 American Journal of Plant Sciences, 2011, 2, 255-261 doi:10.4236/ajps.2011.22027 Published Online June 2011 (http://www.SciRP.org/journal/ajps) Copyright © 2011 SciRes. AJPS Biodiversity in a Tomato Germplasm for Free Amino Acid and Pigment Content of Ripening Fruits Guillermo Raúl Pratta1,2, Gustavo Rubén Rodríguez1,2, Roxana Zorzoli2,3, Liliana Amelia Picardi2,3, Estela Marta Valle1,4 1National Council for Scientific and Technical Research, Buenos Aires, Argentina; 2 Chair of Genetics, Agronomic Sciences Faculty, National University of Rosario, Zavalla, Argentine; 3Council for Research of the National University of Rosario, Zavalla, Argentine; 4 Institute of Molecular and Cell Biology of Rosario, CONICET/Biochemical and Pharmaceutical Sciences Faculty, Suipacha, Rosario, Argentine. Email: gpratta@unr.edu.ar, gpratta@conicet.gov.ar Received March 8th, 2011; revised March 25th, 2011; accepted May 6th, 2011. ABSTRACT Free amino acid and pigment composition in fruits at two ripening stages from a selected tomato germplasm was stud- ied. The aims were contributing to knowledge on variability of ripening metabolism and identifying more consistently the genetic background of the plant material under analysis. Significant differences (p < 0.05) were found among rip- ening stages and among genotypes within ripening stage for all amino acids and pigments except by asparagine, alanine and chlorophyll b contents. The highest relative amino acid content corresponded to glutamate, glutamine, and GABA though some genotypes had relatively high asparagine content. Glutamate, glutamine and GABA performed op- positely: the former increa sed along ripening while the latter two decreased in their relative content. A Principal Com- ponents (PC) analysis was applied, determining that metabolites hav ing the greatest contribution to genera l variability were threonine, serine, glutamate, glutamine, glycine, isoleucine, leucine, tyrosine, phenylalanine, lycopene and beta- carotene, which showed the highest association with PC1. Alanine and chlorophylls a and b were highly associated to PC2. These two first PC explained the 62% of the to tal variation, and genotypes were distributed according to the rip- ening stage in their coordinates. Accordingly, a Hierarchical Clustering resulted in a dendrogram having a relatively high cophenetic correlation (0.70), in which two well defined groups were obtained according to ripening stage. These results verified the existence of variability in the metabolism of ripening fruit for amino acids and pigments, and al- lowed to identify unequivocally a set of selected tomato germplasm according to the fruit metabolic profiles in these two ripening stages. Keywords: Solanum Section Lycopersicon, Plant Breeding , Plant Genetic Resources, Multivariate Analysis 1. Introduction From an evolutionary viewpoint, tomato (Solanum ly- copersicum) ripening could be considered as a transition from a green stage that prevents fruit consumption (hence protecting the developing seeds) to a ripe stage in which its attributes are optimum to attract predators, which consume fruit and help to disperse mature seeds [1]. This transition includes morphological, biochemical and phy- siological changes that lead to acquisition of appropriate color, texture, flavor, among other traits determining fruit quality. Some of these changes are variations in free amino acids and pigment composition [2,3]. Glutamate percent content noticeably increased between mature green and red ripe stages, simultaneously to a reduction in glutamine and GABA levels in tomato varieties [4]. From a productive viewpoint, the conservation of red ripe attributes during a longer time prolongs the opportu- nity of fruit commercialization, especially for the fresh market [5]. Shelf life is a measure for the period of tomato quality adequate maintenance, and has been reported as negatively correlated to glutamate relative molar content of ripe mature fruits [6]. Long shelf life tomatoes have been currently obtained by introgressing spontaneous ripening mutant genes such as nor, rin, alc and Nr, or by genetic transformation [7]. Both strategies have disadvantages,  Biodiversity in a Tomato Germplasm for Free Amino Acid and Pigment Content of Ripening Fruits 256 because spontaneous mutations present pleiotropic effects that diminish fruit quality and transgenic food are not well accepted by public opinion even presently [5]. Exotic germplasm of Solanum section Lycopersicon comprises S. lycopersicum var. cerasiforme, the closest relative S. pimpinellifolium, and other 10 wild species. They are invaluable plant genetic resources contributing abiotic and biotic resistance or tolerance genes but also for increasing fruit quality and prolonging shelf life [8,9]. Seventeen recombinant inbred lines from an interspecific cross S. lycopersicum cv. Caimanta x S. pimpinellifolium LA722 were obtained after six selfing cycles with an- tagonistic-divergent selection for fruit weight and shelf life [10] and characterized by quantitative fruit traits, total pericarp polypeptide profiles and AFLP markers [11,12]. A wide variation was found for all analyzed phenotypic and molecular attributes. The general goal of this research was to study free amino acid and pigment composition in this selected tomato germplasm, with the aims of contributing to know- ledge on variability of ripening metabolism, identifying more consistently the RILs genetic background, and verifying associations between glutamate content and fruit shelf life. 2. Materials and Methods 2.1. Plant Material Ten seeds of RILs 4, 10, 12 and 15 together with the experimental testers, parents Caimanta and LA722 (Fig- ure 1), were sown in seedling trays on August under a glasshouse. Then plants were grown at the experimental field station “José F. Villarino” located at latitude 33°S and longitude 61°W, from October to March under greenhouse conditions. Previous to the transplantation, the soil (a typical argiudol) was fertilized with poultry grit. The crop was watered twice a week, levels of irrigation that were sufficient to avoid water stress during the plants growing period. Mean values of fruit shelf life (in days) were: Caimanta = 13.00, LA722 = 18.60, RIL4 = 16.21, RIL 10 = 21.17, RIL12 = 17.41 and RIL15 = 12.79 (av- eraged from [10] and [12]). 2.2. Determination of Amino Acids and Pigment Composition Samples of tomato fruits (four per line) were harvested at the mature green stage (MG, when fruit is green but stops growing) and red ripe stage (RR, when fruit is completely red but still firm). Pericarp tissue of harvested fruits was obtained by removing the locule tissue and seeds and then they were stored at –80˚C until analysis [2]. Pericarp tissue (0.5 - 1 g fresh weight) was extracted with 5 ml chloro form/methanol (1.5/3.5 v/v) and the amino acids relative Figure 1. Fruits of four tomato Recombinant Inbred Lines (RIL) and their parents Solanum lycopersicum cv. Caimanta and S. pimpinellifolium LA722, the experimental testers. composition was determined in the methanolic phase by derivatization with ninhydrin or o-phthaldialdehyde using an amino acids analyzer following [4]. Pigments were determined according to [3]. 2.3. Statistical Analysis Distribution’s normality of total free amino acids and pigment contents (in μmol·mg–1 fresh weight and mg·100 g–1 fresh weight, respectively) and each amino acid per- cent composition was analyzed following [13]. Com- parisons were made by a hierarchical ANOVA for vari- ables having normal distribution, in which the principal source of variation was ripening stage and the nested source of variation was genotype within ripening stage. Kruskall-Wallis test was applied for variables displaying not normal distribution. Mean values were calculated from three independent experiments in all cases. The Pearson correlation coefficients (r) among all variables (including the averaged shelf life values) were calculated [14]. Multivariate Analysis of Principal Components and Hierarchical Clustering with Ward method and Euclidean distances were used to identify metabolites (amino acids and pigments) mostly contributing to general variability and to assess the importance of each source of variation (genotype and ripening stage) in categorizing the obtained set of results [15] to get a data mining approach on me- tabolic changes occurring during tomato ripening. 3. Results Mean values of each amino acid relative molar content, and total amino acid and pigment contents of the pericarp fruit at MG and RR stages of the four RILs and the parents are in Table 1. All variables, except by total free amino Copyright © 2011 SciRes. AJPS 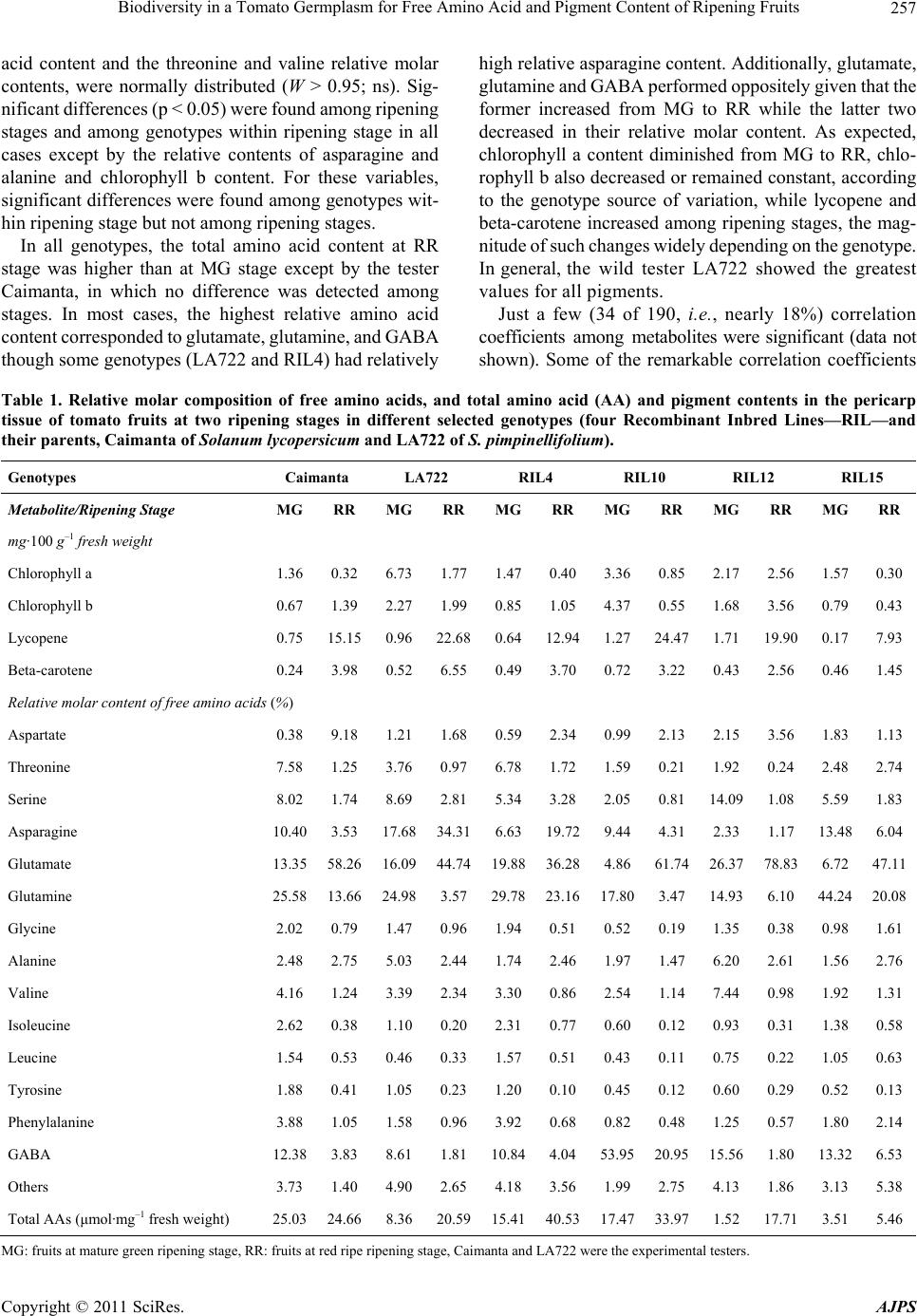 Biodiversity in a Tomato Germplasm for Free Amino Acid and Pigment Content of Ripening Fruits Copyright © 2011 SciRes. AJPS 257 acid content and the threonine and valine relative molar contents, were normally distributed (W > 0.95; ns). Sig- nificant differences (p < 0.05) were found among ripening stages and among genotypes within ripening stage in all cases except by the relative contents of asparagine and alanine and chlorophyll b content. For these variables, significant differences were found among genotypes wit- hin ripening stage but not among ripening stages. In all genotypes, the total amino acid content at RR stage was higher than at MG stage except by the tester Caimanta, in which no difference was detected among stages. In most cases, the highest relative amino acid content corresponded to glutamate, glutamine, and GABA though some genotypes (LA722 and RIL4) had relatively high relative asparagine content. Additionally, glutamate, glutamine and GABA performed oppositely given that the former increased from MG to RR while the latter two decreased in their relative molar content. As expected, chlorophyll a content diminished from MG to RR, chlo- rophyll b also decreased or remained constant, according to the genotype source of variation, while lycopene and beta-carotene increased among ripening stages, the mag- nitude of such changes widely depending on the genotype. In general, the wild tester LA722 showed the greatest values for all pigments. Just a few (34 of 190, i.e., nearly 18%) correlation coefficients among metabolites were significant (data not shown). Some of the remarkable correlation coefficients Table 1. Relative molar composition of free amino acids, and total amino acid (AA) and pigment contents in the pericarp tissue of tomato fruits at two ripening stages in different selected genotypes (four Recombinant Inbred Lines—RIL—and their parents, Caimanta of Solanum lycopersicum and LA722 of S. pimpinellifolium). Genotypes Caimanta LA722 RIL4 RIL10 RIL12 RIL15 Metabolite/Ripening Stage MG RR MGRR MGRR MGRR MG RR MGRR mg·100 g–1 fresh weight Chlorophyll a 1.36 0.32 6.73 1.77 1.47 0.40 3.36 0.852.17 2.56 1.570.30 Chlorophyll b 0.67 1.39 2.27 1.99 0.85 1.05 4.37 0.551.68 3.56 0.790.43 Lycopene 0.75 15.15 0.96 22.68 0.64 12.94 1.27 24.47 1.71 19.90 0.177.93 Beta-carotene 0.24 3.98 0.52 6.55 0.49 3.70 0.723.22 0.43 2.56 0.461.45 Relative molar content of free amino acids (%) Aspartate 0.38 9.18 1.21 1.68 0.59 2.34 0.99 2.132.15 3.56 1.831.13 Threonine 7.58 1.25 3.76 0.97 6.781.72 1.590.21 1.92 0.24 2.482.74 Serine 8.02 1.74 8.69 2.81 5.34 3.28 2.05 0.8114.09 1.08 5.591.83 Asparagine 10.40 3.53 17.6834.316.6319.729.444.312.33 1.17 13.486.04 Glutamate 13.35 58.26 16.09 44.74 19.88 36.284.8661.74 26.37 78.83 6.7247.11 Glutamine 25.58 13.6624.983.5729.7823.1617.803.4714.93 6.10 44.2420.08 Glycine 2.02 0.79 1.47 0.96 1.940.51 0.520.19 1.35 0.38 0.981.61 Alanine 2.48 2.75 5.03 2.44 1.74 2.461.97 1.476.20 2.61 1.562.76 Valine 4.16 1.24 3.39 2.34 3.30 0.862.54 1.147.44 0.98 1.921.31 Isoleucine 2.62 0.38 1.10 0.20 2.310.77 0.600.12 0.93 0.31 1.380.58 Leucine 1.54 0.53 0.46 0.33 1.570.51 0.430.11 0.75 0.22 1.050.63 Tyrosine 1.88 0.41 1.05 0.23 1.20 0.100.45 0.120.60 0.29 0.520.13 Phenylalanine 3.88 1.05 1.58 0.96 3.92 0.680.82 0.481.25 0.57 1.802.14 GABA 12.38 3.83 8.611.81 10.84 4.04 53.9520.9515.56 1.80 13.326.53 Others 3.73 1.40 4.90 2.65 4.18 3.56 1.99 2.75 4.13 1.86 3.135.38 Total AAs (μmol·mg–1 fresh weight) 25.03 24.668.3620.5915.4140.5317.4733.971.52 17.71 3.515.46 M G: fruits at mature green ripening stage, RR: fruits at red ripe ripening stage, Caimanta and LA722 were the experimental testers.  Biodiversity in a Tomato Germplasm for Free Amino Acid and Pigment Content of Ripening Fruits 258 were those between glutamate and glutamine (r = –0.79, p < 0.01), threonine, glycine, isoleucine, leucine, tyrosine and phenylalanine (r > 0.75, p < 0.001 in all cases), serine, alanine and valine (r > 0.72, p < 0.01 in all cases), iso- leucine and lycopene (r = –0.72, p < 0.01), and lycopene and beta-carotene (r = 0.87, p < 0.001). On the other hand, GABA, asparagine, and chlorophyll a and b had no sig- nificant correlation with any metabolite, though chloro- phyll b at MG was positively associated with fruit shelf life (r = 0.90, p < 0.01). Other metabolites associated with this latter trait were leucine at RR (r = –0.92, p < 0.01), alanine at RR (r = –0.85, p < 0.05) and lycopene at RR (r = 0.88, p < 0.05) but the previously reported correlation with glutamate at RR [6] was not confirmed in this ex- periment (r = 0.21, ns). The two first Principal Components (PC1 and PC2) explained the 62% of the total variation in metabolite composition of MG and RR tomato fruits. If PC3 was also considered, this percentage increases to 72% but only PC1 and PC2 will be further analyzed. Metabolite contribu- tions to PC1 and PC2 (i.e., the respective eigenvalues and the association among each original variable and both PCs) are shown in Table 2. The metabolites having the more remarkable correlation coefficients (threonine, serine, glutamate, glutamine, glycine, isoleucine, leucine, tyro- sine, phenylalanine, lycopene and beta-carotene) also had the greatest contribution to general variability and the highest association with PC1. Alanine and chlorophylls a and b were highly associated to PC2. Figure 2 shows the distribution of genotypes by ripening stage (and also of each metabolite) in the PC1 and PC2 coordinates. A clear separation by ripening stage was obtained in the axis of PC1, which explained the greatest proportion of total variability (46%, Table 2). It should be considered as an extent of variability according to ripening metabolism, indicating that threonine, serine, valine, glutamine, gly- cine, isoleucine, leucine, tyrosine, and phenylalanine relative molar content were higher at MG while glutamate relative content and lycopene and beta-carotene contents were higher at RR, though there were some genotype exceptions (RIL10 MG and RIL15 RR, Figure 2). PC2, which explained a lesser proportion of general variability (16%), better accounted for genotype biodiversity, and separations in this axis are more noticeable at the MG stage (Figure 2). Hence, genotypic differences are less important at RR. Hierarchical Clustering confirmed PC results. The dendrogram had a relatively high cophenetic correlation (0.70) and two well defined groups were obtained accord- ing to ripening stage, one including all genotypes at MG and RIL15 at RR, and the other, the remaining genotypes at RR (Figure 3). In the first group, separation among genotypes occurred at a higher distances that in the second Table 2. Eigenvalue composition of Principal Components 1 and 2 (PC1 and PC2, respectively) and associations among metabolites and each Principal Component (a1 and a2, respectively). Metabolite PC1 a1 PC2 a2 Aspartate (%) –0.18 –0.54 –0.05 -–0.09 Threonine (%) 0.29 0.89 –0.20–0.35 Serine (%) 0.24 0.72 0.24 0.43 Asparagine (%) –0.02 –0.07 –0.04 –0.06 Glutamate (%) –0.25 –0.77 –0.11 –0.20 Glutamine (%) 0.24 0.72 –0.10–0.17 Glycine (%) 0.29 0.88 –0.10–0.18 Alanine (%) 0.09 0.27 0.39 0.69 Valine (%) 0.21 0.65 0.27 0.49 Isoleucine (%) 0.30 0.91 –0.18–0.32 Leucine (%) 0.29 0.86 –0.24–0.42 Tyrosine (%) 0.27 0.83 –0.07–0.12 Phenylalanine (%) 0.28 0.85 v0.25–0.45 GABA (%) 0.03 0.09 0.24 0.42 Others (%) 0.21 0.64 0.01 0.02 Total AA (μmol·mg–1) –0.16 –0.48 –0.28 –0.50 Chlorophyll a (mg·100 g–1) 0.09 0.26 0.41 0.72 Chlorophyll b (mg·100 g–1) –0.09 –0.28 0.38 0.68 Lycopene (mg·100 g–1) –0.29 –0.89 –0.13 –0.24 Beta-carotene (mg·100 g–1) –0.26 –0.79 –0.15 –0.26 group, RIL10 being a discrepant genotype at MG. RIL15 had the minor differences between ripening stages, while the most discrepant genotype at RR was the tester Cai- manta, the cultivated parent. 4. Discussion As previously reported, a wide range of variability was found in this experiment for tomato pericarp free amino acid composition, and total amino acids and pigments content [6,7,8]. Greatest variability was detected among mature green and red ripe stages than among genotypes, indicating that these attributes are primarily ripening- regulated [16] and constitute one of the biological changes taking place during such a transition [17]. In fact, as demonstrated by Hierarchical Clustering, ripening stages could be identified by each amino acid relative molar composition, and total free amino acids and pigments content. For the latter attributes, association is visually Copyright © 2011 SciRes. AJPS 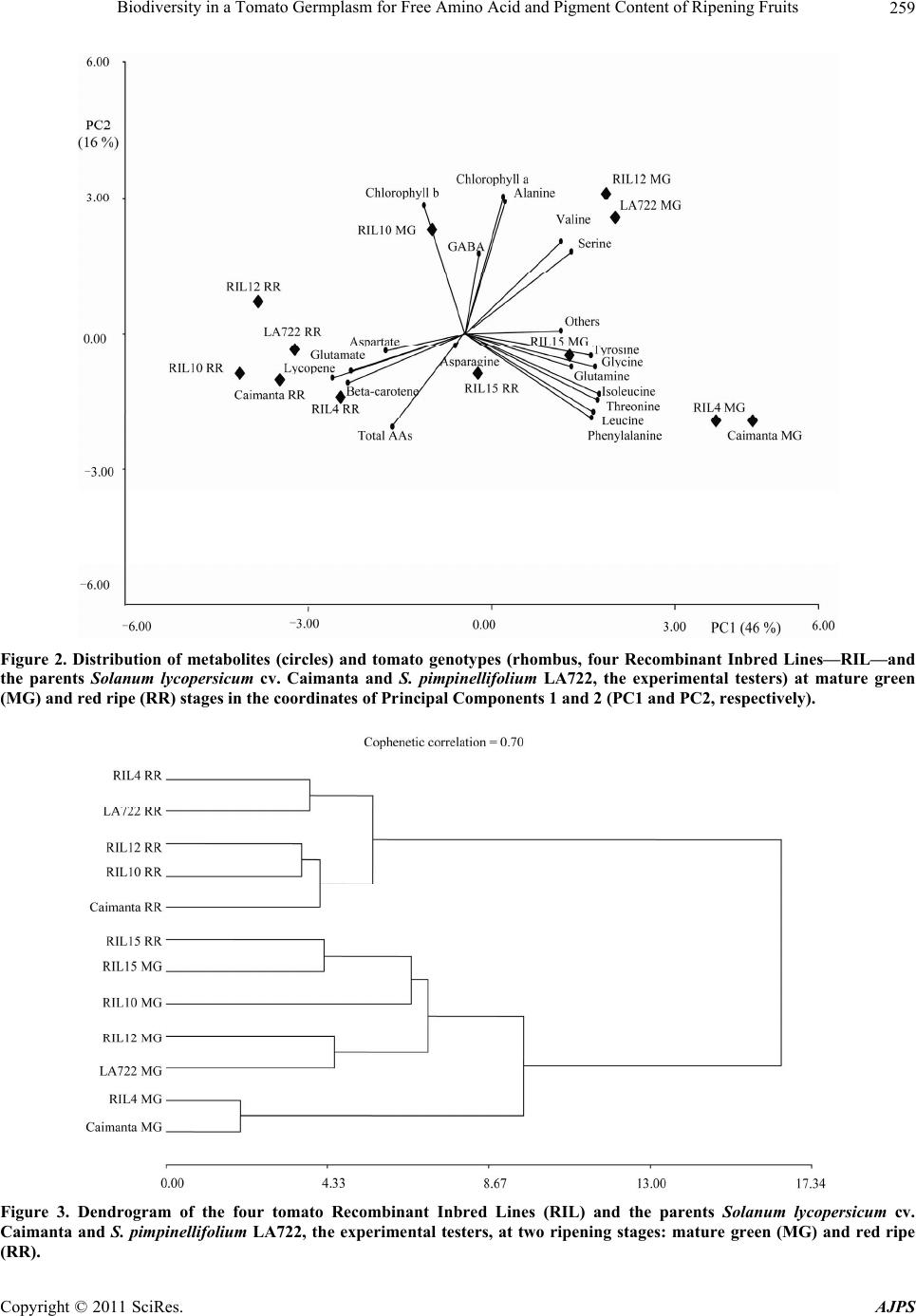 Biodiversity in a Tomato Germplasm for Free Amino Acid and Pigment Content of Ripening Fruits259 Figure 2. Distribution of metabolites (circles) and tomato genotypes (rhombus, four Recombinant Inbred Lines—RIL—and the parents Solanum lycopersicum cv. Caimanta and S. pimpinellifolium LA722, the experimental testers) at mature green (MG) and red ripe (RR) stages in the coordinates of Principal Components 1 and 2 (PC1 and PC2, respectively). Figure 3. Dendrogram of the four tomato Recombinant Inbred Lines (RIL) and the parents Solanum lycopersicum cv. Caimanta and S. pimpinellifolium LA722, the experimental testers, at two ripening stages: mature green (MG) and red ripe (RR). Copyright © 2011 SciRes. AJPS  Biodiversity in a Tomato Germplasm for Free Amino Acid and Pigment Content of Ripening Fruits 260 noticeable, but for amino acids composition, different physiological functions were proposed. Glutamate was proposed as a precursor of chlorophyll [18], hence the increase in its percent composition towards the end of ripening could be due to the cessation of chlorophyll bio- synthesis. On the other hand, another role was claimed for glutamate [17], given that even a mutant defective in chlorophyll degradation also shown the increase at ripe stage [3]. As an “umami taste” is provoked by glutamate, the higher glutamate relative molar content in all tomato germplasm analyzed ([19,20] in addition to the already mentioned authors) could have an attracting to mammal- ian predators function [21]. Interestingly, genotype bio- diversity was higher in fruits at mature green stage than at red ripe stage. As wild germplasm was analyzed here, this finding suggesting an evolutive conservation across dif- ferent genetic backgrounds of optimum attributes to at- tract fruit predators so assuring seed dispersion [7]. Cor- relation between glutamate composition at RR and fruit shelf life proposed by [6] was not confirmed in this ex- periment, probably due to differences in gene frequencies among populations studied in both reports. Spontaneous mutant homozygote genotypes having marked pleiotropic effects on tomato many ripening associated traits were studied in that report, which could explain the significant correlation found in that study. However, significant as- sociations of shelf life with pigments (chlorophyll b at mature green and lycopene at red ripe stages) and other amino acids (leucine and alanine at red ripe stages) were detected in this set of recombinant inbred lines and their parents. This interesting finding should be further ex- plored, since the correlation coefficients were high and positive with pigments, and high and negative with amino acids. Accordingly, the wild parent LA722 was the genotype contributing the highest values of both pigments and fruit shelf life, and the lesser values of both amino acids at the corresponding ripening stages. Other important modification in amino acid metabolism during tomato ripening transition was the increase in asparagine levels in the wild tester LA722, also shown by RIL4. In previous reports [2-4,6], asparagine decreased from mature green to red ripe in different tomato geno- types (normal and mutant for ripening cultivars, the wild variety S. lycopersicum var. cerasiforme) as well as glutamine and GABA. These two latter were the most abundant amino acids together with the previously men- tioned glutamate. The different performance of asparagine in LA722 and RIL4 was similar to the currently observed performance of glutamate in most tomato genotypes. Such an increase in glutamate was even observed in LA722 and RIL4, though the ratio glutamate at red ripe: glutamate at mature green stages was lower in both genotypes than in the remaining ones, hence alternative metabolic functions related to the ammonium assimilation/dissimilation cycle [16] during ripening transition could be taking place in the wild germplasm. The alternative function and parallel performance of glutamate/asparagine in these genotypes would also contribute to identify more consistently the RILs genetic background, since one of them were more similar to the wild parent while the others performed as the cultivated Caimanta, which evidences gene segrega- tion and recombination during the selection process. Ad- ditionally, the lack of association among glutamate con- tent and fruit shelf life reported by [6] could also be ex- plained by this evident variability in ripening amino acids metabolism. Finally, the great contribution of threonine, serine, valine, glycine, isoleucine, tyrosine, and phenylalanine relative molar content to variability among ripening stages, detected by both single correlation and Principal Com- ponent analyses, deserves further studies to elucidate its physiological meaning, since there are no antecedents in their contribution to tomato ripening. REFERENCES [1] J. J. Giovannoni “Genetic Regulation of Fruit Develop- ment and Ripening,” Plant Cell, Vol. 16, Supplement 1, September 2004, pp. 170-180. doi:10.1105/tpc.019158 [2] E. M. Valle, S. B. Boggio and H. W. Heldt, “Free Amino Acids Composition of Phloem Sap and Growing Fruit of Lycopersicon Esculentum,” Plant Cell Ph ysiology, Vol. 39, No. 4, June 1998, pp. 458-461. [3] S. Bortolotti, S. B. Boggio, L. Delgado, E. G. Orellano and E. M. Valle, “Different Induction Patterns of Glutamate Metabolising Enzymes in Ripening Fruits of the Tomato Mutant Green Flesh,” Physiologia Plantarum, Vol. 119, No. 3, May 2003, pp. 384-391. doi:10.1034/j.1399-3054.2003.00184.x [4] S. B. Boggio, J. F. Palatnik, H. W. Heldt and E. M. Valle, “Changes in the Aminoacid Composition and Nitrogen Metabolizing Enzymes in Ripening Fruits of Lycopersicon Esculentum Mill,” Plant Science, Vol. 159, No. 1, March 2000, pp. 125-133. doi:10.1016/S0168-9452(00)00342-3 [5] G. R. Rodríguez, G. R. Pratta, D. R. Liberatti, R. Zorzoli and L. A. Picardi, “Inheritance of Fruit Shelf Life and Other Tomato Fruit Quality Traits Estimated from Crosses and Backcrosses of Selected Parents,” Euphytica, Vol. 176, July 2010, pp. 137-147. [6] G. Pratta, R. Zorzoli, S. B. Boggio, L. A. Picardi and E. M. Valle, “Glutamine and Glutamate Levels and Related Metabolizing Enzymes in Tomato Genotypes with Dif- ferent Shelf-Life,” Scientia Horticulturae, Vol. 100, No. 1-4, April 2004, pp. 341-347. doi:10.1016/j.scienta.2003.08.004 [7] J. A. Labate, S. Grandillo, T. Fulton, S. Muños, A. L. Copyright © 2011 SciRes. AJPS  Biodiversity in a Tomato Germplasm for Free Amino Acid and Pigment Content of Ripening Fruits261 Caicedo, I. Peralta, Y. Ji, R. T. Chetelat, J. W. Scout, M. J. Gonzalo, D. Francis, W. Yang, E. van der Knaap, A. M. Baldo, B. Smith-White, L. A. Mueller, J. P. Prince, N. E. Blanchard, D. V. Storey, M. R. Stevens, M. D. Robbins, J. F. Wang, B. E. Liedl, M. A. O’Connell, J. R. Stommel, K. Auki, Y. Iijima, A. J. Slade, S. R. Hurst, D. Loeffler, M. N. Steine, D. Vafeados, C. McGuire, C. Freeman, A. Amen, J. Goodstal, D. Facciotti, J. van Eck and M. Causse, “To- mato,” In: C. Kole, Ed., Genome Mapping and Molecular Breeding in Plants, Vol. 5: Vegetables, Springer-Verlag, Berlin-Heidelberg, June 2007. [8] N. Schauer, D. Zamir and A. Fernie, “Metabolic Profiling of Leaves and Fruits of Wild Species Tomato: A Survey of the Solanum Lycopersicum Complex,” Journal of Ex- perimental Botany, Vol. 56, No. 410, April 2005, pp. 297-307 (Special Issue). [9] R. Zorzoli, G. Pratta and L. A. Picardi, “Variabilidad Para la Vida Postcosecha y El Peso de Los Frutos en Tomate Para Familias F3 de un HÍBrido InterespecÍFico,” Pes- quisa Agropecuaria Brasileira, Vol. 35, November 2000, pp. 2423-2427. doi:10.1590/S0100-204X2000001200013 [10] G. R. Rodriguez, G. R. Pratta, R. Zorzoli and L. A. Picardi, “Recombinant Lines Obtained from an Interspecific Cross among Lycopersicon Species Selected by Fruit Weight and Fruit Shelf Life,” Journal of the American Society for Horticultural Science, Vol. 131, No. 5, October 2006, pp. 651-656. [11] M. Gallo, G. R. Rodríguez, R. Zorzoli, L. A. Picardi and G. R. Pratta, “Proteómica de la Madurez del Tomate,” Revista de Investigaciones de la Facu lta d de Cien cias Ag rarias de la Universidad Nacional de Cuyo, Vol. 42, December 2010, pp. 119-133. [12] G. R. Pratta, G. R. Rodríguez, R. Zorzoli, E. M. Valle and L. A. Picardi, “Phenotypic and Molecular Characterization of Selected Tomato Recombinant Inbred Lines Derived from a Cross Solanum Lycopersicum × S. Pimpinelli- folium,” Journal of Genetics, 2001, in Press. [13] S. S. Shapiro and M. B. Wilk, “An Analysis of Variance Test for Normality Complete Samples,” Biometrika, Vol. 52, No. 3-4, June 1965, pp. 591-611. [14] M. Kearsey and H. Pooni, “The Genetical Analysis of Quantitative Traits,” Chapman and Hall, London, May 1996. [15] Q. S. Du, Z. Q. Jiang, W. Z. He, D. P. Li and K. C. Chou, “Amino Acid Principal Component Analysis (AAPCA) and Its Applications in Protein Structural Class Predic- tion,” Journal of Biomolecules Str uctucture and Dynamics, Vol. 23, No. 6, August 2006, pp. 635-640. [16] B. G. Forde and P. J. Lea, “Glutamate in Plants: Metabo- lism, Regulation, and Signaling,” Journal of Experimental Botany, Vol. 58, No. 9, November 2007, pp. 2339-2358. doi:10.1093/jxb/erm121 [17] A. Sorrequieta, G. Ferraro, S. B. Boggio and E. M. Valle, “Free Amino Acid Production during Tomato Fruit Rip- ening: A Focus on L-Glutamate,” Amino Acids, Vol. 38, No. 5, September 2009, pp. 1523-1532. doi 10.1007/s00726-009-0373-1. [18] F. Carrari and A. Fernie, “Metabolic Regulation Under- lying Tomato Fruit Development,” Journal of Experi- mental Botany, Vol. 57, No. 9, February 2007, pp. 1883- 1897. [19] F. Mounet, M. Lemaire-Chamley, M. I. Maucourt, C. Cabasson, J. L. Giraudel, C. Deborde, R. Lessire, P. Gal- lusci, A. Bertrand, M. Gaudillere, C. Rothan, D. Rolin and A. Moinga, “Quantitative Metabolic Profiles of Tomato Flesh and Seeds during Fruit Development: Complemen- tary Analysis with ANN and PCA,” Metabolomics, Vol. 3, No. 3, April 2007, pp. 273-287. doi:10.1007/s11306-007-0059-1 [20] Y. Semel, N. Schauer, U. Roessner, D. Zamir and A. R. Ferni, “Metabolite Analysis for the Comparison of Irri- gated and Non-Irrigated Field Grown Tomato of Varying Genotype,” Metabolomics, Vol. 3, No. 3, April 2007, pp. 289-295.doi:10.1007/s11306-007-0055-5 [21] S. D. Roper and N. Chaudhar, “Processing Umami and Other Tastes in Mammalian Taste Buds,” Annals of the New York Academy of Sciences, Vol. 1170, July 2009, pp. 60-65. doi:10.1111/j.1749-6632.2009.04107.x Copyright © 2011 SciRes. AJPS
|