Paper Menu >>
Journal Menu >>
 Journal of Biomaterials and Nanobiotechnology, 2011, 2, 258-266 doi:10.4236/jbnb.2011.23033 Published Online July 2011 (http://www.SciRP.org/journal/jbnb) Copyright © 2011 SciRes. JBNB Rapid and Facile Purification of Apolipoprotein A-I from Human Plasma Using Thermoresponsive Nanoparticles* Martin Lundqvist1, Tord Berggård2, Erik Hellstrand3, Iseult Lynch1, Kenneth A. Dawson1, Sara Linse3, Tommy Cedervall1,3# 1Centre for BioNano Interactions, School of Chemistry and Chemical Biology, University College Dublin, Dublin, Ireland; 2Department of Protein Technology, Lund University, Lund, Sweden; 3Biophysical Chemistry, Lund University Chemical Centre, Lund, Sweden. Email: #tcedervall@yahoo.com Received February 26th, 2011; revised March 30th 2011; accepted April 18th, 2011. ABSTRACT Nanoparticles can be used to purify proteins from plasma. We report here the purification of apolipoprotein A-I (apoA-I) with high specificity from human plasma using copolymeric nanoparticles. We present an optimized protocol using 50:50 NiPAM:BAM copolymer nanoparticles with thermo-responsive properties as an affinity resin. Repeated pelleting and washing of nanoparticle-captured apoA-I is achieved through temperature cycling. The protein is then eluted using urea followed by an ion exchange step for protein concentration and depletion of nanoparticles. Keywords: Apolipoprotein A-I, Nanoparticles, Selective Purification 1. Introduction The isolation and purification of proteins from complex bodily fluids is a challenging task, especially for low abundance proteins. The presence of a very high number (often several thousands) of other proteins means that classical methods like ion exchange chromatography and gel filtration yield only partial purification and a large number of steps may be needed. Affinity purification pro- vides a shorter purification route and can lead to much simplified purification schemes if an entity with high af- finity and high specificity for the desired protein is at hand. Commonly used entities are peptides, proteins, spe- cific antibodies or low Mw ligands coupled to chroma- tography resins. For example, factor IX can be purified with vey high specificity from human plasma using con- formation-specific antibodies that bind solely to the metal-stabilized conformer [1]. An example of a low Mw ligand is p-aminomethyl-benzene-sulfonamide for affin- ity purification of carbonic anhydrase [2]. Examples of peptides and proteins used successfully in single-step purification are the streptococcal IgA-binding peptide for human IgA [3], and an affibody, derived from staphylo- coccal protein A and selected by phage display, for Taq DNA polymerase [4]. While antibodies and other protein ligands offer very high specificity with excellent purity of the isolated protein, the scale-up costs are high be- cause the required amount of ligand scales linearly with batch size. Development of less expensive and more versatile tools for affinity purification is therefore still a tractable goal. Protein purification methods based on nanoparticles have recently been developed, especially nickel-ni-tri- lo- triacetate modified magnetic nanoparticles for purifying poly-His-tagged proteins [5-7]. The magnetism of the particles makes the purification extremely easy. However, this approach requires engineering of the protein to be purified and is not feasible for purification of proteins from bodily fluids. Non-tagged proteins may be purified using magnetic nanoparticles modified with specific high affinity ligands as described above for chromatographic resins. Here we report the use of thermo-responsive copoly- mer nanoparticles for protein purification. The special properties of NiPAM:BAM copolymer particles allow *This work was funded by an Irish Research Council for Science, En- gineering and Technology Postdoctoral Fellowship (M.L.), the Marianne and Marcus Wallenberg Foundation (M.L.), the EU FP6 p roject NanoInteract (NMP4-CT-2006-033231), and the SFI SRC BioNanoInteract (07 SRC B1155), Centre for Nano-Vaccine, Copen- hagen, Denmark, and the Swedish Research Council (VR).  Rapid and Facile Purification of Apolipoprotein A-I from Human Plasma Using Thermoresponsive Nanoparticles Copyright © 2011 SciRes. JBNB 259 for repeated precipitation and re-suspension through tem- perature cycling, and specific interaction makes it po- ssible to purify with high efficiency one specific protein from human plasma, apolipoprotein A-I (apoA-I). ApoA-I is the major protein constituent of high-density lipopro- tein (HDL). HDL is involved in reverse cholesterol transport and is thought to play an important role in low- ering the risk of coronary artery disease [8-10]. The tra- ditional purification of apolipoproteins from plasma is a time consuming process [11-15]. The lipoproteins are normally isolated from plasma by one or several prepara- tive ultracentrifugation steps. The purified lipoproteins contain a high degree of lipids and several proteins. The apolipoproteins are delipified using organic solvents and single apolipoproteins are separated from the mixture of proteins by biophysical and biochemical methods. This process is not protein friendly and takes days if not weeks. In contrast, the method presented here can be completed in less than 12 hours yielding apoA-I of high quality and purity. 2. Material and Methods 2.1. 70 nm NIPAM:BAM 50:50 Polymer Particles N-isopropylacrylamide-N-tert-butylacrylamide (NIPAM: BAM) copolymer particles of 70 nm diameter with 50:50 ratio of the monomers were synthesized in SDS micelles by free radical polymerization as described previously [16], although higher SDS concentration was used in the present work. The procedure for the synthesis was as fo- llows: 2.8 g monomers (in the appropriate wt/wt ratio), and 0.28 g crosslinker (N,N-methylenebisacrylamide) were dissolved in 190 mL MilliQ water with 0.8 g SDS and degassed by bubbling with N2 for 30 min. Polymeri- sation was induced by adding 0.095 g ammonium persul- fate initiator in 10 mL degassed MilliQ water and heating at 70˚C for 4 hours [17]. Particles were extensively dial- ysed against MilliQ water for several weeks, changing the water daily. Particles were lyophilized and stored in the fridge until used. 2.2. Plasma Human blood was withdrawn from healthy humans into vessels pre-treated with EDTA-solution. The vessels where centrifuged for 5 min at 800 Relative Centrifuga- tion Force (RCF). The supernatants (the plasma) were transferred to new vessels and stored in –80˚C freezer until the time of use. Before use the plasma vessel was thawed and centrifuged for 3 min at 16.1 kRCF and the supernatant was transferred to a new vessel, or in the case where more than one plasma vessel was needed the supernatants were pooled. 2.3. Particle and Plasma Mixtures Stock solutions of particles were made by dispersing lyophilized copolymer particles, to a concentration of 10 mg/mL, in 10 mM Tris/HCl, pH 7.5 with 0.15 mM NaCl and 1 mM EDTA or in 10 mM phosphate, pH 7.5 with 0.15 mM NaCl and 1 mM EDTA. The mixture was kept on ice until all the particles were dispersed (1-3 hours) before mixing with plasma. After mixing the particles and plasma the sample was kept on ice except during the centrifugation steps, which were performed at room tem- perature. In one experiment, lyophilized copolymer particles were dissolved directly in plasma by adding plasma to the particles and keeping the mixtures on ice for 1 hour. 2.4. Ion-Exchange Chromatography Ion exchange chromatography was performed on a Hi- Trap DEAE FF 1 mL column from GE Healthcare at room temperature in 10 mM Tris/HCl, pH 7.5 with 1 mM EDTA (buffer A). Urea fractions (eluates from the nano- particles) were pooled and passed through a 0.45 µm syringe filter. After filtration the urea and salt concentra- tions were lowered by ten-fold dilution with buffer A. (Alternatively, the salt and urea concentration may be lowered by dialysis against buffer A). The sample was thereafter loaded onto the column. ApoA-I was eluted by stepwise increasing the NaCl concentration: the first step was elution with 50 mM NaCl in buffer A to remove the nanoparticles; the second step (the elution of apoA-I) is performed with 10 0.15 M NaCl in buffer A. 2.5. SDS-PAGE In this study two different SDS-PAGE gels have been used: PIERCE, PreciseTM Protein Gels 4% - 20% pre- made gel, which are commercially available; and 12% gels which were cast immediately prior to use. 2.6. Commercial ApoA-I ApoA-I (A-0722) was obtained from Sigma-Aldrich and used as obtained. 2.7. Biophysical Analysis by UV-, CD- and Fluorescence Spectrometry The commercial apoA-I (Sigma-Aldrich, A-0722) was diluted 10 times with 10 mM phosphate, 0.15 M NaCl, 1 mM EDTA, pH 7.5, to approximately 0.15 mg/mL which corresponds approximately to the concentration of the purified apoA-I as estimated from the absorbance at 280 nm. The CD signal between 200 and 260 nm was recorded on a Jasco J-720 spectropolarimeter at 25˚C using a 1 mm quartz cuvette. The fluorescence emission spectra, 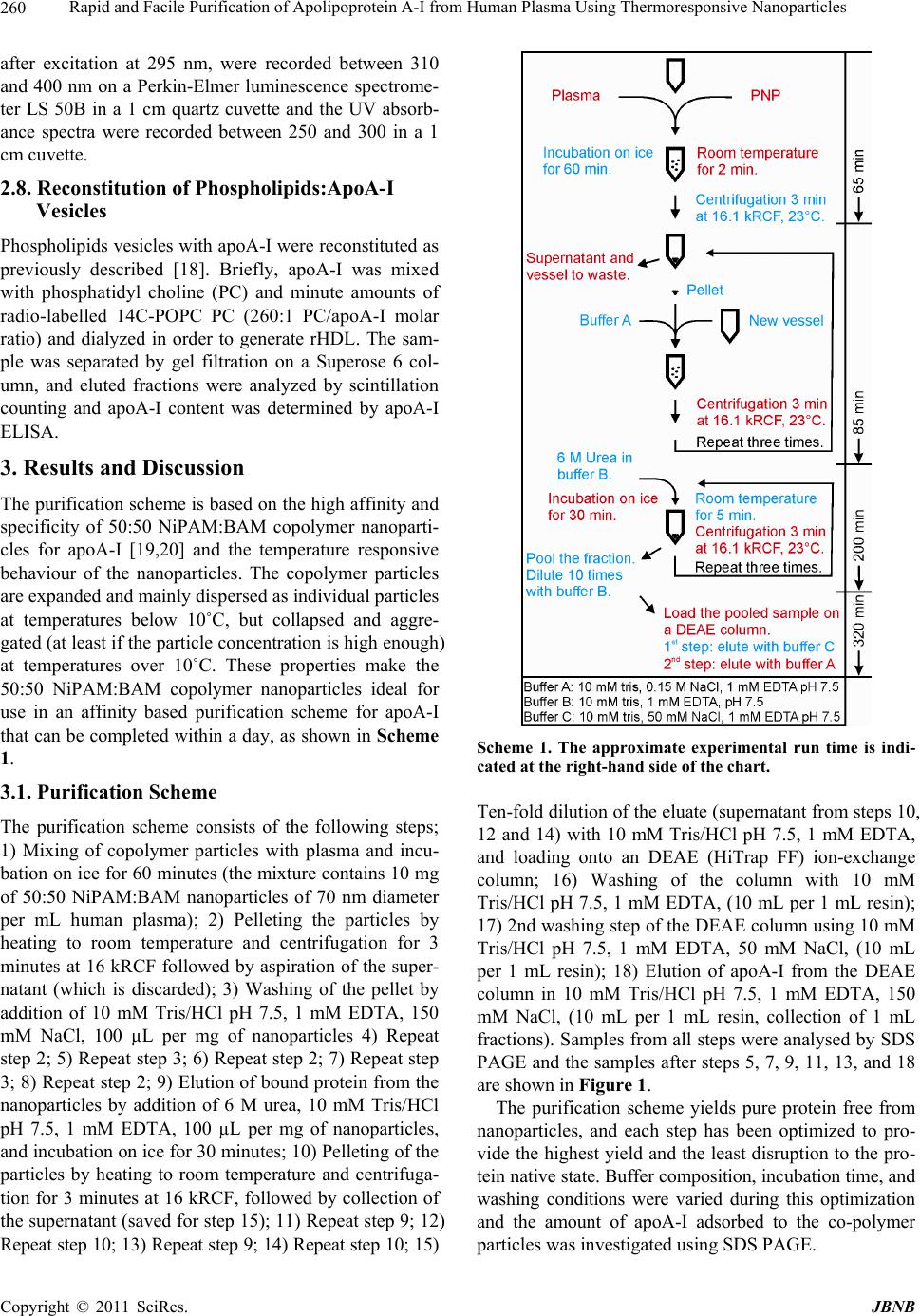 Rapid and Facile Purification of Apolipoprotein A-I from Human Plasma Using Thermoresponsive Nanoparticles Copyright © 2011 SciRes. JBNB 260 after excitation at 295 nm, were recorded between 310 and 400 nm on a Perkin-Elmer luminescence spectrome- ter LS 50B in a 1 cm quartz cuvette and the UV absorb- ance spectra were recorded between 250 and 300 in a 1 cm cuvette. 2.8. Reconstitution of Phospholipids:ApoA-I Vesicles Phospholipids vesicles with apoA-I were reconstituted as previously described [18]. Briefly, apoA-I was mixed with phosphatidyl choline (PC) and minute amounts of radio-labelled 14C-POPC PC (260:1 PC/apoA-I molar ratio) and dialyzed in order to generate rHDL. The sam- ple was separated by gel filtration on a Superose 6 col- umn, and eluted fractions were analyzed by scintillation counting and apoA-I content was determined by apoA-I ELISA. 3. Results and Discussion The purification scheme is based on the high affinity and specificity of 50:50 NiPAM:BAM copolymer nanoparti- cles for apoA-I [19,20] and the temperature responsive behaviour of the nanoparticles. The copolymer particles are expanded and mainly dispersed as individual particles at temperatures below 10˚C, but collapsed and aggre- gated (at least if the particle concentration is high enough) at temperatures over 10˚C. These properties make the 50:50 NiPAM:BAM copolymer nanoparticles ideal for use in an affinity based purification scheme for apoA-I that can be completed within a day, as shown in Scheme 1. 3.1. Purification Scheme The purification scheme consists of the following steps; 1) Mixing of copolymer particles with plasma and incu- bation on ice for 60 minutes (the mixture contains 10 mg of 50:50 NiPAM:BAM nanoparticles of 70 nm diameter per mL human plasma); 2) Pelleting the particles by heating to room temperature and centrifugation for 3 minutes at 16 kRCF followed by aspiration of the super- natant (which is discarded); 3) Washing of the pellet by addition of 10 mM Tris/HCl pH 7.5, 1 mM EDTA, 150 mM NaCl, 100 µL per mg of nanoparticles 4) Repeat step 2; 5) Repeat step 3; 6) Repeat step 2; 7) Repeat step 3; 8) Repeat step 2; 9) Elution of bound protein from the nanoparticles by addition of 6 M urea, 10 mM Tris/HCl pH 7.5, 1 mM EDTA, 100 µL per mg of nanoparticles, and incubation on ice for 30 minutes; 10) Pelleting of the particles by heating to room temperature and centrifuga- tion for 3 minutes at 16 kRCF, followed by collection of the supernatant (saved for step 15); 11) Repeat step 9; 12) Repeat step 10; 13) Repeat step 9; 14) Repeat step 10; 15) Scheme 1. The approximate experimental run time is indi- cated at the right-hand side of the chart. Ten-fold dilution of the eluate (supernatant from steps 10, 12 and 14) with 10 mM Tris/HCl pH 7.5, 1 mM EDTA, and loading onto an DEAE (HiTrap FF) ion-exchange column; 16) Washing of the column with 10 mM Tris/HCl pH 7.5, 1 mM EDTA, (10 mL per 1 mL resin); 17) 2nd washing step of the DEAE column using 10 mM Tris/HCl pH 7.5, 1 mM EDTA, 50 mM NaCl, (10 mL per 1 mL resin); 18) Elution of apoA-I from the DEAE column in 10 mM Tris/HCl pH 7.5, 1 mM EDTA, 150 mM NaCl, (10 mL per 1 mL resin, collection of 1 mL fractions). Samples from all steps were analysed by SDS PAGE and the samples after steps 5, 7, 9, 11, 13, and 18 are shown in Figure 1. The purification scheme yields pure protein free from nanoparticles, and each step has been optimized to pro- vide the highest yield and the least disruption to the pro- tein native state. Buffer composition, incubation time, and washing conditions were varied during this optimization and the amount of apoA-I adsorbed to the co-polymer particles was investigated using SDS PAGE. 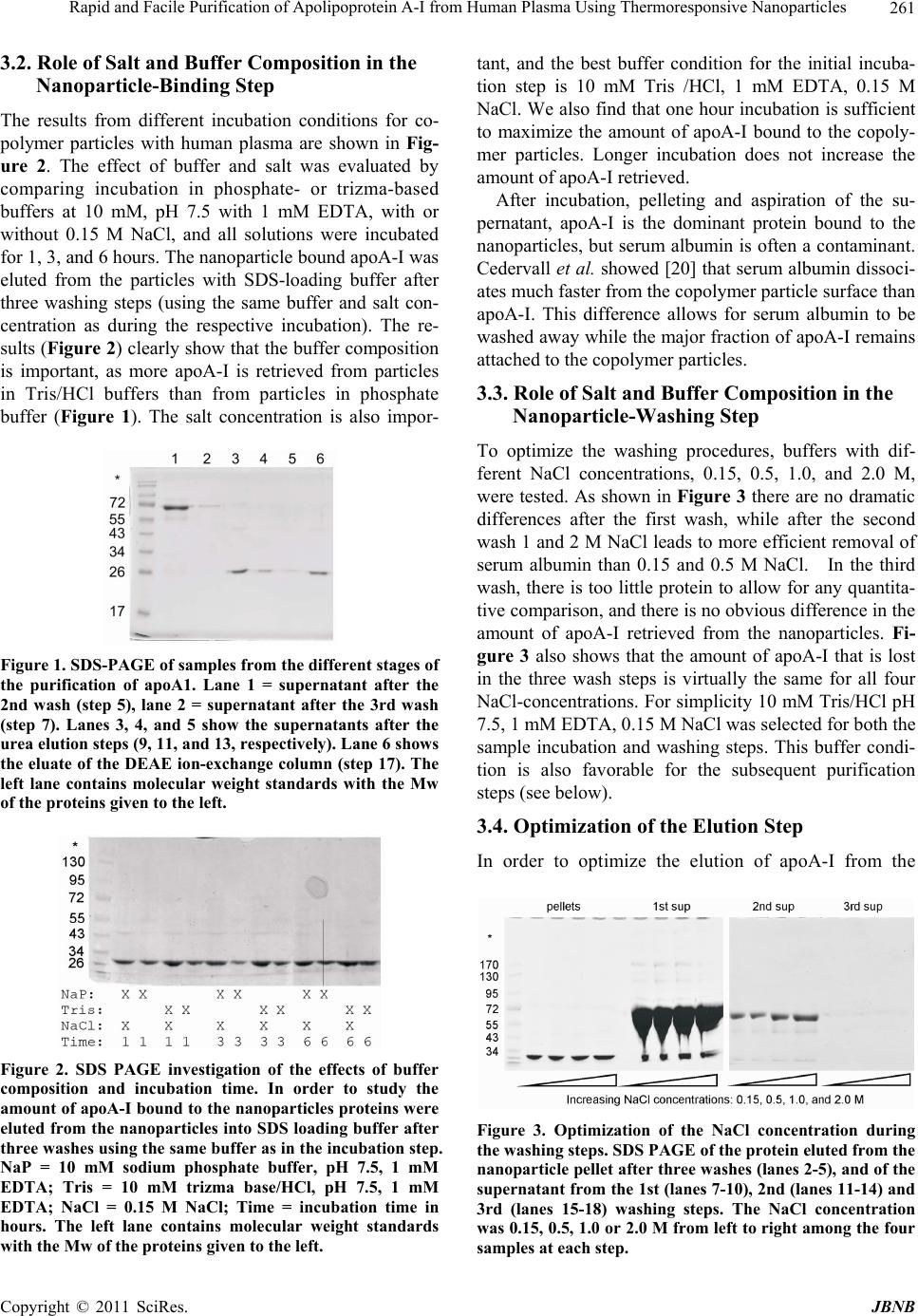 Rapid and Facile Purification of Apolipoprotein A-I from Human Plasma Using Thermoresponsive Nanoparticles Copyright © 2011 SciRes. JBNB 261 3.2. Role of Salt and Buffer Composition in the Nanoparticle-Binding Step The results from different incubation conditions for co- polymer particles with human plasma are shown in Fig- ure 2. The effect of buffer and salt was evaluated by comparing incubation in phosphate- or trizma-based buffers at 10 mM, pH 7.5 with 1 mM EDTA, with or without 0.15 M NaCl, and all solutions were incubated for 1, 3, and 6 hours. The nanoparticle bound apoA-I was eluted from the particles with SDS-loading buffer after three washing steps (using the same buffer and salt con- centration as during the respective incubation). The re- sults (Figure 2) clearly show that the buffer composition is important, as more apoA-I is retrieved from particles in Tris/HCl buffers than from particles in phosphate buffer (Figure 1). The salt concentration is also impor- Figure 1. SDS-PAGE of samples from the different stages of the purification of apoA1. Lane 1 = supernatant after the 2nd wash (step 5), lane 2 = supernatant after the 3rd wash (step 7). Lanes 3, 4, and 5 show the supernatants after the urea elution steps (9, 11, and 13, respectively). Lane 6 shows the eluate of the DEAE ion-exchange column (step 17). The left lane contains molecular weight standards with the Mw of the proteins given to the left. Figure 2. SDS PAGE investigation of the effects of buffer composition and incubation time. In order to study the amount of apoA-I bound to the nanoparticles proteins were eluted from the nanoparticles into SDS loading buffer after three washes using the same buffer as in the incubation step. NaP = 10 mM sodium phosphate buffer, pH 7.5, 1 mM EDTA; Tris = 10 mM trizma base/HCl, pH 7.5, 1 mM EDTA; NaCl = 0.15 M NaCl; Time = incubation time in hours. The left lane contains molecular weight standards with the Mw of the proteins given to the left. tant, and the best buffer condition for the initial incuba- tion step is 10 mM Tris /HCl, 1 mM EDTA, 0.15 M NaCl. We also find that one hour incubation is sufficient to maximize the amount of apoA-I bound to the copoly- mer particles. Longer incubation does not increase the amount of apoA-I retrieved. After incubation, pelleting and aspiration of the su- pernatant, apoA-I is the dominant protein bound to the nanoparticles, but serum albumin is often a contaminant. Cedervall et al. showed [20] that serum albumin dissoci- ates much faster from the copolymer particle surface than apoA-I. This difference allows for serum albumin to be washed away while the major fraction of apoA-I remains attached to the copolymer particles. 3.3. Role of Salt and Buffer Composition in the Nanoparticle-Washing Step To optimize the washing procedures, buffers with dif- ferent NaCl concentrations, 0.15, 0.5, 1.0, and 2.0 M, were tested. As shown in Figure 3 there are no dramatic differences after the first wash, while after the second wash 1 and 2 M NaCl leads to more efficient removal of serum albumin than 0.15 and 0.5 M NaCl. In the third wash, there is too little protein to allow for any quantita- tive comparison, and there is no obvious difference in the amount of apoA-I retrieved from the nanoparticles. Fi- gure 3 also shows that the amount of apoA-I that is lost in the three wash steps is virtually the same for all four NaCl-concentrations. For simplicity 10 mM Tris/HCl pH 7.5, 1 mM EDTA, 0.15 M NaCl was selected for both the sample incubation and washing steps. This buffer condi- tion is also favorable for the subsequent purification steps (see below). 3.4. Optimization of the Elution Step In order to optimize the elution of apoA-I from the Figure 3. Optimization of the NaCl concentration during the washing steps. SDS PAGE of the protein eluted from the nanoparticle pellet after three washes (lanes 2-5), and of the supernatant from the 1st (lanes 7-10), 2nd (lanes 11-14) and 3rd (lanes 15-18) washing steps. The NaCl concentration was 0.15, 0.5, 1.0 or 2.0 M from left to right among the four samples at each step.  Rapid and Facile Purification of Apolipoprotein A-I from Human Plasma Using Thermoresponsive Nanoparticles Copyright © 2011 SciRes. JBNB 262 nanoparticles, at least two factors are necessary to con- sider in order to maximize the protein yield. The effi- ciency of the elution is, of course, important for the final protein yield. However, the buffer composition after the elution is also important for the protein yield as it will affect the subsequent purification steps. Buffers with high salt concentration, high and low pH, different de- tergents, or urea were tested for their elution efficiency (Figure S3 in supplementary material), determined as the highest amount of protein in the gel bands. The highest amount of apoA-I was eluted using detergents, followed by 3 M NaCl, followed by 6 M urea. At lower NaCl concentration, or at pH 4 or 9.2, little or no elution could be observed. Urea was chosen as eluant for the purifica- tion scheme as it allows the eluted proteins to be loaded directly onto an ion exchange column without changing the buffer. The detergents, although they are the best eluants, were not chosen as it could be difficult to remove them from the protein, resulting in a lower purity of the final product. Figure 1 shows the different stages in the purification of apoA-I schematically. The amount of eluted apoA-I decreases in each step of elution with 6 M urea (Figure 1, Lanes 3-5). The small amount of apoA-I remaining in the third urea eluate does not motivate a fourth urea step. 3.5. Ion Exchange Chromatography After elution of apoA-I from the copolymer particles, ion-exchange (DEAE) chromatography is used to sepa- rate apoA-I from impurities, mainly the proteins apoli- poprotein E and serum albumin [19] and copolymer par- ticle fragments and to remove urea and change the buffer. Before loading onto the DEAE-column the pooled sam- ple is diluted 10 times with 10 mM phosphate buffer pH 7.5, 1 mM EDTA, to reduce the NaCl concentration. Bound molecules are eluted from the DEAE-column in a stepwise manner. Non-protein substances are eluted in buffer with 50 mM NaCl, apoA-I is eluted in buffer with 0.15 M NaCl, and serum albumin elutes in 3 M NaCl, which regenerates the column (serum albumin would elute much earlier if a gradient elution profile were used). If the first step (elution in 50 mM NaCl) is not in- cluded, something that scatters light under 270 nm is co- eluted with apoA-I (data not shown). The fraction from the first elution step does not contain protein (no visible bands in a coomassie stained gel). Analysis by NMR spectroscopy (data not shown) indicates an interaction between buffer molecules and very large molecules and/ or a slight increase in the solution viscosity evidenced as a slower tumbling of the molecules and a broader peak [21]. The most probable explanation for the observed phe- nomena is that the eluted fraction consists of fragments of the copolymer particles. However, this has to be in- vestigated further. 3.6. Maximizing ApoA-I Yield Figure 3 shows that even though apoA-I has a high af- finity for the copolymer particle surface, a large fraction of the apoA-I is washed away in the first washing step. Two experiments were conducted to maximize the yield of purified apoA-I. The effect of the copolymer particle concentration was checked by keeping the total sample volume and the total plasma concentration constant while the concentration of particles was varied. After incuba- tion, the same amount (by weight) of copolymer particles was removed from each sample and the amount of bound apoA-I analyzed by SDS PAGE after elution in SDS loading buffer. The results show that the total binding of apoA-I increases with the particle concentration (Figure S1, supplementary material), hence, one limiting factor in the yield is the available particle surface area. To con- firm this result a second experiment was conducted in which different amounts of lyophilized copolymer parti- cles were dispersed in the same volume of plasma. The trend of increasing yield of apoA-I with increasing amount of available surface area was confirmed (Figure S2, supplementary material). Table 1 shows the amounts of purified apoA-I for two experiments with different particle concentrations. The purification efficiency, in per- cent, is calculated assuming that the concentration of apoA-I in the original plasma is ~1 mg/ml. These results show that it is not the apoA-I concentration in blood that is the limiting factor for the yield of purified apoA-I, but the solubility (dispersability) of the copolymer particles Table 1. Comparison of the efficiency of the copolymer nanoparticles at purifying apoA-1 from plasma with two different dispersion methods—with TBS as an intermediate step, or direct dispersion in plasma. 1 a 2b Theoretical max apoA-I (mg)c 3.6 2.2 Particles (mg)d 45 22 Plasma/particles (ml/mg)e 0.08 0.1 Amount purified apoA-I (μg)f 224 277 Purification efficiency (%)g 6 13 Purified apoA-I/amount particles (μg/mg)h 5 13 aCopolymer particles first dispersed in TBS before mixing with plasma; bCopolymer particles directly dispersed in plasma; cCalculated at a nominal apoA-I concentration of 1 mg/ml plasma; dTotal amount of copolymer parti- cles used in the experiment; eRatio between plasma and copolymer particles in the experiment; fTotal amount of purified apoA-I in the experiment; gPurification efficiency in percent purified apoA-I compared to the nominal apoA-I concentration; hAmount purified apoA-I per mg copolymer particles.  Rapid and Facile Purification of Apolipoprotein A-I from Human Plasma Using Thermoresponsive Nanoparticles Copyright © 2011 SciRes. JBNB 263 Figure S1. ApoA1 retrieved with different amounts of polymer particles added to 210 μl plasma in a total volume of 910 μl. Lanes 1, 2, 3, 4, 5, 6, 7, and 8 represent 0.007, 0.07, 0.14, 0.35, 0.7, 1.4, 2.1, and 7.0 mg co-polymer particles, respectively. The 0.35, 0.7, 1.4, 2.1 and 7.0 mg samples con- tain protein from an identical amount of polymer particles. Figure S2. Lanes 1, 2, 3, and 4 represent 10, 5, 2.5 and 1 mg co-polymer nanoparticles, respectively, that were dissolved in 1 ml plasma. Washed with 3 x 1000, 500, 250 and 100 μL, respectively, of 10 mM trizma/HCl pH 7.5, 1 mM EDTA, 0.15 M NaCl. The pellets were dissolved in 100, 50, 25 and 10 μL SDS-loading buffer, respectively, and 10 μL of each sample was loaded on the electrophoresis gel. Figure S3. Elution of apoA1 from the co-polymer particles using different agents. Lanes 1, 2, and 3 are eluates in 1, 2, and 3 M NaCl, respectively, lane 4 the eluate with 2% triton, lane 5 with 2% tween-20, lane 6 with 6 M urea, lane 7 10 mM sodium acetate buffer, pH 4, lane 8 10 mM Tris/HCl pH 9.2, and lane 9 buffer with 2% SDS. All eluates except in lanes 7 and 8 contain 10 mM Tris/HCl pH 7.5. in plasma and thus the amount of available surface area. The highest concentration of particles that can be dis- persed in plasma is ~10 mg per ml, as above at this con- centration particle aggregation occurs, and thus the over- all surface area available for bind decreases again. 3.7. Lipid-Free ApoA-I Preliminary results indicate that the particles bind apoA-I together with lipids in the first step of the purification scheme. As the goal was to purify lipid free apoA-I, the final purified sample was checked for cholesterol and triglycerides with commercial enzyme-based detection kits (see SI). No lipids were detected in the purified apoA-I samples suggesting that the purified apoA-I is lipid free. 3.8. Comparison with Commercial ApoA-I Integrity, Structure and Function The purified apoA-I was compared to commercially available apoA-I. On a coomassie stained 1D SDS-PAGE gel, the commercial protein gives rise to three bands, while the protein purified by the method described here gives rise to one band on the same gel with similar amounts of sample loaded, as shown in Figure 4(a). The identity of the purified apoA-I has been confirmed re- peatedly by mass spectrometry [21,22]. The apparent differences in size between the three bands in the com- mercial sample of apolipoprotein A-1 may be due to modifications or proteolysis. Another explanation may be that the copolymer particles specifically target a sub- population of apoA-I, for example, the fraction with oxi- dized side chains. This is less likely as the apoA-I found in the wash fractions has the same mobility as the puri- fied protein. Thus, the three bands in commercial apoA-I are most likely due to the harsh conditions used during the existing purification process, whereas the protocol presented here leads to a homogeneous protein prepara- tion. Three different spectroscopic methods, UV absorbance, intrinsic tryptophan fluorescence and far UV circular dichroism spectroscopy, are used to evaluate and com- pare the structure of apoA-I purified via the method pre- sented here with commercial apoA-I. The UV absorb- ance spectra, Figure 4(b), gives information on local structural changes around aromatic amino acids; the in- trinsic tryptophan fluorescence spectra, Figure 4(c), gives information on changes in the tertiary structure; and the far UV circular dichroism spectra, Figure 4(d), reports on changes in the secondary structure. In all of the spec- tra there are no or very small deviations between the commercial and purified apoA-I, indicating that the terti- ary and secondary structure of apoA-I are essentially the same in the two samples. The phospholipids binding function of the purified apoA-I was examined by reconstitution of phospholip- ids:apoA-I vesicles. Figure S4 shows that the copolymer particle purified apoA-I interacts with and forms vesicles together with phospholipids in a similar way to that shown previously for apoA-I purified with a traditional purification protocol [18].  Rapid and Facile Purification of Apolipoprotein A-I from Human Plasma Using Thermoresponsive Nanoparticles Copyright © 2011 SciRes. JBNB 264 (a) (b) (c) (d) Figure 4. Comparison between commercial apoA-I and the apoA-I purified by the method described here. (a) Lane 1 = mo- lecular weight standards with the Mw of the proteins given to the left, lane 2 = commercial apoA-I, lane 3 = apoA-I purified in this work. In (b), (c) and (d), dark grey = commercial apoA-I and light grey = purified apoA-I. (b) absorbance, (c) fluores- cence and (d): circular dichroism spectra. Figure S4. ApoA1 was mixed with labelled PL in order to generate rHDL. The sample was analyzed by gelfiltration. Eluted fractions were analyzed by scintillation counting and apoA1 ELISA. 4. Conclusions Copolymer nanoparticles offer efficient and facile puri- fication of apoA-I from complex protein mixtures, such as plasma. The purification scheme presented here dra- matically reduces the time needed for purification. The final yield of pure apoA-I is up to 13 mg/g nanoparticles, which is comparable to other affinity chromatography systems. The particles also bind apolipoprotein E, A-IV, and A-II with high specificity. These apolipoproteins are in much lower concentrations in plasma and are therefore a challenge to purify. However, in a scaled up purifica- tion protocol it should be possible to purify also these proteins using the copolymer particles. Additionally, slight modification of the comonomer ratio could be used to optimize the particles for selective binding of these proteins. The copolymer particles are easy and inexpensive to produce and their physical and chemical characteristics (including surface charge and/or hydrophobicity or sur- face energy) are easy to vary. Additionally, the particles can be modified with ligands with high specificity for selected target proteins. These features make the co- polymer particles an attractive choice for purification of proteins, not limited to apolipoproteins. 5. Acknowledgments We thank Professor Björn Dahlbäck and Dr Cecilia Oslakovic for help with the reconstitution of phosphol- ipid:apoA-I vesicles and for valuable comments on the manuscript. Access to and use of UCD Conway Mass Spectrometry Resource instrumentation is gratefully ac- knowledged.  Rapid and Facile Purification of Apolipoprotein A-I from Human Plasma Using Thermoresponsive Nanoparticles Copyright © 2011 SciRes. JBNB 265 6. Supporting Information Available Supplementary information regarding the optimization of the purification protocol is presented. Figures showing the effects of particle concentration on apoA-I purity (Figure S1), and yield (Figure S2) and the effectiveness of various agents to elute the purified apoA-I from the particles (Figure S3), and the reconstituted phospholip- ids:apoA-I vesicles (Figure S4). REFERENCES [1] H. A. Liebman, S. A. Limentani, B. C. Furie and B. Furie, “Immunoaffinity Purification of Factor-Ix (Christmas Factor) by Using Conformation-Specific Antibodies Di- rected against the Factor-Ix - Metal-Complex,” Proceed- ings of the National Academy of Sciences of the United States of America, Vol. 82, No. 11, 1985, pp. 3879-3883. doi:10.1073/pnas.82.11.3879 [2] S. O. Falkbring, P. O. Göthe, P. O. Nyman, L. Sundberg and J. Porath, “Affinity Chromatography of Carbonic An- hydrase,” FEBS Letters, Vol. 24, No. 2, 1972, pp. 229- 235. doi:10.1016/0014-5793(72)80773-7 [3] C. Sandin, S. Linse, T. Areschoug, J. M. Woof, J. Rein- holdt and G. Lindahl, “Isolation and Detection of Human IgA Using a Streptococcal IgA-binding Peptide,” Journal of Immunology, Vol. 169, No. 3, 2002, pp. 1357-1364. [4] K. Nord, E. Gunneriusson, J. Ringdahl, S. Ståhl, M. Uhlen and P.-A. Nygren, “Binding Proteins Selected from Combinatorial Libraries of an [alpha]-Helical Bacterial Receptor Domain,” Nature Biotechnology, Vol. 15, No. 8, 1997, pp. 772-777. doi:10.1038/nbt0897-772 [5] H. W. Gu, K. M. Xu, C. J. Xu and B. Xu, “Biofunctional Magnetic Nanoparticles for Protein Separation and Patho- gen Detection,” Chemical Communications, Vol. 9, 2006, pp. 941-949. doi:10.1039/b514130c [6] D. B. Shieh, C. H. Su, F. Y. Chang, Y. N. Wu, W. C. Su, J. R. Hwu, J. H. Chen and C. S. Yeh, “Aqueous Nickel- nitrilotriacetate Modified Fe3O4-NH3+ Nanoparticles for Protein Purification and Cell Targeting,” Nanotechnology, Vol. 17, No. 16, 2006, pp. 4174-4182. doi:10.1088/0957-4484/17/16/030 [7] C. C. You, A. Verma and V. M. Rotello, “Engineering the Nanoparticle-Biomacromolecule Interface,” Soft Matter, Vol. 2, No. 3, 2006, pp. 190-204. [8] N. E. Miller, “Associations of High-Density-Lipoprotein Subclasses and Apolipoproteins with Ischemic-Heart- Di- sease and Coronary Atherosclerosis,” American Heart Journal, Vol. 113, No. 2, 1987, pp. 589-597. doi:10.1016/0002-8703(87)90638-7 [9] W. P. Castelli, R. D. Abbott and P. M. McNamara, “Sum- mary Estimates of Cholesterol Used to Predict Coronary Heart-Disease,” Circulation, Vol. 67, No. 4, 1983, pp. 730-734. doi:10.1161/01.CIR.67.4.730 [10] G. G. Rhoads, C. L. Gulbrandsen and A. Kagan, “Serum- Lipoproteins and Coronary Heart-Disease in a Population Study of Hawaii Japanese Men,” New England Journal of Medicine, Vol. 294, No. 6, 1976, pp. 293-298. doi:10.1056/NEJM197602052940601 [11] C. Edelstein, D. Pfaffinger and A. M. Scanu, “Advantages and Limitations of Density Gradient Ultracentrifugation in the Fractionation of Human-Serum Lipoproteins—Role of Salts and Sucrose,” Journal of Lipid Research, Vol. 25, No. 6, 1984, pp. 630-637. [12] R. J. Havel, H. A. Eder and J. H. Bragdon, “The Distribu- tion and Chemical Composition of Ultracentrifugally Se- parated Lipoproteins in Human Serum,” The Journal of Clinical Investigation, Vol. 34, No. 9, 1955, pp. 1345- 1353.doi:10.1172/JCI103182 [13] D. W. Swinkels, H. L. M. Haklemmers and P. N. M. De- macker, “Single Spin-Density Gradient Ultracentrifuga- tion Method for the Detection and Isolation of Light and Heavy Low-Density-Lipoprotein Subfractions,” Journal of Lipid Research, Vol. 28, No. 10, 1987, pp. 1233-1239. [14] T. Tadey and W. C. Purdy, “Chromatographic Tech- niques for the Isolation and Purification of Lipoproteins,” Journal of Chromatography B: Biomedical Sciences and Applications, Vol. 671, No. 1-2, 1995, pp. 237-253. doi:10.1016/0378-4347(95)00051-J [15] K. H. Weisgraber, T. P. Bersot, R. W. Mahley, G. Franceschini and C. R. Sirtori, “A-I Milano Apoprotein Isolation and Characterization of a Cysteine-Containing Variant of the A-I Apoprotein from Human High Density Lipoproteins,” The Journal of Clinical Investigation, Vol. 66, No. 5, 1980, pp. 901-907. doi:10.1172/JCI109957 [16] X. Wu, R. H. Pelton, A. E. Hamielec, D. R. Woods and W. McPhee, “The Kinetics of Poly(N-Isopropylacryla- mide) Microgel Latex Formation,” Colloid and Polymer Science, Vol. 272, No. 4, 1994, pp. 467-477. doi:10.1007/BF00659460 [17] I. Lynch, I. Miller, W. M. Gallagher and K. A. Dawson, “Novel Method to Prepare Morphologically Rich Poly- meric Surfaces for Biomedical Applications via Phase Separation and Arrest of Microgel Particles,” Journal of Physical Chemistry B, Vol. 110, No. 30, 2006, pp. 14581- 14589.doi:10.1021/jp061166a [18] C. Oslakovic, M. J. Krisinger, A. Andersson, M. Jauhiai- nen, C. Ehnholm and B. Dahlback, “Anionic Phospholip- ids Lose Their Procoagulant Properties When Incorpo- rated into High Density Lipoproteins,” Journal of Bio- logical Chemistry, Vol. 284, No. 9, 2009, pp. 5896-5904. doi:10.1074/jbc.M807286200 [19] T. Berggård, et al., “140 Mouse Brain Proteins Identified by Ca2+-Calmodulin Affinity Chromatography and Tan- dem Mass Spectrometry,” Journal of Proteome Research, Vol. 5, No. 3, 2006, pp. 669-687. [20] T. Cedervall, I. Lynch, M. Foy, T. Berggård, S. C. Don- nelly, G. Cagney, S. Linse and K. A. Dawson, “Detailed Identification of Plasma Proteins Adsorbed on Copolymer Nanoparticles,” Angewandte Chemie International Edi- tion, Vol. 46, No. 30, 2007, pp. 5754-5756. doi:10.1002/anie.200700465 [21] T. Cedervall, I. Lynch, S. Lindman, T. Berggard, E. Thu- lin, H. Nilsson, K. A. Dawson and S. Linse, “Understand-  Rapid and Facile Purification of Apolipoprotein A-I from Human Plasma Using Thermoresponsive Nanoparticles Copyright © 2011 SciRes. JBNB 266 ing the Nanoparticle-Protein Corona Using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 104, No. 7, 2007, pp. 2050-2055.doi:10.1073/pnas.0608582104 [22] M. Lundqvist, I. Sethson and B. H. Jonsson, “Protein Adsorption onto Silica Nanoparticles: Conformational Changes Depend on the Particles’ Curvature and Protein Stability,” Langmuir, Vol. 277, No. 20, 2004, pp. 10639- 10647. ABBREVIATIONS ApoA-I: Apolipoprotein A-I; RCF: Relative Centrifugal Force; PC: Phosphatidyl Choline. |

