Paper Menu >>
Journal Menu >>
 Vol.1, No.1, 1-7 (2011) Open Journal of Immunology doi:10.4236/oji.2011.11001 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/OJI/ Successful expression and purification of dppd, using a codon optimized synthetic gene* Suely S. Kashino, Antonio Campos-Neto# The Forsyth Institute, Global Infectious Disease Research Center, Boston, USA; #Corresponding Author: acampos@forsyth.org Received 14 May 2011, revised 8 June 2011, accepted 17 June 2011. ABSTRACT DPPD (Rv0061) is a difficult to express protein of Mycobacterium tuberculosis that elicits strong and specific delayed type hypersensitivity reac- tions in humans infected with M. tuberculosis. Therefore DPPD is a molecule that can improve the specificity of the tuberculin skin test, which is widely used as an aid for the diagnosis of tu- berculosis. However, a pitfall of our initial stud- ies was that the DPPD molecule used to perform the skin tests was engineered as fusion mole- cule with another Mycobacterium protein. This approach was used because no expression of DPPD could be achieved either as a single molecule or as a fusion protein using a variety of commercially available expression systems. Here, we report the production and purification of rDPPD using a synthetic gene engineered to contain E. coli codon bias. The gene was cloned into pET14b expression vector, which was sub- sequently used to transform Rosetta 2(DE3) pLysS or BL-21(DE3)pLysS host cells. The re- combinant protein was over-expressed after in- duction with IPTG and its purification was easily achieved at levels of 5 – 10 mg/l of bacterial broth cultures. The purified protein was con- firmed to be DPPD by Mass Spectroscopy se- quencing analysis. Moreover, purified rDPPD stimulated peripheral blood mononuclear cells of PPD positive blood donors to produce high levels of IFN-γ, thus confirming that this mole- cule is biologically active. Because of the DPPD gene is restricted to the tuberculosis-complex organisms of Mycobacterium genus, this highly purified molecule should be useful for the iden- tification of individuals sensitized with tubercle bacilli. Keywords: Mycobacterium tuberculosis; DPPD; Codon Bias 1. INTRODUCTION Over-expression of recombinant proteins in Esche- richia coli host cells followed by purification in affinity resins has become a routine and useful procedure to produce a variety of antigenic molecules. However, not seldom and for not entirely known reasons, the bacterial host cells fail to properly express the recombinant pro- teins coded for by the genes cloned into a number of different expression vectors. Several possibilities have been raised to explain this difficult-to-express proteins puzzle. These include protein toxicity to E. coli, plasmid or protein instability, inefficient transcription or transla- tion, inefficient post-translational modification, and pre- sence in the cloned gene of inadequate or non-used co- don sequences by the E. coli host cells [1,2]. A typical example of difficult-to-express protein is DPPD, a Mycobacterium tuberculosis molecule that we have described several years ago [3,4]. The name DPPD represents the single letter code of the first four amino acids of the N-terminus sequence of the mature form of the protein (aspartic acid, proline, proline, aspartic acid). DPPD is a small protein composed of 84 amino acids and is a major component of the complex protein mix- ture present in purified protein derivative (PPD), which is the antigenic preparation used in the skin test per- formed in humans and cattle for the diagnosis of tuber- culosis [5-9]. Therefore, DPPD is interesting candidate molecule to be used as a purified antigen for the diagno- sis of tuberculosis. However, expression of recombinant DPPD in E. coli host cells either as a single molecule or as a fusion protein, using an exhaustive list of commer- cially available expression systems for this host cells, consistently failed. Moreover, rDPPD could not be ex- pressed in other cell systems such as the yeast Pichia pastoris and Streptococcus lividans. In addition, at- tempts to produce a synthetic DPPD polypeptide through several protein manufacturing services were also unsuc- *Financial support: This work was supported by the National Institutes of Health g rant R01AI076425.  S. S. Kashino et al. / Open Journal of Immunology 1 (2011) 1-7 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/OJI/ 2 cessful. Partial success to express rDPPD as a single molecule was achieved using Mycobacterium smegmatis as host cells transformed with the plasmid pSMT3 [10]. Unfortunately, the yields ofpurified protein obtained with this system was extremely low (~500 μg/liter of bacterial culture medium), which precludes the production of the protein for biological or clinical studies. However, be- cause in this system both the host cells and the cloned gene belong to the same bacterial genus (Mycobacte- rium), these studies suggested that shared preferential codon usage between the two species facilitated the ex- pression of rDPPD in M. smegmatis. In the present work we tested this possibility. A syn- thetic DPPD gene containing a sequence designed to have optimized E. coli preferential codons was obtained and cloned into pET14b. Transformation of E .coli host cells with this plasmid followed by induction with IPTG resulted in consistent and successful over-expression rDPPD, which could be purified by affinity chromatog- raphy (Ni-NTA resin) yielding 5 - 10 mg of purified re- combinant protein per liter of bacterial culture. 2. MATERIAL AND METHODS 2.1. DPPD Codon Optimization and Gene Subcloning The codon optimization for maximum expression in E. coli was performed using iteratively sampling from a codon usage table to find a low free energy solution, which typically results in decreased secondary structure of the mRNA. The DPPD optimized sequence was syn- thesized at Blue Heron (https://www.blueheronbio.com) using a proprietary technology which allows 100% ac- curacy. An Nde I and a Bam HI restriction enzyme se- quences flanking the 5’ and 3’ ends respectively were incorporated in the DPPD gene sequence. The synthe- sized DNA was cloned into pUCminusMCS (Blue Heron) followed by sequencing, which confirmed the designed optimized sequence. DPPD synthetic gene was then sub- cloned into the pET14b expression vector (Novagen EMD Chemicals, Gibbstown, NJ) using the two restric- tion enzymes. Successful subclones were confirmed by PCR using Taq Plus DNA polymerase (Invitrogen) and DNA sequencing. 2.2. rDPPD Protein Expression and Purification Plasmid pET14b harboring the DPPD optimized gene was used to transform Escherichia coli BL21(DE3) pLysS or Rosetta 2 (DE3)pLysS (Novagen) expression hosts. 100 ml LB cultures of IPTG-induced E. coli were processed under native or denaturing conditions followed by affinity chromatography using nickel-nitrilotriacetic acid (Ni-NTA) agarose matrix (Qiagen, Valencia, CA). Purified rDPPD was analyzed by SDS- PAGE and con- centration was determined by BCA (Pierce Thermo Sci- entific, Waltham, MA). 2.3. Western Blot Purified recombinant DPPD protein was electropho- resed on SDS-PAGE (4% - 20% gradient, BioRad, Her- cules, CA) under reducing conditions and transferred to a PVDF membrane (Immobilon-P, Millipore, Billerica, MA). The blot was blocked for one hour at room tem- perature with Tris-HCl buffer pH 7.4 containing 0.1% Tween 20 and 1% BSA, andsubsequently incubated with HRP-labeled anti-His tag monoclonal antibody (Invitro- gen, Carlsbad, CA). Bound conjugate was detected by using the LumiGlo chemiluminescent system (Cell Sig- naling Technology, Danvers, MA), and visualized by ex- posing the membrane to an autoradiography film (Kodak BioMax, Rochester, NY). 2.4. Human Interferon-Gamma Response Blood samples (10 ml) were collected with informed consent from 6 healthy, confirmed PPD positive donors. Human peripheral blood mononuclear cells (PBMC) were isolated by gradient centrifugation (Histopaque, Sigma- Aldrich, St. Louis, MO). 2 × 105 cells PBMC in com- plete RPMI supplemented with 10% human AB serum were incubated (37˚C, 5% CO2) with different concen- trations of rDPPD (0.4 to 10 μg/ml) or PPD (2.5 μg/ml to 10 μg/ml); PHA (1/500) or medium alone was in- cluded as positive and negative controls, respectively. Supernatants were collected after 72 h of culture and as- sayed for IFN-γ content using commercial capture and detection mAb pairs for sandwich ELISA (BD Biosci- ences, Rockville, MD). 3. RESULTS 3.1. Codon Optimization The M. tuberculosis DPPD full length gene (Rv0061) codes for a typical secreted protein, which includes a signal peptide sequence of 39 amino acids followed by the signal peptidase recognition sequence Ala-Ser-Ala (Figure 1). Because in our original studies [3,4,10] we found that M. tuberculosis produces only the mature sequence of the protein we opted to evaluate the codon optimization designed to include only this form of the molecule. To facilitate expression an initiation ATG codon was added to 5’ end of the gene sequence. Figure 2 depicts the DPPD optimized gene sequence compared to that of M. tuberculosis.  S. S. Kashino et al. / Open Journal of Immunology 1 (2011) 1-7 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/OJI/ 3 Figure 1. Deduced amino acid sequence of the protein coded by Mycobacterium tuberculosis Rv0061. The deduced amino acid sequence of the 112 residues of the protein, reveals that the full length gene codes for a typical secretory protein, which includes a signal peptide sequence (illustrated in red letters), the signal peptidase recognition sequence ASA (illustrated in green, bold, italic letters) and the mature sequence of protein (DPPD), which is depicted in black bold letters. 3.2. Protein Expression and Purification The DNA fragment containing the synthetic optimized DPPD sequence was cut from pUCminusMCS and sub -cloned into pET-14b vector. This expression vector con- tains a Histidine tag (His-tag) sequence before the Nde I cloning site thus generating a recombinant protein con- taining a sequence of six His residues at the N-terminus to facilitate its purification by affinity binding to a Ni-NTA agarose matrix. The expression vector was used to transform Rosetta 2(DE3)pLysS or BL21 (DE3)pLysS host cells which were then induced with IPTG. Recom- binant protein expression was assessed by SDS-PAGE with Coomassie blue staining and is illustrated in Figure 3A (lanes 1 and 2 respectively). Over- expressed pro- teins bands of MW ~12 kDa and ~25 kDa can be seen in the lane corresponding to the IPTG induced culture. No such strongly stained bands are seen in the lane corre- sponding to the non-induced culture (lane 1). The recombinant protein was purified as soluble pro- tein from BL21(DE3)pLysS or from the inclusion bodies in Rosetta 2(DE3)pLysS using affinity chromatography. Yields of purification ranged from 5-10mg of purified protein per liter of induced culture. Figure 3 (lane 3B) shows the purified recombinant protein. Two major band aggregates are seen at a positions matching the over- expressed protein seen in the induce cultures (Figure 3A, lane 2). The lower bands (MW ~8 - 15 kDa) are within the range of the predicted MW of the native mature form of DPPD (9.02 kDa). This pattern of migration is usually observed with proteins with high content in proline residues [11-14] and is consistent with our former ob- servations for both native and recombinant molecules [4,10]. In addition, three other bands of higher MW (~20 - 30 kDa) are also clearly seen. At first, many of these Figure 2. DPPD DNA sequence with E. coli optimized codon usage. Codon optimization for protein expression was done using the Blue Heron Expression Optimization tool which mini- mizes secondary mRNA structure to reduce translational impediments. Optimized DPPD se- quence is displayed in bold (upper sequence). Note that an Nde I and a Bam HI restriction en- zyme sequences (underline sequences) flanking the 5’ and 3’ ends respectively were incorpo- rated in the optimized sequence. For comparison purposes the Rv0061 gene sequence (coding the mature form of the protein only) is also shown (lower sequence). 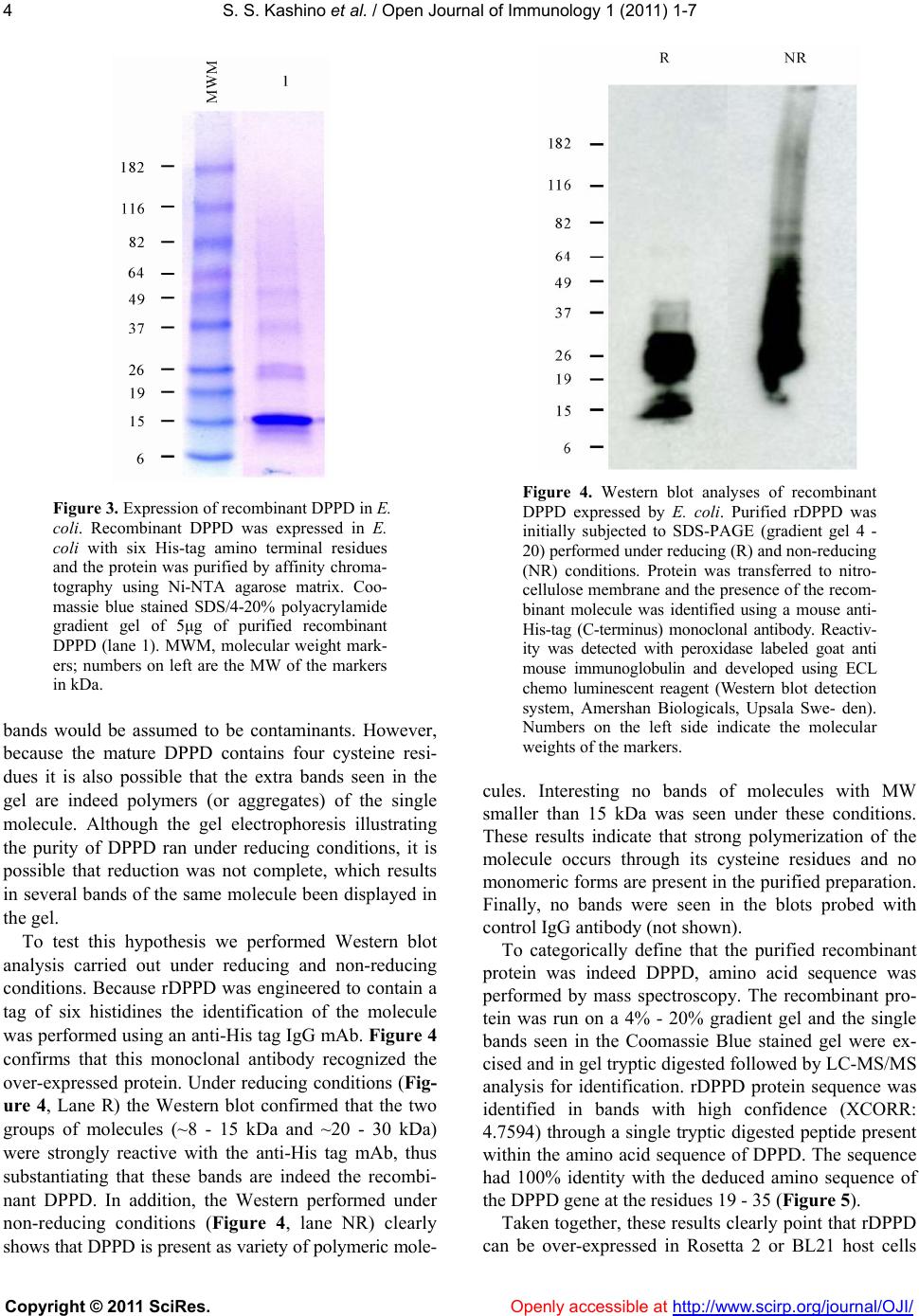 S. S. Kashino et al. / Open Journal of Immunology 1 (2011) 1-7 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/OJI/ 4 Figure 3. Expression of recombinant DPPD in E. coli. Recombinant DPPD was expressed in E. coli with six His-tag amino terminal residues and the protein was purified by affinity chroma- tography using Ni-NTA agarose matrix. Coo- massie blue stained SDS/4-20% polyacrylamide gradient gel of 5μg of purified recombinant DPPD (lane 1). MWM, molecular weight mark- ers; numbers on left are the MW of the markers in kDa. bands would be assumed to be contaminants. However, because the mature DPPD contains four cysteine resi- dues it is also possible that the extra bands seen in the gel are indeed polymers (or aggregates) of the single molecule. Although the gel electrophoresis illustrating the purity of DPPD ran under reducing conditions, it is possible that reduction was not complete, which results in several bands of the same molecule been displayed in the gel. To test this hypothesis we performed Western blot analysis carried out under reducing and non-reducing conditions. Because rDPPD was engineered to contain a tag of six histidines the identification of the molecule was performed using an anti-His tag IgG mAb. Figure 4 confirms that this monoclonal antibody recognized the over-expressed protein. Under reducing conditions (Fig- ure 4, Lane R) the Western blot confirmed that the two groups of molecules (~8 - 15 kDa and ~20 - 30 kDa) were strongly reactive with the anti-His tag mAb, thus substantiating that these bands are indeed the recombi- nant DPPD. In addition, the Western performed under non-reducing conditions (Figure 4, lane NR) clearly shows that DPPD is present as variety of polymeric mole- Figure 4. Western blot analyses of recombinant DPPD expressed by E. coli. Purified rDPPD was initially subjected to SDS-PAGE (gradient gel 4 - 20) performed under reducing (R) and non-reducing (NR) conditions. Protein was transferred to nitro- cellulose membrane and the presence of the recom- binant molecule was identified using a mouse anti- His-tag (C-terminus) monoclonal antibody. Reactiv- ity was detected with peroxidase labeled goat anti mouse immunoglobulin and developed using ECL chemo luminescent reagent (Western blot detection system, Amershan Biologicals, Upsala Swe- den). Numbers on the left side indicate the molecular weights of the markers. cules. Interesting no bands of molecules with MW smaller than 15 kDa was seen under these conditions. These results indicate that strong polymerization of the molecule occurs through its cysteine residues and no monomeric forms are present in the purified preparation. Finally, no bands were seen in the blots probed with control IgG antibody (not shown). To categorically define that the purified recombinant protein was indeed DPPD, amino acid sequence was performed by mass spectroscopy. The recombinant pro- tein was run on a 4% - 20% gradient gel and the single bands seen in the Coomassie Blue stained gel were ex- cised and in gel tryptic digested followed by LC-MS/MS analysis for identification. rDPPD protein sequence was identified in bands with high confidence (XCORR: 4.7594) through a single tryptic digested peptide present within the amino acid sequence of DPPD. The sequence had 100% identity with the deduced amino sequence of the DPPD gene at the residues 19 - 35 (Figure 5). Taken together, these results clearly point that rDPPD can be over-expressed in Rosetta 2 or BL21 host cells  S. S. Kashino et al. / Open Journal of Immunology 1 (2011) 1-7 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/OJI/ 5 Figure 5. DPPD peptide sequence identified by mass spec- troscopy in purified recombinant protein. The peptide sequence and positioning within the peptide donor protein (DPPD) is highlighted in red/bold/underline. The trypsin cleavage sites (right side of the amino acids R and K) that generated the pep- tide are illustrated above the molecule. transformed with pET-14b containing the DPPD gene with optimized sequence for E. coli. 3.3. Biological Validation of Purified rDPPD One important requirement to validate a microbial molecule as a diagnostic tool or as a vaccine candidate is that the immune response of a host sensitized or infected with the microbe donor of that protein recognizes that molecule. Our former studies using rDPPD expressed as fusion protein have confirmed that guinea pigs and hu- mans sensitized or infected with M. tuberculosis develop specific T cell response to DPPD. Therefore, it became important to verify that the rDPPD molecule expressed and purified in the present work would also be recog- nized by T cells from M. tuberculosis sensitized indi- viduals. To test this requirement, PBMC were obtained from six healthy volunteers who were known to be re- sponders to purified protein derivative (PPD) of tubercu- lin, the antigen that is used in the human skin test for the diagnosis of tuberculosis. Recognition of rDPPD was tested by antigen-induced production of IFN-γ by the donors’ PBMC. As it can be seen in Figure 6, rDPPD stimulated the PBMC of all six tuberculin sensitive do- nors to produce high levels of IFN-γ. No response was observed with PBMC obtained from three PPD non- responder controls (not shown). Due to the limited number of individuals analyzed these results cannot at this point be correlated with clinical validation of rDPPD as a tool to be used in the diagnosis of tuberculosis. However, because rDPPD isreadily recognized by PBMC of tuberculosis sensitized individuals the results clearly indicate that the newly expressed and purified recombinant molecule is biologically active. 4. DISCUSSION I early studies we showed that a recombinant fusion molecule composed of rRa12-DPPD elicited delayed type hypersensitivity in humans comparable to that elic- ited by standard PPD antigen [3]. Unfortunately, this fusion protein contains a 14 kDa polypeptide from M. tuberculosis which, in contrast to DPPD, is broadly dis- Figure 6. Recognition of purified recombinant DPPD by hu- man PBMC. IFN-γ production by PBMC from PPD positive healthy donors following stimulation for 72 h with medium, rDPPD (10 μg/ml) or PPD (10 μg/ml) was measured by sand- wich ELISA in the culture supernatants. Bars represent the SD of the means calculated from the results of triplicate cultures. tributed among the Mycobacterium genus. If on one hand we were successful to generate a purified recom- binant molecule, on the other hand it introduced an un- desired property to the rDPPD i.e. , a fusion protein that is no longer specific for M. tuberculosis. However, this “homemade” fusion protein expression system [15] was the only procedure that we found to be successful to produce rDPPD. A variety of commercially available systems that uses E. coli as host cells consistently failed to express rDPPD. These included various vectors such pET, pQ30 (Qiagen), pThioHis (Invitrogen), and pGEX- 2T (GE Healthcare, Piscataway, New Jersey). Host E. coli cells tested included BL21(DE3), BL21(DE3)pLysS, and Rosetta 2(DE)pLysS (Novagen). In addition, rDPPD could not be expressed using other host cells such as the yeast Pichia pastoris or Streptococcus lividans (unpub- lished observations). However, as mentioned before, low levels of rDPPD could be expressed and purified by using the Mycobac- terium expression vector pSMT3 and Mycobacterium smegmatis as host cell [10]. This observation prompted us to hypothesize that preferential codon usage could have been the condition that facilitated the synthesis of a M. tuberculosis protein in a Mycobacterium host cell. Here, we tested this possibility using a standard expres- sion vector (pET14b) and standard E. coli host cells (Rosetta 2(DE3)pLysS or BL21(DE3)pLysS), which are a well known systems designed for high levels of protein production. The technological advent of achieving ro- bust and automated synthesis of large DNA molecules permitted us to produce a DPPD gene containing a se- quence designed to match the preferential codon usage of E. coli instead of that of Mycobacterium. The E. coli  S. S. Kashino et al. / Open Journal of Immunology 1 (2011) 1-7 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/OJI/ 6 optimized DPPD gene was synthesized by BlueHeron Biotechnology, Seattle WA. Blue Heron uses proprietary codon utilization databank, geneassembly instruments, and other technologies to accurately and rapidly engi- neer and assemble oligonucleotides into full-length con- structs. Using BL21 or Rosetta 2 E. coli transformed with pET14b containing the optimized DPPD gene we were able to successfully express and purify rDPPD in yields never before achieved. Approximately 5 - 10 mg of purified recombinant was consistently obtained per liter of bacterial culture. The deduced MW of mature DPPD molecule (9,022.9 Da) does not agree with the molecular mass of ~12 kDa of the major band of the purified molecule estimated by SDS-PAGE. However, this same pattern of migration was observed with the native molecule when we first discovered DPPD [4]. This phenomenon of abnormal gel migration has been described for several proteins that have an unusually high proline content and a low iso- electric point caused by high contents of aspartic and glu- tamic acid residues [12-14]. In general, the classical SDS- PAGE method often overestimates molecular weights of molecules if the proline content is >10% in a given pro- tein [11]. The general consensus is that this altered mi- gration pattern is caused by the amino acid composition, and not post-translational modification [14]. Coinciden- tally DPPD fits all these predictive parameters. Out of the 84 amino acids that compose the mature form of the molecule, 16 (19%) are prolines, 7 (8.3%) are aspartic acid and 3 (3.6%) are glutamic acid. Moreover, the theo- retical pI of the mature DPPD protein is 4.2, which was obtained using the ExPASy Proteomics algorithm of The Swiss Institute of Informatics (http://au.exp asy.org/tools/ protparam.html). Therefore, these unique molecular cha- racteristics of the mature rDPPD are in consonance with its pattern of migration observed in the PAGE analysis. Importantly, the 12 kDa band seen in the PAGE was confirmed to be DPPD by mass spectroscopy. Also interesting was the confirmation that the purified recombinant DPPD was readily recognized by the PBMC obtained from tuberculosis sensitized healthy individuals. These observations point to the potential use of this sin- gle and defined molecule as a potential reagent for the tuberculin skin test. Alternatively, rDPPD can be also an interesting molecule to be tested as component of the recently developed whole blood IFN-γ release assay for the diagnosis of tuberculosis. As mentioned before, this molecule is unique to members of the M. tuberculosis complex only, therefore an attractive specific antigen for test development. Finally, it is important to emphasize that the procedure described in this manuscript to achieve workable con- centrations of rDPPD per liter of bacterial culture, uses conventional standard operating procedures for produc- tion of recombinant proteins. Therefore, if rDPPD, in future experiments, proves to be useful for the diagnosis of tuberculosis, no hurdles should exist to upscalethe production of this molecule under GLP or GMP condi- tions for clinical use. 5. ABBREVIATIONS DPPD, a difficult to express protein of Mycobacte- rium tuberculosis. 6. CONFLICT OF INTEREST None of the authors has any financial conflict of in- terest. REFERENCES [1] Jana, S. and Deb, J.K. (2005) Strategies for efficient production of heterologous proteins in Escherichia coli. Applied Microbiology and Biotechnology, 67, 289-298. doi:10.1007/s00253-004-1814-0 [2] Brondyk, W.H. (2009) Selecting an appropriate method for expressing a recombinant protein. Methods in Enzy- mology, 463, 131-147. doi:10.1016/S0076-6879(09)63011-1 [3] Campos-Neto, A., Rodrigues-Junior, V., Pedral-Sampaio, D.B., Netto, E.M., Ovendale, P.J., Coler, R.N., Skeiky, Y.A., Badaro, R., Reed. S.G., (2001) Evaluation of DPPD, a single recombinant Mycobacterium tuberculosis protein as an alternative antigen for the Mantoux test. Tuberculo- sis (Edinb), 81, 353-358. doi:10.1054/tube.2001.0311 [4] Coler, R.N., Skeiky, Y.A., Ovendale, P.J., Vedvick, T.S., Gervassi, L., Guderian, J., Jen, S., Reed, S.G. and Cam- pos-Neto, A. (2000) Cloning of a Mycobacterium tuber- culosis gene encoding a purified protein derivative pro- tein that elicits strong tuberculosis-specific delayed-type hypersensitivity. Journal of Infectious Diseases; 182, 224-233. doi:10.1086/315677 [5] I. Schiller, H.M. Vordermeier, W.R. Waters, A.O. Whelan, M. Coad, E. Gormley, B.M. Buddle, M. Palmer, T. Thacker, J. McNair, M. Welsh, R.G. Hewinson, B. Oesch. (2010) Bovine tuberculosis: Effect of the tuberculin skin test on in vitro interferon gamma responses. Veterinary Immunology and Immunopathology, 136, 1-11. doi:10.1016/j.vetimm.2010.02.007 [6] Lee, E. and Holzman, R.S. (2002) Evolution and current use of the tuberculin test. Clinical Infectious Diseases, 34, 365-370. doi:10.1086/338149 [7] Rangel-Frausto, M.S., Ponce-De-Leon-Rosales, S., Mar- tinez-Abaroa, C. and Haslov, K. (2001) Tuberculosis and tuberculin quality: Best intentions, misleading results. Infection Control and Hospital Epidemiol, 22, 481-484. doi:10.1086/501937 [8] Villarino, M.E., Brennan, M.J., Nolan, C.M., Catanzaro, A., Lundergan, L.L., Bock, N.N., Jones, C.L., Wang, Y.C. and Burman, W.J. (2000) Comparison testing of current (PPD-S1) and proposed (PPD-S2) reference tuberculin standards. American Journal of Respiratory and Critical  S. S. Kashino et al. / Open Journal of Immunology 1 (2011) 1-7 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/OJI/ 7 Care Medicine, 161, 1167-1171. [9] Whipple, D.L., Palmer, M.V., Slaughter, R.E. and Jones, S.L. (2001) Comparison of purified protein derivatives and effect of skin testing on results of a commercial gamma interferon assay for diagnosis of tuberculosis in cattle. Journal of Veterinary Diagnostic Investigation, 13, 117-122. doi:10.1177/104063870101300204 [10] Liu, C., Flamoe, E., Chen, H.J., Carter, D., Reed, S.G., Campos-Neto, A., (2004) Expression and purification of immunologically reactive DPPD, a recombinant Myco- bacterium tuberculosis skin test antigen, using Mycobac- terium smegmatis and Escherichia coli host cells. Cana- dian Journal of Microbiology, 50, 97-105. doi:10.1139/w03-109 [11] Furthmayr, H. and Timpl, R. (1971) Characterization of collagen peptides by sodium dodecylsulfate-poly- acry- lamide electrophoresis. Analytical Biochemistry, 41, 510- 516. doi:10.1016/0003-2697(71)90173-4 [12] Marrs, J.A. and Bouck, G.B. (1992) The two major mem- brane skeletal proteins (articulins) of Euglena gracilis de- fine a novel class of cytoskeletal proteins. The Journal of Cell Biology, 11 8,1465-1475. doi:10.1083/jcb.118.6.1465 [13] Ogle, K.F., Lee, K.K. and Krause, D.C. (1992) Nucleo- tide sequence analysis reveals novel features of the phase-variable cytadherence accessory protein HMW3 of Mycoplasma pneumoniae. Infection and Immunity, 60, 1633-1641. [14] Proft, T., Hilbert, H., Layh-Schmitt, G. and Herrmann, R. (1995) The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insolu- ble fraction and exhibits size polymorphism in the strains M129 and FH. The Journal of Bacteriology, 177, 3370-3378. [15] Wang, A., Clapper, J., Guderian, J.A., Foy, T.M., Fanger, G.R., Retter, M.W. and Skeiky, Y.A. (2003) A novel method for increasing the expression level of recombi- nant proteins. Protein Expression and Purification, 30, 124-133. doi:10.1016/S1046-5928(03)00075-5 |

