one uses visual observation to make judgment on a cow's lameness or soundness. Visual observation heavily depends on the observer's experience, hence is subjective or ob- server-dependent. And even worse, it is inconsistent. It's reported that the agreement between veterinari- ans can be as low as 45% [1]. It is necessary and ur- gent to develop an objective detection method that can automatically detect lameness when it occurs. In this paper, we describe how statistical models can be used to develop such methods and how well the sta- tistical models perform. Keywords: Statistical Modeling; Bovine Lameness Detection 1. INTRODUCTION Bovine lameness is one of the major dairy health prob- lems for dairy industry. It can occur even in well- ma- naged herds. Lameness may be described as a deviation from the “normal walking pattern” due to injury or disease, usually accompanied by pain (Black’s Veteri- nary Dictionary, 1985 [2]). It can be caused by injuries or genetic factors. Severe lameness will lead to reduced productivity and profitability. Lameness causes mil- lions of dollars in loss of revenue to the dairy industry in America every year. Current method of lameness detection is based on observers making a subjective judgment on the gaits of the cows and has been docu- mented to be unreliable. Thomsen et al. (2008) [1] used kappa statistics to evaluate intra- and inter-observer agreement. They reported the weighted kappa values ranging from 0.38 to 0.78 for intra-observer agreement, with mean kappa values across all observers of 0.60 and 0.53 before and after training, respectively. For inter-observer agreement, the weighted kappa values ranged from 0.24 to 0.68 with mean kappa values across all pairs of observers of 0.48 and 0.52 before and after training, respectively. Training had only a limited positive effect on intra- and inter-observer agreement. Rajkondawar et al. (2002) [3] proposed an innova- tive technology for directly measuring the ground reac- tion forces (GRF) exerted by the cows while walking. Certain limb movement variables (LMV) are derived from the GRF measurements and are combined with actual clinical evaluations by an experienced veterinar- ian. The system has been commercialized by BouMatic as StepMetrixTM (see Figure 1) and was permanently installed in two dairy farms. Ground reaction forces (GRF) exerted by the cows while walking is measured every time. We examined 14 - 15 cows per week. Gaits in all four limbs were evaluated for each animal. Mod- eling, however, was only attempted across the hind limbs due to an insufficient number of fore limb lame- ness. The majority of lameness occurs in the hind limbs but the front limbs do on occasion have lameness problems (5% of the time). Locomotion scores, claw and soft tissue pain were determined on 356 Holstein dairy cows from two commercial dairy farms. The LMVs were generated by StepMatrixTM each time when the cows walked on the system. An ID system is also incorporated to associate the derived LMVs with the animals that walk through. Measurements correspond- ing to the hind limbs, left hind (LH) and right hind (RH), are AGRF(LH/RH), PGRF(LH/RH), ENERGY (LH/RH) and STIME(LH/RH). PGRF and AGRF are the ground reaction force (GRF) normalized by the animal’s dynamic weight of a tested limb. LHSTIME (or RHSTIME) is defined as proportion of stance time corresponding to left hind limb (or right hind limb) relative to the sum of left and right hind limbs. There- fore the two stance times LHSTIME and RHSTIME  Y. K. Wu et al. / J. Biomedical Science and Engineering 4 (2011) 419-425 Copyright © 2011 SciRes. JBiSE 420 Figure 1. A photograph of stepMetrixTM. sum to one. Only left hind will be kept for building the model and the other will be redundant. A brief descrip- tion of each LMV is shown in Table 1. There are a number of statistical issues related to the modeling of bovine lameness using LMVs. LMV meas- urements from each of the two hind limbs will be clearly influenced by the lameness status of both limbs. In par- ticular we will expect high correlation among the LMVs both within and across limb. Furthermore, a higher sus- ceptibility to lameness will increase the probability of lameness on either limb; one would expect a high degree of association between the responses from the two limbs. Therefore, predictive modeling of lameness in terms of the LMVs will need to address both limbs simultane- ously. Note that we can consider the data to be clustered, the two limbs belonging to a cow forming a cluster of size two, and responses from different cows are assumed to be independent. Statistical challenges are arising in modeling categorical data from clustered observations and some approaches are given in Dean (1992) [4] and Morel and Neerchal (1997) [5]. Table 1. Description of limb movement variables. LMV Description Unit PGRF Peak Ground Reaction Force nondimensiona AGRF Average Ground Reaction Force nondimensiona STIME Stance Time nondimensiona ENERGY Fourier Transformation of GRF 1/second The other issue relates to the model selection. We may either treat the response for each cow as a multinomial (both limbs sound, at least one lame limb, and both limbs lame) or as two correlated binomial responses. Statistical methods are available for both approaches. We provide both analyses and compare their predictive per- formances. Table 2 provides basic summary statistics for LMV. The means and standard deviations computed are ob- tained from the LMV measurements for each limb. The analysis of variance of each LMV on the effect of each limb’s lameness is given in appendix Table (B). We found the lameness of each limb has a higher significant effect on the LMVs of the same limb, but has less sig- nificant effect on the opposite limb. Table (A) in appen- dix provides the correlations and indicates strong corre- lations between some pairs of LMV. There exists strong collinearity between the LMVs. Especially, the correla- tion coefficients between LH PGRF and LH AGRF and between RH PGRF and RH AGRF both exceed 0.94. And there are other four correlation coefficients that exceed 0.83. The study was conducted in two farms. The cows walked freely across the StepMetrixTM machine after every milking (with three milkings per day). Between 14 - 15 cows were randomly selected for clinical examina- tion every week. The database consists of clinical diagnosis for each cow, and the corresponding LMV measurements on its two hind limbs. An experienced veterinarian examined each claw and made a clinical assessment whether or not each hind limb is lame (L), sound (S) or mildly lame (M) according to its locomotion score, pain index and lesion severity score. Locomotion and lesion scores were es- tablished for each cow as previously described by Raj- kondawar et al. (2002) [3]. Claws and inter-digital areas were brushed with water and soap and examined for lesions after locomotion scores were established. Claw pain was determined by compression using a hoof tester instrumented to transfer the compression force through a Dillon force gauge (see Figure 2) as described by Dyer et al. (2007) [6]. Increasing levels of claw compression Table 2. Summary statitsics for the LMV. variables MEANSTD DEV CV min max LH_PGRF 0.453 0.085 0.187 0.13 0.9 RH_PGRF 0.449 0.086 0.192 0.17 0.92 LH_AGRF 0.369 0.067 0.180 0.12 0.69 RH_AGRF 0.366 0.067 0.183 0.15 0.79 LH_ENERGY0.414 0.088 0.212 0.09 0.86 RH_ENERGY0.409 0.090 0.220 0.15 0.9 LH_STIME 50.4697.590 0.150 22 89  Y. K. Wu et al. / J. Biomedical Science and Engineering 4 (2011) 419-425 Copyright © 2011 SciRes. JBiSE 421 Figure 2. An instrumented hoof tester is used to assess the force that elicits hoof pain reaction. Sound cattle can sustain 75 Kg with no reaction. The ratio of the force eliciting hoof pain reaction and 75 Kg is proposed as a hoof pain index. were applied to attain 75 Kg pressure or until the cow no longer tolerated the compression by showing a with- drawal response. Pressure attained at the onset of foot withdrawal was recorded only after animals reacted to three repeated compression tests. Pain associated with lesions of the integument was assessed using an algo- meter (4.5 Kg scale) with a blunt probe pressed against the integument (see Figure 3). The probe was placed on the lesion surface or on the junction of the inter-digital integument with the volar integument. Increasing levels of pressure was applied to the integument or lesion to attain 4.5 Kg pressure or until the cow showed a with- drawal response. Pressure attained at the onset of foot withdrawal was recorded only after animals reacted to three repeated compression tests. Since most of the Figure 3. An algometer is used to assess the force that elicits pain reaction of the soft interdigital tissue. Sound cattle can sustain 4.5 Kg with no reaction. The ratio of the force eliciting pain reaction of the soft interdigital tissue and 4.5 Kg is pro- posed as a pain index for the soft interdigital tissue. lameness occurs in the hind limbs, only the left hind(LH) and right hind(RH) limbs were examined. 2. MODEL BUILDING The bovine lameness database described in section 1 offers several challenges for a model builder. First and foremost, the lameness statuses for the two hind limbs of each cow need to be modeled simultaneously while ac- counting for correlation between the limb movement variables collected within one cow. Another challenge is that the clinical diagnosis can be difficult to categorize. Sound limbs are those which have no significant pain and have no lesions. Severely lame limbs are those with lesions and pain. However, diagnosis of mildly lame cases can be difficult, depending veterinarian’s experi- ence and the severity of lameness. It turns out that the dairy farmers are more concerned about identifying those who are not perfectly sound. Therefore, we com- bined the mildly and severely lame cases into a single category and called it “lame”. Thus we have a binary response variable, for each limb, “1” representing lame- ness and “0” representing soundness. Similarly we can define a cow-level lameness outcome. If at least one of her hind limbs is lame, this cow is lame, otherwise is sound. A few basic notations are developed to help us define the models considered here. Let Yij denote the observa- tion on the jth limb (j = 1 if limb is left hind, j = 2 if limb is right hind) of the ith cow, Yij = 1 if jth limb of cow i is lame and Yij = 0 otherwise. There are primarily two ap- proaches for modeling: cow-level models and limb-level models, depending on each observation representing either a cow or a limb. 2.1. Cow-Level Model Since the outcome for each cow is binary (either lame- ness or soundness), we assume the response variable and covariates for one cow are independent of those for an- other cow. A logistic regression model was built for the cow-level model as follows: Pr1log ii Yitx (1) where Yi is the outcome variable for the ith cow, Yi = 1 if at least one of the hind limbs of the ith cow is lame and Yi = 0 if neither of her hind limbs is lame; β is the regres- sion coefficient vector; xi is the limb movement variable vector for the ith cow, and the logit function is written as 1 log 1u it ue . 2.2. Limb-Level Model Similarly, Limb-level models can be built depending on different assumptions. Hence we have:  Y. K. Wu et al. / J. Biomedical Science and Engineering 4 (2011) 419-425 Copyright © 2011 SciRes. JBiSE 422 12 345 12 345 Pr1 logij ij ijij ij ijj ijij ij ij ij Yitxx xxx (2) where Yij is the outcome variable for the ith cow's jth limb. j = 1 if it is left hind and j = 2 if it is right hind. In model (2), we need to consider the following two possible con- ditions: (a) there does not exist correlation between left hind and right hind limbs within one cow, and (b) there exists correlation between left hind and right hind limbs within one cow. In the first case, ordinal logistic regres- sion can be applied directly (SAS PROC LOGISTIC [7]). In the second case, we need to consider within subject correlation and GEE model can be applied (SAS PROC GENMOD [7]). If we neglect the difference between the left hind and right hind limb-level models, assuming they have com- mon intercept, we have: 12 345 12 345 Pr1 logij ij ijij ij ijij ij ij ij ij Yitxx xxx (3) Similarly, we need to consider the existence of within subject correlation in model (3). The selection between the competent limb-level models can be obtained through the likelihood ratio test for model (2) against model (3). 3. LIKELIHOOD MODELS (Limb-level) It is reasonable to assume that the responses of the two limbs are correlated within each cow. Hence, overdis- persion may exist. If we are allowed to assume the cor- relation coefficient between the two hind limbs is the same for each cow, in the likelihood framework, there are two basic approaches to deal with overdispersion. One is beta-binomial model [4], and the other is finite- mixture model [5]. Here we demonstrate the likelihood function with fi- nite-mixture approach. Let ρ2 be the correlation coeffi- cient between the two limbs for each cow, and let piLH and piRH be the probability of occurrence of lameness in left hind and right hind limbs respectively for the ith cow. The relationships of the lameness within each cow can be summarized as follows in Table 3, where 2 11 11 , iiLH iLHiRH iRH iLH iRH PPPPP PP 10 211, iiRHiLH iRH iLH iLHiRH iRH PP PP PPPP 01 211, iiLHiLHiRH iLHiLH iRHiRH PPPP PPPP Table 3. Probability distribution of occurrence of lameness within ith cow. LH 1 0 Marginal 1 Pi11 P i10 P iRH RH 0 Pi01 P i00 1-PiRH Marginal PiLH 1-PiLH 1 ,11 1 2 01 iRHiRHiLHiLH iRHiLHiRHiLHi PPPP PPPPP and PiLH and PiRH are the probability of lameness in left hind limb and right hind limb respectively for the ith cow. The likelihood function can be expressed as: n ii LL 1, where 00011011 00011011 iiii I i I i I i I ii PPPPL , where IiLH is the indicator function of lameness status of LH and RH limbs such that Ii11=1 if both LH and RH limbs are lame, =0 otherwise; Ii10=1 if RH limb is lame and LH is sound, =0 otherwise; Ii01=1 if both RH limb is sound and LH is lame, =0 otherwise; Ii00=1 if both LH and RH limbs are sound, =0 otherwise. The log-likelihood function can be maximized (locally) to find the maximal likelihood estimator (MLE) of the parameters with appropriately chosen initial values (SAS PROC NLP [8]). The estimated correlation coefficient is ρ2 = 0.0861563, which indicates that the correlation be- tween the two hind limbs is relatively small, and rea- sonably neglectable. The beta-binomial model gives a similar result. 4. NUMERICAL RESULTS The modeling results with respect to sensitivity and spe- cificity are summarized as follows in Tabl e s 4 , 5, 6 and 7. Here the lameness status indicator means the sound category if its value is 0 and lame category if its value is 1, respectively. 5. CONCLUSION AND DISCUSSION In this paper we discussed different approaches to model bovine lameness with limb movement variables (LMVs), generated by the StepMetrixTM system when a cow walked through the machine. An experienced veterinar- ian carefully examined and made a clinical assessment of each hind claw of those randomly chosen cows in two participating dairy farms. The lameness statuses assessed by the veterinarian were used as “Gold Standard” to evaluate the performance of several statistical models. 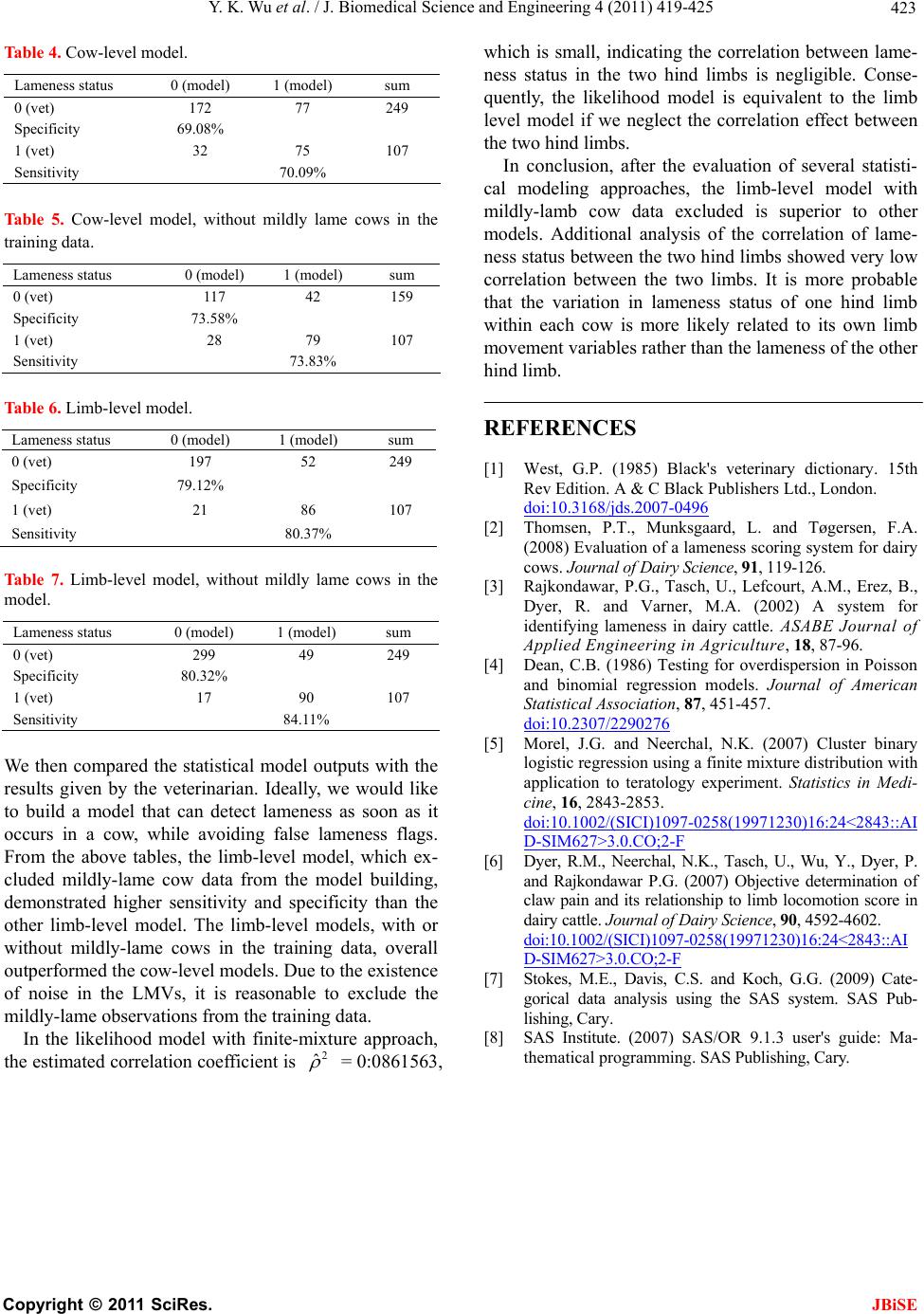 Y. K. Wu et al. / J. Biomedical Science and Engineering 4 (2011) 419-425 Copyright © 2011 SciRes. JBiSE 423 Table 4. Cow-level model. Lameness status 0 (model) 1 (model) sum 0 (vet) 172 77 249 Specificity 69.08% 1 (vet) 32 75 107 Sensitivity 70.09% Table 5. Cow-level model, without mildly lame cows in the training data. Lameness status 0 (model) 1 (model) sum 0 (vet) 117 42 159 Specificity 73.58% 1 (vet) 28 79 107 Sensitivity 73.83% Table 6. Limb-level model. Lameness status 0 (model) 1 (model) sum 0 (vet) 197 52 249 Specificity 79.12% 1 (vet) 21 86 107 Sensitivity 80.37% Table 7. Limb-level model, without mildly lame cows in the model. Lameness status 0 (model) 1 (model) sum 0 (vet) 299 49 249 Specificity 80.32% 1 (vet) 17 90 107 Sensitivity 84.11% We then compared the statistical model outputs with the results given by the veterinarian. Ideally, we would like to build a model that can detect lameness as soon as it occurs in a cow, while avoiding false lameness flags. From the above tables, the limb-level model, which ex- cluded mildly-lame cow data from the model building, demonstrated higher sensitivity and specificity than the other limb-level model. The limb-level models, with or without mildly-lame cows in the training data, overall outperformed the cow-level models. Due to the existence of noise in the LMVs, it is reasonable to exclude the mildly-lame observations from the training data. In the likelihood model with finite-mixture approach, the estimated correlation coefficient is 2 ˆ = 0:0861563, which is small, indicating the correlation between lame- ness status in the two hind limbs is negligible. Conse- quently, the likelihood model is equivalent to the limb level model if we neglect the correlation effect between the two hind limbs. In conclusion, after the evaluation of several statisti- cal modeling approaches, the limb-level model with mildly-lamb cow data excluded is superior to other models. Additional analysis of the correlation of lame- ness status between the two hind limbs showed very low correlation between the two limbs. It is more probable that the variation in lameness status of one hind limb within each cow is more likely related to its own limb movement variables rather than the lameness of the other hind limb. REFERENCES [1] West, G.P. (1985) Black's veterinary dictionary. 15th Rev Edition. A & C Black Publishers Ltd., London. doi:10.3168/jds.2007-0496 [2] Thomsen, P.T., Munksgaard, L. and Tøgersen, F.A. (2008) Evaluation of a lameness scoring system for dairy cows. Journal of Dairy Science, 91, 119-126. [3] Rajkondawar, P.G., Tasch, U., Lefcourt, A.M., Erez, B., Dyer, R. and Varner, M.A. (2002) A system for identifying lameness in dairy cattle. ASABE Journal of Applied Engineering in Agriculture, 18, 87-96. [4] Dean, C.B. (1986) Testing for overdispersion in Poisson and binomial regression models. Journal of American Statistical Association, 87, 451-457. doi:10.2307/2290276 [5] Morel, J.G. and Neerchal, N.K. (2007) Cluster binary logistic regression using a finite mixture distribution with application to teratology experiment. Statistics in Medi- cine, 16, 2843-2853. doi:10.1002/(SICI)1097-0258(19971230)16:24<2843::AI D-SIM627>3.0.CO;2-F [6] Dyer, R.M., Neerchal, N.K., Tasch, U., Wu, Y., Dyer, P. and Rajkondawar P.G. (2007) Objective determination of claw pain and its relationship to limb locomotion score in dairy cattle. Journal of Dairy Science, 90, 4592-4602. doi:10.1002/(SICI)10 97-0258(19 971230)1 6:24<28 43::AI D-SIM627>3. 0.CO;2 -F [7] Stokes, M.E., Davis, C.S. and Koch, G.G. (2009) Cate- gorical data analysis using the SAS system. SAS Pub- lishing, Cary. [8] SAS Institute. (2007) SAS/OR 9.1.3 user's guide: Ma- thematical programming. SAS Publishing, Cary.  Y. K. Wu et al. / J. Biomedical Science and Engineering 4 (2011) 419-425 Copyright © 2011 SciRes. JBiSE 424 Appendix Table A. Correlations between the LMVs. RH_PGRF LH_AGRF RH_AGRF LH_ENERGY RH_ENERGY LH_STIME LH_PGRF 0.317 0.948 0.282 0.864 0.363 0.300 RH_PGRF 1 0.288 0.944 0.322 0.868 -0.382 LH_AGRF 1 0.287 0.843 0.348 0.296 RH_AGRF 1 0.306 0.838 -0.354 LH_ENERGY 1 0.385 0.190 RH_ENERGY 1 -0.263 ANOVA tables of LMVs on Lameness status: Table B1. ANOVA Table of LHLAMESCORE on LH_PGRF. Source DF Sum of Squares Mean Square F ValuePr > F Model 1 0.352144 0.352144 58.38 <.0001 Error 354 2.135168 0.006032 Total 355 2.487311 Table B2. ANOVA Table of LHLAMESCORE on LH_AGRF. Source DF Sum of Squares Mean Square F ValuePr > F model 1 0.267368 0.267368 72.84 <.0001 error 354 1.29931 0.00367 total 355 1.566677 Table B3. ANOVA Table of LHLAMESCORE on LH_STIME. Source DF Sum of Squares Mean Square F Value Pr > F Model 1 2873 2873 54.88 <.0001 Error 354 18532.1 52.35057 Total 355 21405.1 Table B4. ANOVA Table of LHLAMESCORE on LH_ENERGY. Source DF Sum of Squares Mean Square F ValuePr > F model 1 0.317513 0.317513 43.75<.0001 error 354 2.569063 0.007257 total 355 2.886576 Table B5. ANOVA Table of RHLAMESCORE on LH_AGRF. Source DF Sum of Squares Mean Square F ValuePr > F model 1 0.005813 0.005813 1.32 0.2517 error 354 1.560864 0.004409 total 355 1.566677 Table B6. ANOVA Table of RHLAMESCORE on LH_PGRF. SourceDFSum of SquaresMean Square F ValuePr > F Model 1 0.005795 0.005795 0.83 0.3638 Error 3542.481516 0.00701 Total 3552.487311 Table B7. ANOVA Table of RHLAMESCORE on LH_PGRF. Source DF Sum of SquaresMean Square F ValuePr > F Model 1 0.001595 0.001595 0.2 0.6585 Error 3542.884981 0.00815 Total 355 2.886576 Table B8. ANOVA Table of RHLAMESCORE on RH_PGRF. Source DFSum of Squares Mean Square F ValuePr > F Model 1 0.029742 0.029742 4.16 0.042 Error 3542.528093 0.007142 Total 3552.557835 Table B9. ANOVA Table of LHLAMESCORE on RH_AGRF. Source DF Sum of Squares Mean Square F ValuePr > F Model 1 0.017994 0.017994 4.41 0.0364 Error 3541.443479 0.004078 Total 3551.461473 Table B10. ANOVA Table of LHLAMESCORE on RH_ENERGY. SourceDFSum of SquaresMean Square F ValuePr>F Model 10.00409 0.00409 0.52 0.4734 Error 3542.810259 0.007939 Total 3552.814349 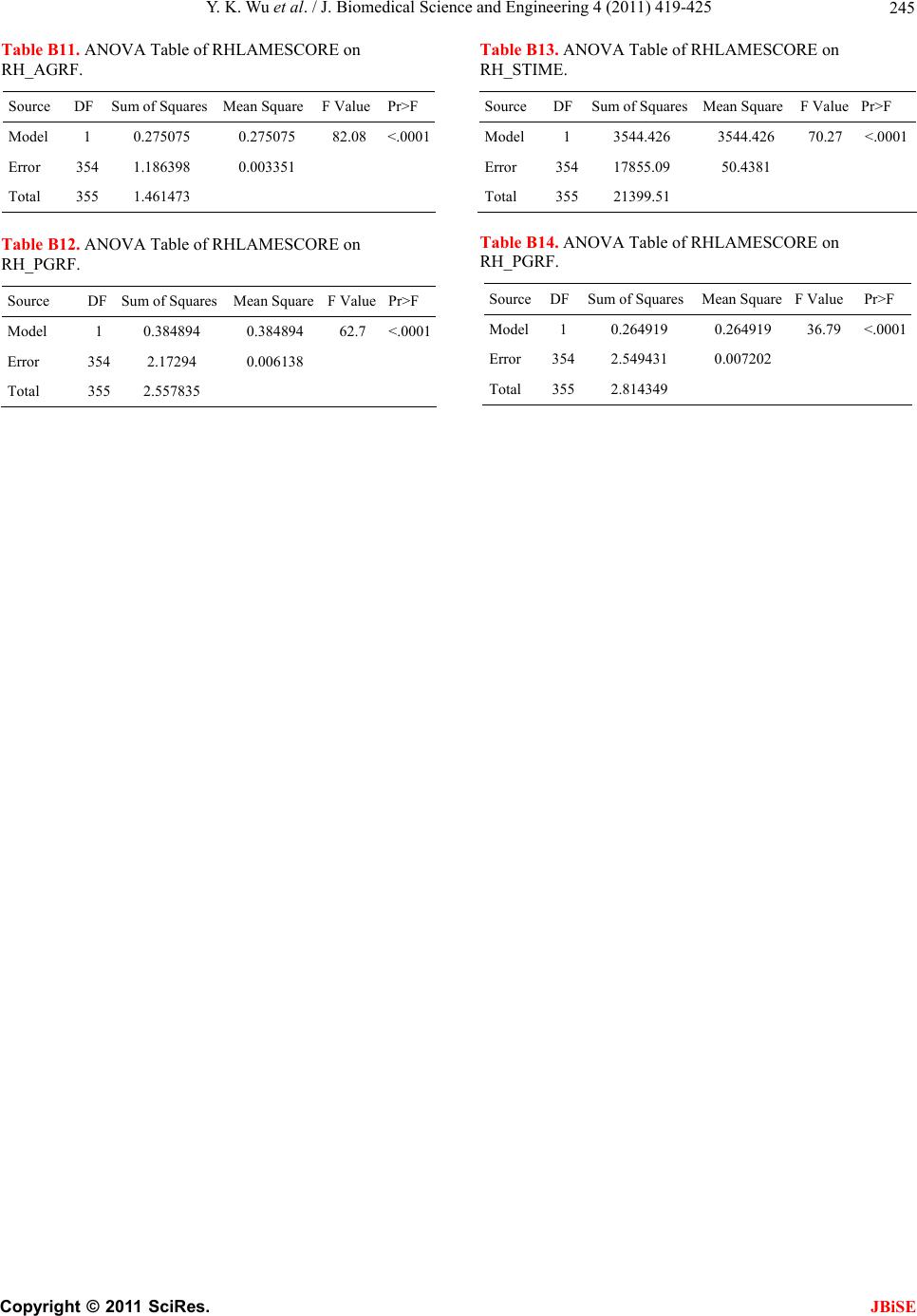 Y. K. Wu et al. / J. Biomedical Science and Engineering 4 (2011) 419-425 Copyright © 2011 SciRes. JBiSE 245 Table B11. ANOVA Table of RHLAMESCORE on RH_AGRF. Source DF Sum of Squares Mean Square F ValuePr>F Model 1 0.275075 0.275075 82.08 <.0001 Error 354 1.186398 0.003351 Total 355 1.461473 Table B12. ANOVA Table of RHLAMESCORE on RH_PGRF. Source DF Sum of Squares Mean Square F ValuePr>F Model 1 0.384894 0.384894 62.7 <.0001 Error 354 2.17294 0.006138 Total 355 2.557835 Table B13. ANOVA Table of RHLAMESCORE on RH_STIME. Source DFSum of SquaresMean Square F ValuePr>F Model 1 3544.426 3544.426 70.27<.0001 Error 35417855.09 50.4381 Total 35521399.51 Table B14. ANOVA Table of RHLAMESCORE on RH_PGRF. SourceDFSum of SquaresMean Square F Value Pr>F Model 1 0.264919 0.264919 36.79 <.0001 Error 3542.549431 0.007202 Total 3552.814349 |

