Natural Science
Vol.5 No.6A(2013), Article ID:33201,6 pages DOI:10.4236/ns.2013.56A005
Decolorization of triphenyl methane dyes by Fomitopsis feei
![]()
Department of Microbiology, Kakatiya University, Warangal, India; *Corresponding Author: bindunidadavolu@gmail.com
Copyright © 2013 S. V. S. S. S. L. Hima Bindu Nidadavolu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 21 April 2013; revised 22 May 2013; accepted 31 May 2013
Keywords: Triphenyl Methane Dyes; Fomitopsis feei; Dye Decolorization; Lignolytic Enzymes; Triphenyl Methane Reductase
ABSTRACT
Six triphenylmethane dyes viz., bromophenol blue, basic fuchsin, methyl violet, methyl green, ethyl violet and malachite green were studied for their decolorization by Fomitopsis feei. Among, basic fuchsin (98%) was maximum decolorized followed by bromophenol blue (96.8%). However, the rate of bromophenol blue decolorization was very fast. There was no correlation between the lignolytic activity and dye decolurization of the dyes. The highest laccase and lignin peroxidase activities were observed in basic fuchsin (46 U/mL) and methyl green (44 U/mL) respectively after 21 days of incubation, which were poor dye degraders. The triphenylmethane reductase enzyme was the responsible enzyme for the decolorization of these triphenyl methane dyes. The treatment by using fungal organisms was considered to be the cost-effective and ecofriendly method of decolourization of effluents discharged from the dye industries.
1. INTRODUCTION
Synthetic dyes are extensively used in the textile industries and significant proportion appears in the form of wastewater and is spilled into the environment. More than 10,000 synthetic dyes include the azo, anthroquinone and triphenylmethane dyes, all of which are generally considered as the xenobiotic compounds, which are very recalcitrant to biodegradation [1]. Triphenylmethane dyes are aromatic xenobiotic compounds that are used extensively in many industrial processes, such as textile dyeing, paper printing, and food and cosmetic manufacture [2]. They are known to be highly toxic to mammalian cells and mutagenic and carcinogenic to humans [3-6]. Based on their potential for adverse human health effects, most countries have nominated triphenylmethane dyes as hazardous material and prohibited the use of them in aquaculture and food industry. However, they are still used in some areas due to their relatively low cost, ready availability and efficacy [7]. Since triphenylmethane dyes occur as contaminants and potential human health hazards, there is concern about the fate of them in aquatic and terrestrial ecosystems [8,9]. Considerable amounts of these toxic and mutagenic dyes are discharged into wastewater treatment facilities and thus impose a selective pressure on the microbial flora residing in wastewater habitats. Several physicochemical methods include adsorption, chemical precipitation and flocculation, photolysis, chemical oxidation and reduction, electrochemical treatment and ion pair extraction have been used to eliminate the colored effluents from wastewater [10] but these methods are expensive, of limited applicability and produce large amounts of sludge. As a better alternative, therefore, the development of biological processes using microorganisms for the treatment of dye-containing wastewater has become increasingly important.
Several triphenylmethane dye-decolorizing microorganisms have been reported and their characteristics were reviewed [11]. Only a few studies have reported the bioremoval potentials of brown rot fungi for different dyes [12-17]. Further research work on such fungal strains is much needed in bioremediation perspective. Fomitopsis feei is a basidiomyceteous fungi belongs to the family Fomitopsidaeceae. Brown rot fungus F. feei has been found to have decolorization capacity but only limited literature is available [18]. The present work was aimed at evaluating basidiomycetous fungi for its ability to decolorize several synthetic dyes in shake condition. An attempt has also been made to analyze the concomitant association of ligninolytic enzymes (lignin peroxidase and laccase) production during the decolorization by this fungus.
2. MATERIAL AND METHODS
2.1. Chemicals
Triphenyl methane dyes i.e. Basic fuchsin (BF), Methyl green (MetG), Ethyl violet (EV), Methyl violet (MV) were procured from Hi Media Laboratories Pvt. Ltd. (Mumbai, India) and Malachite green (MG) and Bromophenol blue (BB) were purchased from Merck Specialities Private Limited (Mumbai, India) and Qualigens Fine Chemicals Ltd. (Mumbai, India) respectively. All media components and chemicals used were of highest purity grade.
2.2. Isolation of Microorganism and Culture Conditions
The wood decaying fungus F. feei was collected from the infected wood logs at Pakhal forest (latitude 18˚1'18.82''N and longitude 80˚9'22.16''E) Warangal district, Andhra Pradesh, India. The tentatively identified fungus was phylogenetically confirmed by molecular analysis of D2 region of 28S rDNA as Fomitopsis feei (AY 515327.1). All along the duration of the experiment, fungus was maintained on Malt extract agar medium (MEA (g/L): Malt extract 15, Dipotassium hydrogen phosphate 1, Ammonium chloride 1, 1 N Citric acid 15 mL; Agar 20) and stored at 4˚C and sub cultured every fortnight.
2.3. Decolorization in Liquid Media
For this basidiomycetous fungus, decolorization was tested in a 100-mL Erlenmeyer flask with 25 mL sterile production broth (g/L): Peptone—1, Yeast extract—2, Dipotassium hydrogen phosphate—1, Magnesium sulphate hepta hydrate—0.2, Ammonium sulphate—5, Glucose—20 and pH—6.0, containing 0.001% each triphenyl methane dye. Flasks were inoculated with single 10 mm diameter fungal disk taken from the periphery of the 7-day-old fungal culture in MEA plate and incubated at 28˚C in shaking incubator at 150 rpm speed. After regular intervals include 7, 14 and 21 days of incubation, the fungal culture was harvested by filtering through Whatman No.1 filter paper and the culture filtrate was centrifuged (12,000 × g; 20 min) at 4˚C. Decolorization and ligninolytic enzymes activity was determined from the cell-free supernatant. pH of the supernatant was measured every time using Elico pH meter. The uninoculated dye-containing medium was used as control. The decolorization efficiency was determined using the following equation [19]:
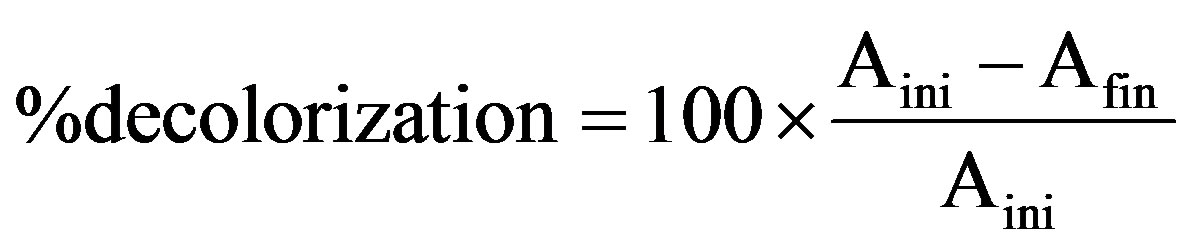
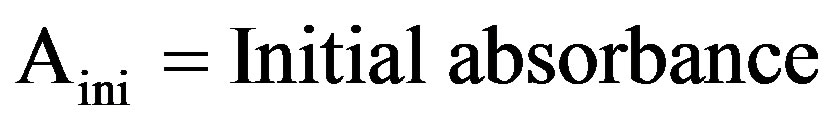
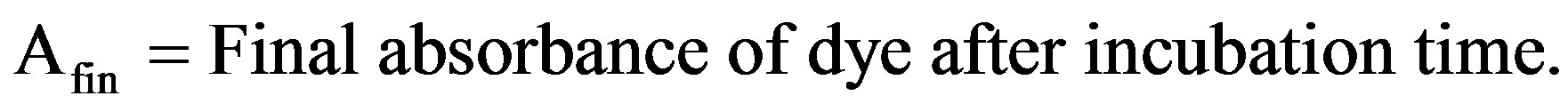
2.4. Dry Mycelium Weight
Dry mycelium weight (DMW) of the fungal mass was obtained by filtering the contents of each flask through pre-weighed Whatmann No.1 filter paper and then it was left to dry to a constant weight at 70˚C. DMW was expressed in terms of grams of biomass per liter of culture.
2.5. Lignolytic Enzyme Assays
Laccase activity was measured [20] by taking the optical density of reaction mixture, prepared by mixing 0.5 mL of distilled water, 1 mL of sodium acetate buffer (pH 4.5), 0.5 mL of substrate solution (46 mM guaiacol) to 0.5 mL of crude enzyme (cell free supernatant) at 440 nm up to 90 sec with 30 sec of time interval. Lignin peroxidase activity was evaluated [21] by following the same procedure of laccase but 0.5 mL of hydrogen peroxide was added in addition to that mixture. For these two enzymes, one activity unit was defined as the amount of enzyme necessary to oxidize one μmol of substrate per minute.
2.6. Triphenylmethane Reductase (TMR) Assay
The standard assay [22] system for TMR comprised 250 μL of 20 mM sodium phosphate buffer (pH 7.0), 250 μL of 20 μM basic fuchsin, 250 μL of 0.1 mM NADH, and 250 μL of the enzyme in a total volume of 1 mL. Each reaction was initiated by the addition of the enzyme, and the initial reaction rate was determined by monitoring the decrease in absorbance at 544 nm in the first 2 min in a temperature-controlled cuvette in a 1.0-cm light path at 40˚C. One unit of enzyme activity was defined as the amount that catalyzed the reduction of 1 μmol of basic fuchsin per min per mL. The corresponding wavelengths were used when other triphenylmethane dyes were tested in place of basic fuchsin.
3. RESULT
Table 1 indicates the result showing pH, dry mycelial weight (DMW), percentage of decolorization and lingolytic enzyme activities of F. feei in shaking condition. With increasing incubation time, pH of the fungal culture broth in all dyes was decreased, which is inversely proportional to dry mycelial weight and percentage of de-
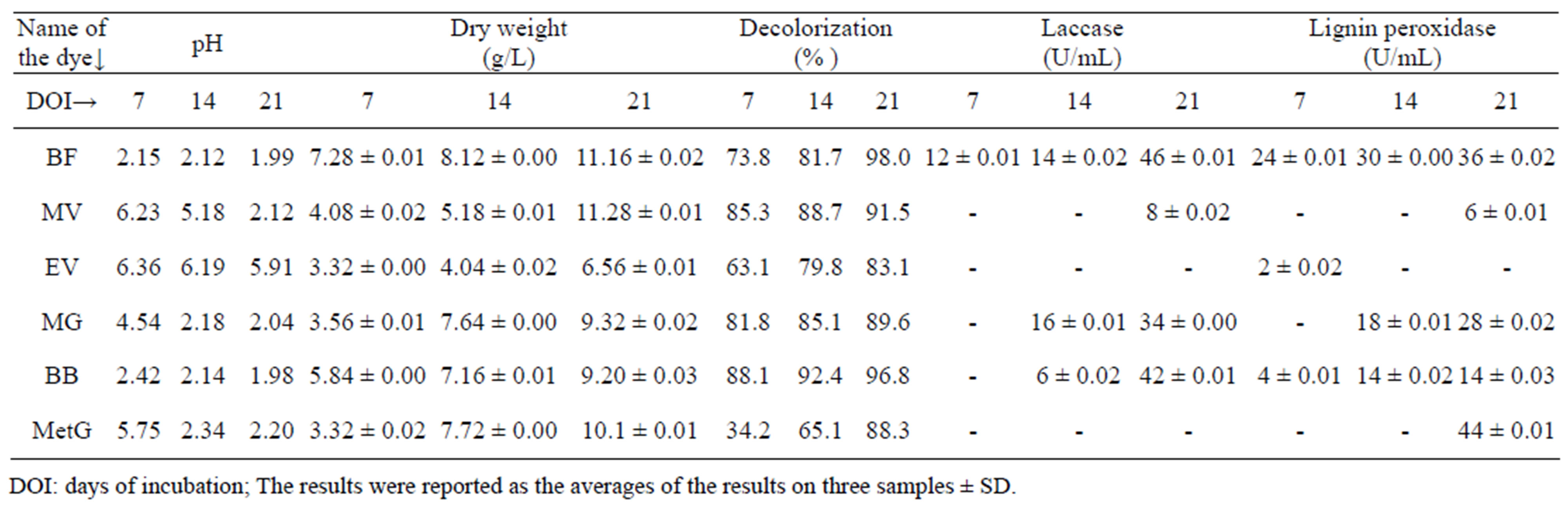
Table 1. Decolorization of triphenyl methane dyes at 0.001% by Fomitopsis feei and their effect on growth and lignolytic enzyme production.
colorization. The growth of F. feei was not inhibited by the dye solution and was increased with the days of incubation, with all dyes. The maximum growth was observed in basic fuchsin after 21 days of incubation (11.16 g/L).
Maximum decolorization with F. feei was observed in basic fuchsin after 21 days of incubation (98%) followed by bromophenol blue (96.8%). The rate of bromophenol blue decolorization was very fast (Figure 1) may be due to the less complex structure of bromophenol blue compared to the other two dyes tested. The rate of decolorization in basic fuchsin (Figure 2) was increased from 73.8% (7 days) to 98% (21 days). 85.3% of decolorization was observed in methyl violet (Figure 3) after 7 days of incubation, which was then increased to 91.5% after 21 days of incubation. Although the initial decolorization of methyl green (Figure 4) was less in 7 days (34.2%), it could reach upto 88.3% after 21 days of incubation but there was no laccase activitiy. However, it is strange that the decolorization of ethyl violet (Figure 5) was increased from 63% to 83% from 7 to 21 days without showing laccase activity. There was no much difference in the rate of decolorization of malachite green (Figure 6) viz. 81.8%, 85.1% and 89.6% in 7, 14 and 21 days respectively. From these results, it is revealed that the percentage of decolorization was increased with incubation time (Figure 7).
Production of lignolytic enzymes during the decolorization varied depending upon the incubation time (Figure 8). The highest lignolytic activity (46 U/mL) was shown in basic fuchsin followed by bromopheonol blue (42 U/mL) after 21 days of incubation. 44 U/mL of lignin peroxidase was observed with methyl green followed by basic fuchsin (36 U/mL) after 21 days of incubation. The presence of both laccase and lignin peroxidase activities were observed in basic fuchsin. Although laccase, lignin peroxidase activities were not observed in methyl violet initially, 8 U/mL and 6 U/mL activities were observed

Figure 1. Decolorization of bromophenol blue by Fomitopsis feei.

Figure 2. Decolorization of basic fuchsin by Fomitopsis feei.

Figure 3. Decolorization of methyl violet by Fomitopsis feei.
respectively after 21 days of incubation. Although there was no laccase and lignin peroxidase activity in ethyl violet and methyl green, 2 U/mL lignin peroxidase (7

Figure 4. Decolorization of methyl green by Fomitopsis feei.

Figure 5. Decolorization of ethyl violet by Fomitopsis feei.

Figure 6. Decolorization of malachite green by Fomitopsis feei.
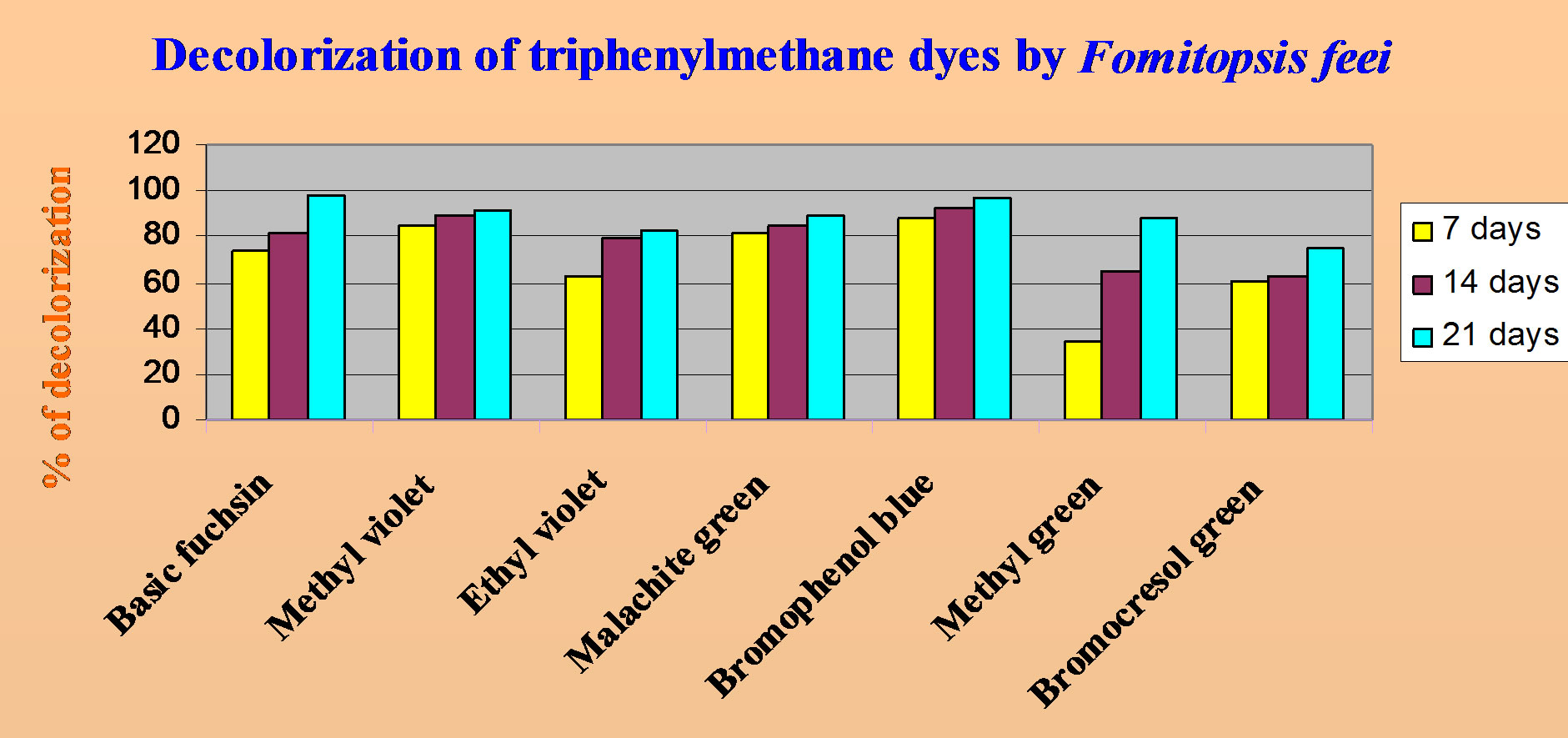
Figure 7. Comparision of decolorization of triphenyl methane dyes by Fomitopsis feei.
days) was showed in ethyl violet which was not observed thereafter. Laccase, lignin peroxidase were not released upto 7 days in malachite green but afterwards were increased with incubation time. The presence of lignolytic activity was not correlated with the decolorization in all dyes since laccase and lignin peroxidase activities were not resulted in almost all dyes. TMR assay was done using NADH co-factor (Table 2). The presence of this
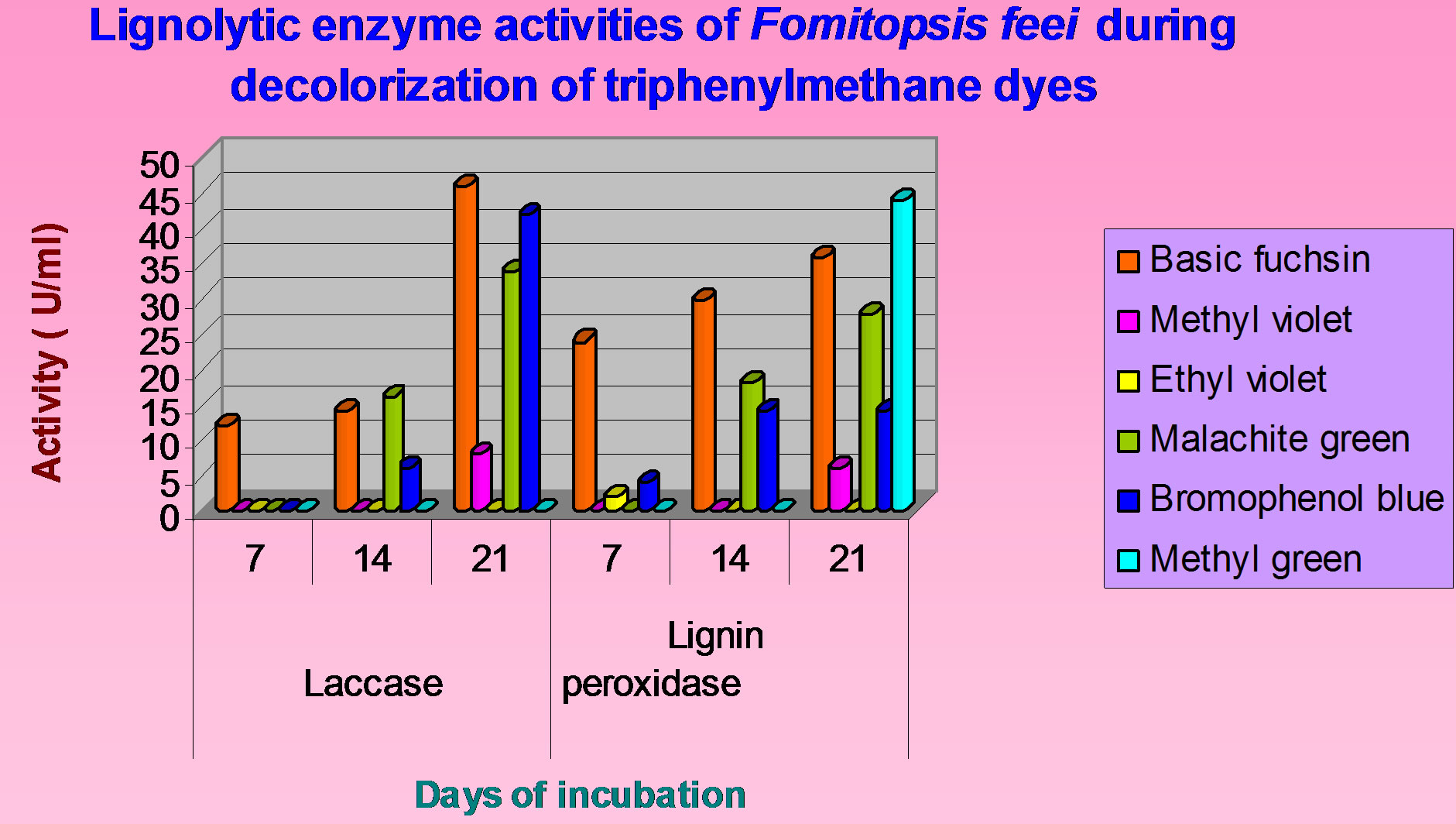
Figure 8. Lignolytic enzyme activities of Fomitopsis feei during decolorization of triphenylmethane dyes.
enzyme with decolorization all these dyes showed the specific activity of this enzyme towards decolorization. The highest activity of TMR was observed in malachite green (0.447 µmole/min/mL) followed by methyl violet (0.312 µmole/min/mL).
4. DISCUSSION
There are very few reports on the biological decolorization and degradation of textile and dye-stuff industrial wastes containing triphenylmethane dyes. All the reports so far available are on a small scale. Triphenylmethane dyes have been reported to undergo extensive biodegradation by ligninolytic cultures of P. chrysosporium [23, 24]. The structural genes encoding lignin peroxidase and laccase have been cloned and characterized [25,26]. The biochemical mechanism underlying the decolorization of triphenylmethane dyes has been elucidated in fungi [11]. Several species of fungi are known to decolorize triphenylmethane dyes by a variety of enzymes as laccase [27] lignin peroxidase [24,28]. After 18 days Lentinula (Lentinus) edodes completely decolorized methyl green, methyl violet, ethyl violet [29].
Ollika et al. [30] purified iso-enzymes of lignin peroxidases from P. chrysosporium and studied the decolorization efficiencies for several dyes including azo and triphenylmethane dyes. Synthetic dyes crystal violet and
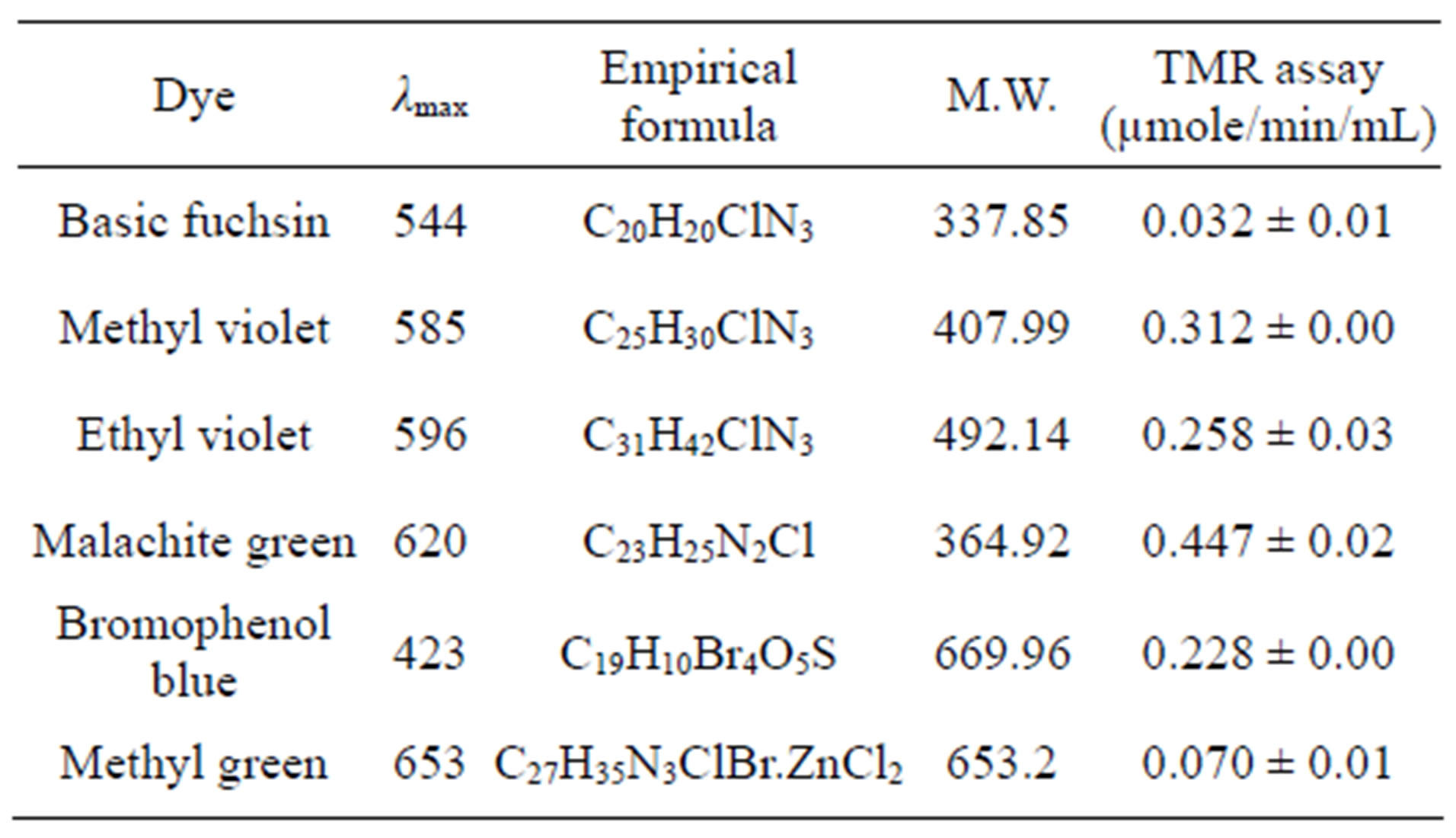
Table 2. Details of triphenyl methane dyes and TM reductase assay of Fomitopsis feei.
malachite green showed 100% and 90% of decolorization on the PDA media [31]. In liquid culture, P. calyptratus was able to decolorize crystal violet and malachite green during 14 days of cultivation in Kirk medium after 14 days the extents of decolorization were only 5 and 27% of the dye, respectively [32]. Decolorization of triphenylmethane dyes (crystal violet, bromophenol blue and malachite green) by three birds nest fungi—Cyathus bulleri, C. stercoreus, C. striatus was reported [27]. Among the three organisms, C. buleri was found to be the most efficient in decolorization.
Particularly, the usefulness of the brown rot fungi for decolorization of wastewater has not been probed at all. Polyporus ostreiformis produced lignin peroxidase along with manganese peroxidase, acid protease, a-amylase in nitrogen-limited liquid media for decolorization [33]. Studies with the brown-rot fungi Gloeophyllum trabeum and Fomitopsis pinicola on liquid media revealed no lignin peroxidase or manganese-dependent peroxidase activity, although nonspecific peroxidase activity was detected [34]. No induction in laccase, lignin peroxidase and tyrosinase activity was observed during decolorization process and significant induction in malachite green reductase (73%) activities were observed during decolorization of malachite green by K. rosea [35]. An enzyme that decolorizes triphenylmethane dyes is designated triphenylmethane reductase (TMR) purified from Citrobacter sp. strain KCTC 18061P and characterized, cloned of the gene encoding this enzyme and its heterologous expression in Escherichia coli was reported by [22].
From our results also it is confirmed that triphenyl methane reductase enzyme plays specific role in the decolorization process compared to lignolytic enzymes. But lignolytic enzyme production was also supported that they might be definitely useful in the degradation process. In contrast to the previous research [18] in which methyl green was not decolorized by F. feei, our result showed the decolorization of methyl green upto 88.3%. In their research they reported that bromophenol blue was decolorized upto 64.8% but in our present research it was decolorized upto 96.8% after 21 days of incubation period. This difference might be in days of incubation and the concentration of dye used. Our findings could contribute to a better knowledge of the properties of F. feei, which has as yet not been studied in detail and its decolorization abilities could be promising for further biotechnological applications.
5. CONCLUSION
It may be concluded from the result that it is possible to decolorize or degrade triphenylmethane dyes by biological means. Biological treatment is the only way for ultimate controlling of pollution generated by textile and dye-stuff industries. Fungal decolorization is a promising alternative to replace or supplement present treatment processes. In conclusion, our results showed that F. feei is able to decolorize efficiently several synthetic dyes belonging to triphenylmethane group.
6. ACKNOWLEDGEMENTS
We thank the Head, Department of Microbiology, Kakatiya University, Warangal for providing necessary laboratory facilities and gratefully acknowledge the funding from University Grants Commission, New Delhi.
REFERENCES
- US Environmental Protection Agency (2005) Waste from the production of dyes and pigments listed as hazardous. Factsheet 530-F-05-004. http://www.epa.gov/epawaste/hazard/wastetypes/wasteid/dyes/dyes-ffs.pdf
- Gregory, P. (1993) Dyes and dyes intermediates. In: Kroschwitz, J.I., Ed., Encyclopedia of Chemical Technology, John Wiley & Sons, New York, 544-545.
- Case, R.A.M. and Pearson, J.T. (1954) Tumours of the urinary bladder in workmen engaged in the manufacture and the use of certain dyestuff intermediates in the British chemical industry. British Journal of Indian Medicine, 11, 213-221.
- Fernandes, C., Lalitha, V.S. and Rao, K.V.K. (1991) Enhancing effect of malachite green on the development of hepatic pre-neoplastic lesions induced by N-nitrosodiethylamine in rats. Carcinogenesis, 12, 839-845. doi:10.1093/carcin/12.5.839
- Littlefield, N.A., Blackwell, B.N., Hewitt, C.C. and Gaylor, D.W. (1985) Chronic toxicity and carcinogenicity studies of gentian violet in mice. Fundamental and Applied Toxicology, 5, 902-912. doi:10.1016/0272-0590(85)90172-1
- Rao, K.V.K. (1995) Inhibition of DNA synthesis in primary rat hepatocyte cultures by malachite green: A new liver tumor promoter. Toxicology Letters, 81, 107-113. doi:10.1016/0378-4274(95)03413-7
- Schnick, R.A. (1988) The impetus to register new therapeutants for aquaculture. Progressive Fish-Culturist, 50, 190-196. doi:10.1577/1548-8640(1988)050<0190:TITRNT>2.3.CO;2
- Burchmore, S. and Wilkinson, M. (1993) “Department of the Environment, Water Research Center, Marlow, Buckinghamshire, United Kingdom,” Report no. 316712, Carcinogenesis, 12, 839-845.
- Nelson, C.R. and Hites, R.A. (1980) Aromatic amines in and near the Buffalo River. Environmental Science and Technology, 14, 1147-1149. doi:10.1021/es60169a020
- Banat, I.M., Nigam, P., Singh, D. and Marchant, R. (1996) Microbial decolorization of textile-dye-containing effluxents: A review. Bioresource Technology, 58, 217-227. doi:10.1016/S0960-8524(96)00113-7
- Azmi, W., Sani, R.K. and Banerjee, U.C. (1998) Biodegradation of triphenylmethane dyes. Enzyme Microbial Technology, 22, 185-191. doi:10.1016/S0141-0229(97)00159-2
- Ali, N., Hameed, A. and Ahmed, S. (2008) Decolorization of structurally different textile dyes by Aspergillus niger SA1. World Journal of Microbiology and Biotechnology, 24, 1067-1072. doi:10.1007/s11274-007-9577-2
- Ambrosio, S.T. and Campos-Takaki, G.M. (2004) Decolorization of reactive azo dyes by Cunninghamella elegans UCP 542 under co-metabolic conditions. Bioresource Technology, 91, 69-75. doi:10.1016/S0960-8524(03)00153-6
- Fahraeus, G. (1952) Formation of laccase by Polyporus versicolor in different culture media. Physiology Plant, 5, 284-291. doi:10.1111/j.1399-3054.1952.tb07716.x
- Husseiny, S.M. (2008) Biodegradation of the reactive and direct dyes using Egyptian isolates. Journal of Applied Science Research, 4, 599-606. http://www.aensiweb.com/jasr/jasr/2008/599-606.pdf
- Sumathi, S. and Manju, B.S. (2000) Uptake of reactive textile dyes by Aspergillus foetidus. Enzyme Microbial Technology, 27, 347-355. doi:10.1016/S0141-0229(00)00234-9
- Zhang, X., Youxun, L., Keliang, Y. and Hangjun, W. (2007) Decolorization of anthraquinone-type dye by bilirubin oxidase producing nonligninolytic fungus Myrothecium spp. IMER1. Journal of Bioscience and Bioengineering, 104, 104-110. doi:10.1263/jbb.104.104
- Lyra, E.S., Moreira, K.A., Porto, T.S., Carneiro da Cunha, M.N., Paz Jr., F.B., Neto, B.B., Lima-Filho, J.L., Cavalcanti, M.A.Q., Converti, A. and Porto, A.L.P. (2009) Decolorization of synthetic dyes by basidiomycetes isolated from woods of the Atlantic Forest (PE). Brazilian World Journal of Microbiology and Biotechnology, 25, 1499- 1504. doi:10.1007/s11274-009-0034-2
- López, M.J., Guisado, G., Vargas-García, M.C., SuárezEstrella, F. and Moreno, J. (2006) Decolorization of industrial dyes by ligninolytic microorganisms isolated from composting environment. Enzyme Microbial Technology, 40, 42-45. doi:10.1016/j.enzmictec.2005.10.035
- Coll, M.P., Fernandez-Abalos, J.M., Villanueva, JR., Santamaria, R. and Perez, P. (1993) Purification and characterization of phenoloxidase (Laccase) from the lignin— Degrading basidiomycete PM I (CECT 2971). Applied Environmental Microbiology, 59, 2607-2613. http://aem.highwire.org/content/59/8/2607.full.pdf+html
- Sarakanen, S., Razal, R.A., Piccariello, T., et al. (1999) Lignin peroxidase: Toward a classification of its role in vivo. Journal of Biological Chemistry, 266, 3636-3643. http://www.jbc.org/content/266/6/3636.long
- Jang, M.-S., Lee, Y.-M., Kim, C.-H., Lee, J.-H., Kang, D.-W., Kim, S.-J. and Lee, Y.-C. (2005) Triphenylmethane reductase from Citrobacter sp. Strain KCTC 18061P: Purification, characterization, gene cloning, and overexpression of a functional protein in Escherichia coli. Applied Environmental Microbiology, 71, 7955-7960. doi:10.1128/AEM.71.12.7955-7960.2005
- Field, A.J., de Jong, E., Feijoo-Costa, F.E. and de Bont, J.A.M. (1993) Screening for ligninolytic fungi applicable to the biodegradation of xenobiotics. Trends in Biotechnology, 11, 44-48. doi:10.1016/0167-7799(93)90121-O
- Bumpus, J.A. and Brock, B.J. (1988) Biodegradation of crystal violet by white rot fungus Phanerochaete chrysosporium. Applied Environmental Microbiology, 54, 1143- 1149. http://aem.asm.org/content/54/5/1143
- Cullen, D. (1997) Recent advances on the molecular genetics of ligninolytic fungi. Journal of Biotechnology, 53, 273-289. doi:10.1016/S0168-1656(97)01684-2
- Gold, M.H. and Alic, M. (1993) Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiology Review, 57, 605-622.
- Vasdev, K., Kuhad, R.C. and Saxena, R.K. (1995) Decolorization of triphenylmethane dyes by the birds nest fungus Cyathus bulleri. Current Microbiology, 30, 269-272. doi:10.1007/BF00295500
- Shin, K.S. and Kim, C.J. (1998) Decolorization of artificial dyes by peroxidase from the white-rot fungus Pleurotus ostreatus. Biotechnology Letters, 20, 569-572. doi:10.1023/A:1005301812253
- Boer, C.G., Obici, L., de Souzam C.G.M. and Peralta, R.M. (2004) Decolorization of synthetic dyes by solid state cultures of Lentinula (Lentinus) edodes producing manganese peroxidase as the main ligninolytic enzyme. Bioresource Technology, 94, 107-112. doi:10.1016/j.biortech.2003.12.015
- Ollikka, P., Alhonmaki, K., Leppanen, V.M., Glumoff, T., Raijola, T. and Suominen, I. (1993) Decolorization of azo, triphenylmethane, heterocyclic, and polymeric dyes by lignin peroxidase isozymes from Phanerochaete chrysosporium. Applied Environment and Microbiology, 59, 4010- 4016.
- Krishnaveni, M. and Kowsalya, R. (2011) Characterization and decolorization of dye and textile effluent by laccase from Pleurotus florida—A white-rot fungi. International Journal of Pharma and Bio Sciences, 2, B117- B123.
- Eichlerova, I., Homolka, L. and Nerud, F. (2006) Ability of industrial dyes decolorization and ligninolytic enzymes production by different Pleurotus species with special attention on Pleurotus calyptratus, strain CCBAS 461. Process Biochemistry, 41, 941-946. doi:10.1016/j.procbio.2005.10.018
- Dey, S., Maiti, T.K. and Bhattacharyya, B.C. (1994) Production of some extracellular enzymes by a lignin peroxidase-producing brown rot fungus, Polyporus ostreiformis and its comparative abilities for lignin degradation and dye decolorization. Applied Environment and Microbiology, 60, 4216-4218. http://aem.asm.org/content/60/11/4216.full.pdf
- Freitag, M. and Morrell, J.J. (1992) Decolorization of the polymeric dye Poly R-478 by wood-inhabiting fungi. Cannadian Journal of Microbiology, 38, 811-822. doi:10.1139/m92-133
- Parshetti, G., Kalme, S., Saratale, G. and Govindwar, S. (2006) Biodegradation of malachite green by Kocuria rosea MTCC 1532. Acta Chimica Slovenica, 53, 492-498. http://acta.chem-soc.si/53/53-4-492.pdf

