Natural Science
Vol.5 No.4A(2013), Article ID:30834,15 pages DOI:10.4236/ns.2013.54A005
Why antibiotics: A comparative evaluation of different hypotheses for the natural role of antibiotics and an evolutionary synthesis
![]()
1Anujeeva Biosciences Pvt Ltd, Pune, India
2Department of Microbiology, Abasaheb Garware College, Pune, India
3Indian Institute of Science Education and Research, Pune, India;
*Corresponding Author: milind@iiserpune.ac.in
Copyright © 2013 Charushila Kumbhar, Milind Watve. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 30 November 2012; revised 30 December 2012; accepted 15 January 2013
Keywords: Secondary Metabolites; Antibiotics; Evolutionary Synthesis; Streptomyces
ABSTRACT
Although secondary metabolites with antimicrobial and other bioactivities are explored extensively, the natural or ecological role(s) of secondary metabolites is not yet clearly known. We review here the different hypotheses for the ecological role of antibiotics, with particular focus on the genus Streptomyces which is unparalleled in the richness of secondary metabolites. We first lay down our expectations from an ecological hypothesis for antibiotics and then weigh the six predominant hypotheses against them including antibiotics as weapons in competition, as aid in sporulation, as bartered benefits in symbioses, as signal molecules in community homeostasis, as weapons in predation and as metabolic waste or bi-products. The analysis shows that no single hypothesis meets all the expectations. While the waste or bi-product hypothesis can safely be eliminated all others have some evidence in support. It is possible therefore that antibiotics serve a multitude of ecological functions and it is possible to visualize a pathway for the radiating functions. According to this synthesis antibiotics evolved primarily as weapons in predation on other microorganisms. The inevitable co-evolution with prey species led to diversification of the genes and pathways. Some of the secondary metabolites eventually radiated to acquire other functions such as competition between predators. Some secondary metabolites evolved animal toxicity as a mutualistic barter to protect the symbiotic partner from grazing/predation by animals. Transcription modulation primarily evolved as activation of defense mechanisms by the prey which may have later radiated to serve interspecies signaling functions. The synthesis successfully links different functions of antibiotics with logical coherence.
1. INTRODUCTION
The single genus that has contributed to a large proportion of antibiotics discovered so far is the genus Streptomyces [1-3]. Using trend analysis, Watve et al [1] showed that the reduction in discovery rate was not due to exhaustion of the resource. Fitting a model to the trend of antibiotic discovery they made an estimate of the total number of secondary metabolites the Streptomyces group may possess and argued that majority of antibiotics from this genus were yet to be discovered. The Streptomyces coelicolor genome published in the following year supported the inference further [4]. After having genomic data on a few other members of this genus we know now that the Streptomyces group has a large number of gene clusters responsible for secondary metabolites which are not expressed at least during growth in conventional culture. Further there is considerable beta diversity across different strains of this group as well [2,5-7], pointing to a large fraction of untapped total diversity of secondary metabolites in this group [8,9]. Genomic information across the bacterial world also asserts us that the apparent majority of known antibiotics coming from mycelial actinomycetes is not a bias introduced by screening methods. Genomically the richness of secondary metabolites in Streptomyces and other mycelial actinomycetes remains unparalleled in the microbial world. Although we have extensive information on the chemistry, bioactivity and genetics of a few thousand secondary metabolites from this group, the fundamental questions as to their ecological role and evolutionary origins remain unanswered [10].
There have been a number of hypotheses suggested from time to time which we review and evaluate here. But before we could see the hypotheses we need to delineate our expectations from the hypotheses. Any hypothesis or a combination of them should explain the following known patterns in the biology of antibiotics.
1.1. The Time of Production
Almost all secondary metabolites are produced by the end or growth phase and beginning of stationary phase. Any hypothesis needs to explain the adaptive significance of this pattern.
1.2. The Genomic and Physiological Costs of Secondary Metabolism
Secondary metabolite gene clusters occupy a significant portion of the genome, (e.g. about 4.5% in S. coelicolor). Also the synthesis involves operation of complex pathways involving several enzymes and endergonic reactions [2]. Any benefit of antibiotics should be sufficiently large to justify the costs.
1.3. The Alpha and Beta Diversity
A large number of compounds have been discovered so far from Streptomyces and the number of undiscovered compounds are claimed to be order of magnitude larger. The large diversity of secondary metabolites within species (alpha diversity) and the turnover across species (beta diversity) in the mycelial actinomycetes is astonishing. This is unlikely to have evolved by chance and needs specific explanation.
1.4. The Unexpressed Majority
Out of the extant diversity, a substantial fraction is not expressed in conventional culture. This needs to be accounted for by the hypothesis.
1.5. The Overexpressed Minority
A relatively small fraction of the compounds are produced in large quantities. This minority has been a major resource of drug discovery for us. But the reasons why only a minority are overexpressed needs to be clarified and the hypothesis should help in this direction.
1.6. Difference in Expression in Solid Surface versus Fluid Environment
It was known from the early days of screening that not all isolates giving zones of inhibition in primary screening give antibiotics at the shake flask or fermenter level. There is a difference in antibiotic production on solid surface and in liquid environment [11-15]. The transcriptional response to sublethal doses of antibiotics is also different in solid and liquid environments [16] and this needs a logical adaptive explanation too.
1.7. The Spectrum of Biological Action
Secondary metabolites of Streptomyces have a variety of biological activities not restricted to antimicrobial action. There are immunomodulatory and other actions on mammalian physiology as well. They are unlikely to be coincidental and need an adaptive explanation.
1.8. The Transcription Modulation Activity in Susceptible Organisms
Almost all antibiotics tested so far have shown transcription modulation activity specifically in susceptible organisms in low doses [17,18]. Resistance to an antibiotic also reduces the transcription modulation action substantially [16]. Any hypothesis needs to account for this adequately.
1.9. The Mycelial Bias
Across the microbial world as well as within Actinobacteria the mycelial forms are richer in secondary metabolite diversity. As we will show below, this association is strong and therefore is likely to have an adaptive reason leading to this association.
2. THE HYPOTESES
In addition to being able to explain the above, the hypothesis needs to have experimental evidence and/or should make experimentally testable predictions. The spectrum of distribution of the hypothesized function and the strength of its association with antibiotic production within the genus Streptomyces and also across the microbial world can strengthen the hypothesis further. Having specified our expectations from an ecological hypothesis, let us now see the hypotheses suggested so far along with their major strengths and weaknesses.
2.1. Competition
Since antibiotics inhibit or kill a spectrum of other microbial species that can be potential competitors, antibiotics are implicated as weapons used in interference competition [18-28]. The potential problem in this hypothesis is that most of the secondary metabolites are produced towards the end of the growth phase and therefore are unlikely to be useful during competition for growth. Attempts to test whether antibiotic producers have an advantage in experimental competition are few and have yielded mixed results [29-31] that have possible alternative interpretations. Other products such as microcins that have been clearly demonstrated to give a competitive advantage are produced during the exponential phase [32]. Therefore the competitive role of antimicrobials produced past the exponential phase is questionable.
2.2. Waste or Bi-Products
Selman Waksman, the discoverer of streptomycin, observed that antibiotics played no real part in modifying or influencing living processes of the producer. It was suggested subsequently that secondary metabolites of plant, animal or microbial origins are metabolic spin offs from primary metabolism, or aberrant and valueless products inevitably formed in the systems [33-43]. It was suggested that some of them happen to have other biological activities coincidently. However, this viewpoint was out of favor soon since the large majority of secondary metabolites were found to have some or the other biological activity. They are produced through various metabolic pathways controlled by different gene clusters, incurring costly investment for the cell [2]. Also the waste product or spin off hypothesis does not explain the great diversity of these compounds produced by a relatively smaller proportion of microorganisms. If secondary metabolite production was an inevitable metabolic demand, then it would have been distributed throughout the living world more or less evenly.
2.3. Spore Development and/or Survival
Majority of organisms that produce antibiotics go through a life cycle in which spores are produced. Both spores and antibiotic production are initiated towards the end of growth phase. It has therefore been suggested that antibiotics may be regulatory molecules that function to coordinate the developmental process or maintain the dormancy of the spore [44]. However, non-antibioticproducing strains appear to produce spores normally and their spores do not seem to be inferior in any way. Mutants with blocked antibiotic production produce normal spores [45]. Although there is a strong association between the time course of sporulation and antibiotic production the association is non-obligate since the signals for sporulation and antibiotic production are largely nonoverlapping [46].
2.4. Symbiosis
Many species of Streptomyces appear to have entered a mutualistic relationship with other organisms in which they protect the organism by their antibiotics and get nutrition in turn. For example Streptomyces symbionts in the beewolf digger wasp protect their cocoon from fungal infection by producing a cocktail of 9 antibiotics and significantly enhance the larva’s chances of survival during hibernation in the soil [47,48]. Non leguminous angiosperm species having actinomycetes in root-nodule have also been noted [49-51]. Some of the fungus-growing ants use antibiotic-producing actinomycetes to control garden parasites [52-59]. Marine soft bodied nonmotile invertebrates such as sponges do not have active defenses and therefore appear to have depended on chemical defense made possible by the secondary metabolites of symbiotic actinomycetes [60-62]. Less known is the symbiosis with lichens and honey bees [63]. It is not yet known how common such symbioses are, what fraction of Streptomyces species are symbionts and therefore whether the symbiotic associations are sufficient to explain the richness of secondary metabolites in the genus.
2.5. Antibiotics as Signaling Molecules
One of the relatively recent suggestions has been that antibiotics are signaling molecules that may regulate the homeostasis of multispecies microbial communities in a coordinated manner [18,64-69]. So far there are no specific suggestions as to the precise meaning of “homeostasis of microbial communities” [18] and the exact role of the signaling molecules. At low concentrations, antibiotics can perhaps be beneficial for the behavior of susceptible bacteria in natural environment. The response of susceptible bacteria is suggested to be “low dose stimulation and high dose inhibition” [66] or hormesis. The evidence for this suggestion is that a number of antibiotics tested have been shown to alter transcription of several genes in susceptible organisms. The precise nature and adaptive function of this signaling is not yet speculated. For the producing organisms it may serve a quorum sensing function, but the transcription regulation activity is seen in susceptible species as well whose function is not yet clearly known. If it serves an essential adaptive function then the unanswered question is why the diversity of secondary metabolites is biased towards some specific groups of microorganisms. Since the compounds are produced by specific groups of organisms, it is necessary to ask why some organisms bear the cost for a trait that is beneficial to a multispecies community.
2.6. Predation
A number of taxonomically diverse genera of bacteria have been shown to exhibit epibiotic mode of predation. Interestingly all the genera whose epibiotic predatory activity has been demonstrated are also shown to express
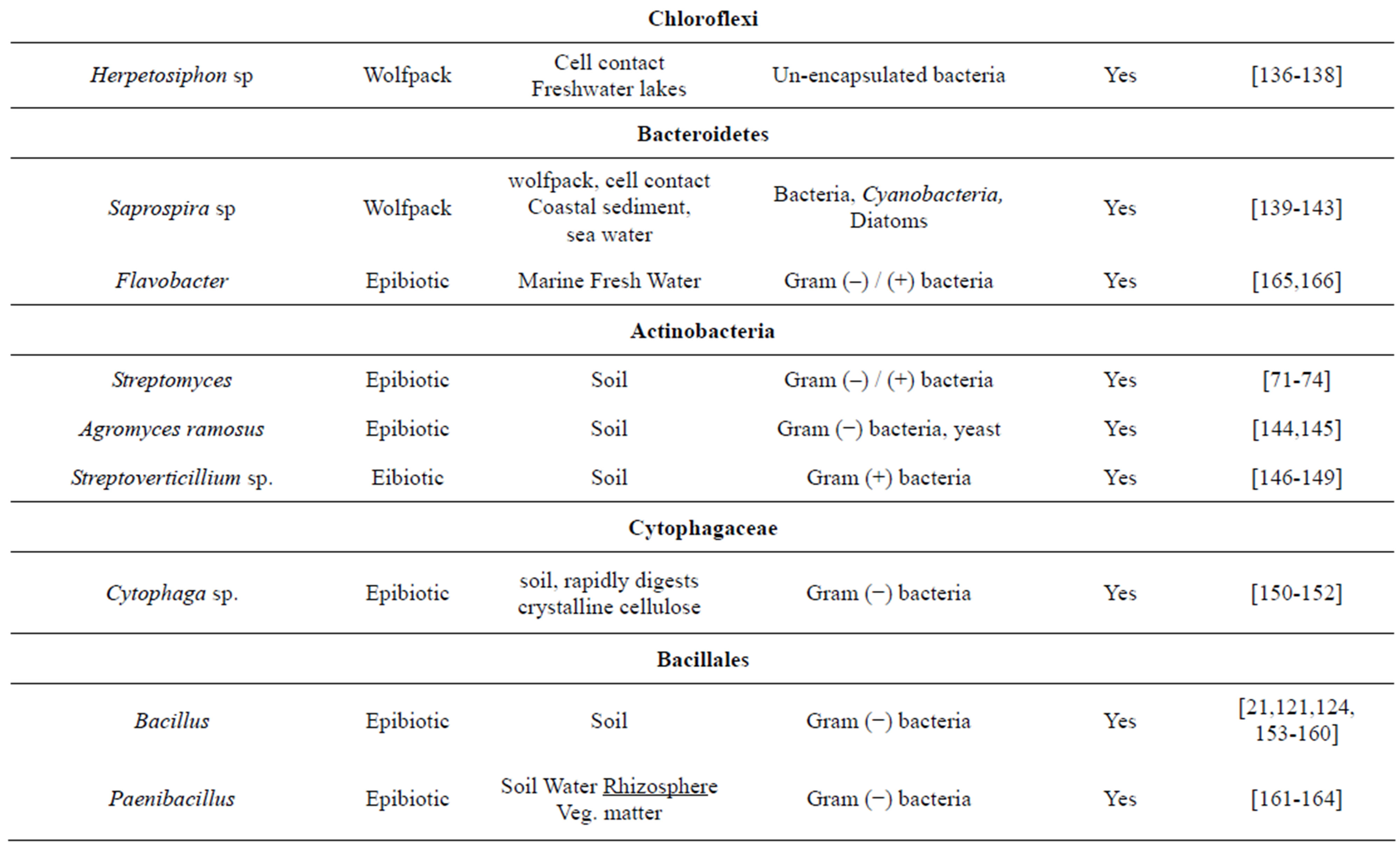
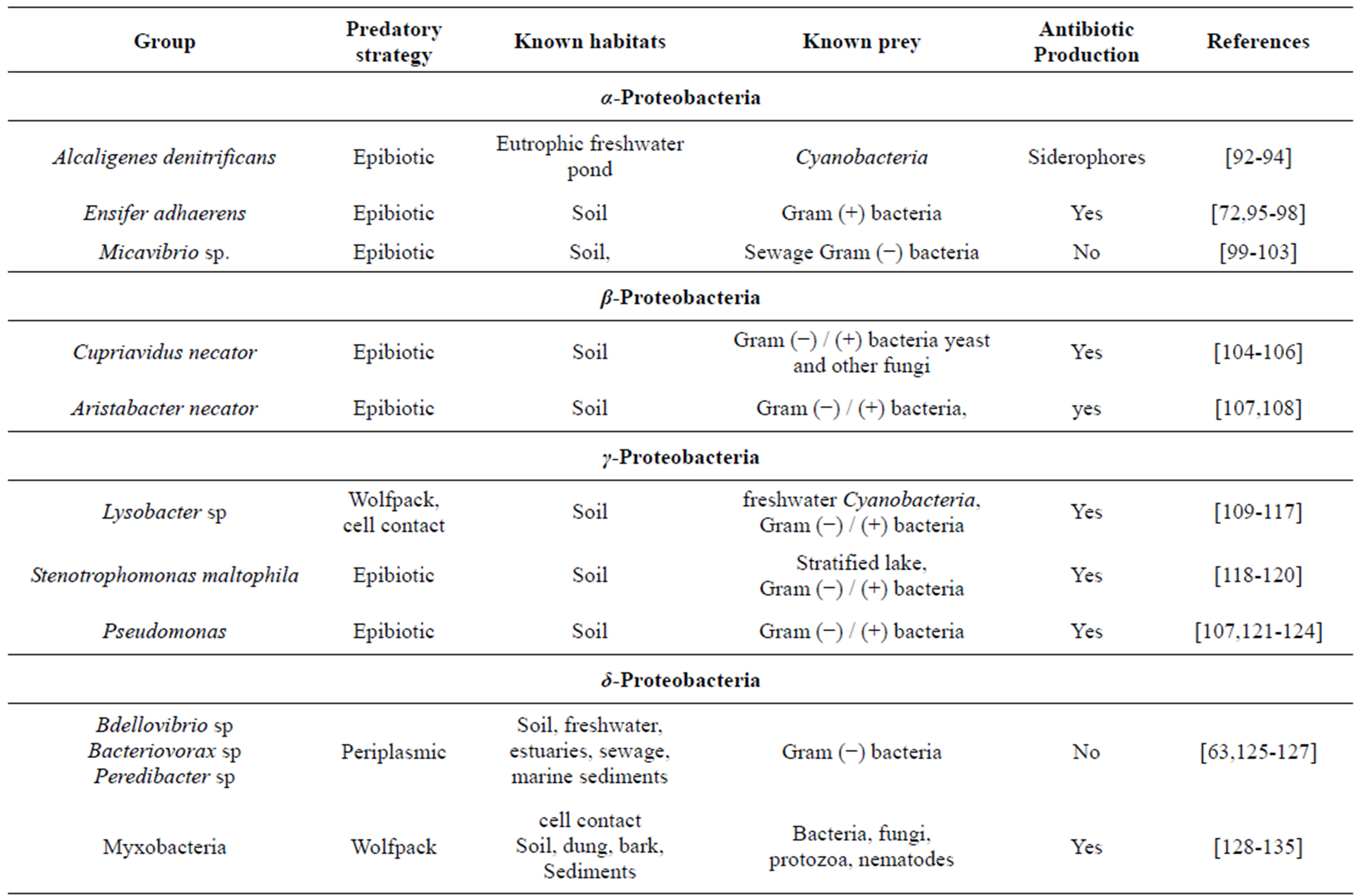
Table 1. Meta-analysis of predatory bacteria and their secondary metabolites. We searched literature for predatory bacterial genera. each of the genera was searched for evidence of secondary metabolites with antimicrobial action. It appears from this analysis that all epibiotic and wolfpack predatory bacteria have antibiotic activity whereas the parasite like forms do not.
secondary metabolites having antimicrobial activity (Table 1). Myxobacteria is an important group of predatory bacteria rich in antibiotics although not as much explored as actinomycetes. It is further demonstrated that mutants unable to express antibiotic activity failed to grow by predatory mode on prey cells [70]. This is perhaps the most direct demonstration of the involvement of antibiotics in predation. Predatory abilities are widespread in the genus Streptomyces too [71-73, Kumbhar et al, manuscript under review]. If predation is defined as the ability to lyse live cells and exhibit growth when no other nutrients are available, the initial experiments for antibiotic screening demonstrated predation although it was not interpreted that way. Waksman and Woodruff screened actinomycetes on a lawn of bacteria on a non-nutrient plate and observed zones of clearance of susceptible cells. This is nothing other than predation [74]. Predation is needed when other nutrients are scanty. This explains why antibiotics are produced by the end of growth phase or under nutrient stress. The strong association between predatory activity and antibiotic production across taxa, the demonstration that predatory abilities are extremely common in soil Streptomyces and the ability of the predation hypothesis to explain the time course of antibiotic production makes it a strong candidate hypothesis.
3. HOW DO THE HYPOTHESES EXPLAIN THE PATTERNS
We listed earlier a number of known patterns in the biology of antibiotics which any hypothesis needs to explain. We will see how each of them is accounted for by the competing hypotheses. The possible stand of each hypothesis on these patterns is summarized in Table 2 and discussed elaborately below.
3.1. Time of Production
Most of the secondary metabolites are secreted by the end of the growth phase or under conditions of nutrient stress [75]. This has been a main argument against the competition hypothesis but is also inconvenient for many other hypotheses. If antibiotics are useful in facilitating growth by reducing competition, modulating transcripttion or any other means then they should be produced during active growth. If they are bi-products of normal metabolism they should be produced when the metabolism is most active, i.e. during the growth phase. The time of production and nutrient stress is supportive of the sporulation hypothesis since this is the right time and stimulus for sporulation. The predation hypothesis also expects antibiotic production on exhaustion of other nutrients. It is also possible that inclusion of some antibiotic in the spore may help survival of the germinating spore by making predatory activity possible immediately after germination. There is some evidence that stored antibiotics are released by germinating spores [21]. The signal hypothesis may argue that the gene expression altered in response to antibiotics facilitates sporulation. Thus the sporulation, signal and predation hypotheses are compatible with the time and nutrient conditions of antibiotic production whereas other hypotheses face a difficulty here.
3.2. The Genomic and Physiological Cost of Secondary Metabolism
A costly trait has evolutionary stability if the individuals incurring the cost get a specific benefit relative to others which is greater than the cost incurred. The signaling hypothesis fails on this ground since the proposed benefit is shared by the multispecies community and the cost is borne by the producer alone. If the cost of producing the signaling molecules is borne by some species and the benefit goes to the entire multispecies community, the evolutionary stability of such a system is questionable. Therefore secondary metabolites are most unlikely to have evolved primarily for signaling purpose. The cost factor is also unfavorable for the waste product hypothesis. Other hypotheses namely competition, symbiosis and predation confer direct benefits to the producer. Here too there is a chance of cheater mutants invading. As explained below, mycelial forms have tackled the cheater problem in a different way.
3.3. Alpha and Beta Diversity of Antibiotics
Co-evolutionary arms race is a process that can lead to such extraordinary characters. A good example is the diversity of immunity related genes in higher vertebrates which has evolved by a coevolutionary arms race with pathogens and parasites. Competition, symbiosis and predation hypotheses imply such co-evolutionary processes whereas no other hypothesis offers a convincing explanation. Predation has been shown to boost diversity [76] and specialization on prey species by predators which is demonstrated in myxobacterial predation [77] as well as apparent in our studies with Streptomyces is sufficient to drive the co-evolutionary path to increasing diversity. A coevolutionary process implies evolution of counter-strategies too. This is evident by the wide variety of mechanisms of antibiotic resistance.
3.4. The Unexpressed Majority of Secondary Metabolite Gene Clusters
The most likely explanation for the wide diversity of unexpressed secondary metabolites is that their unexpressed state is an artifact of studying them under restricted and unnatural cultural conditions. This is compatible with the competition, signaling, symbiosis and
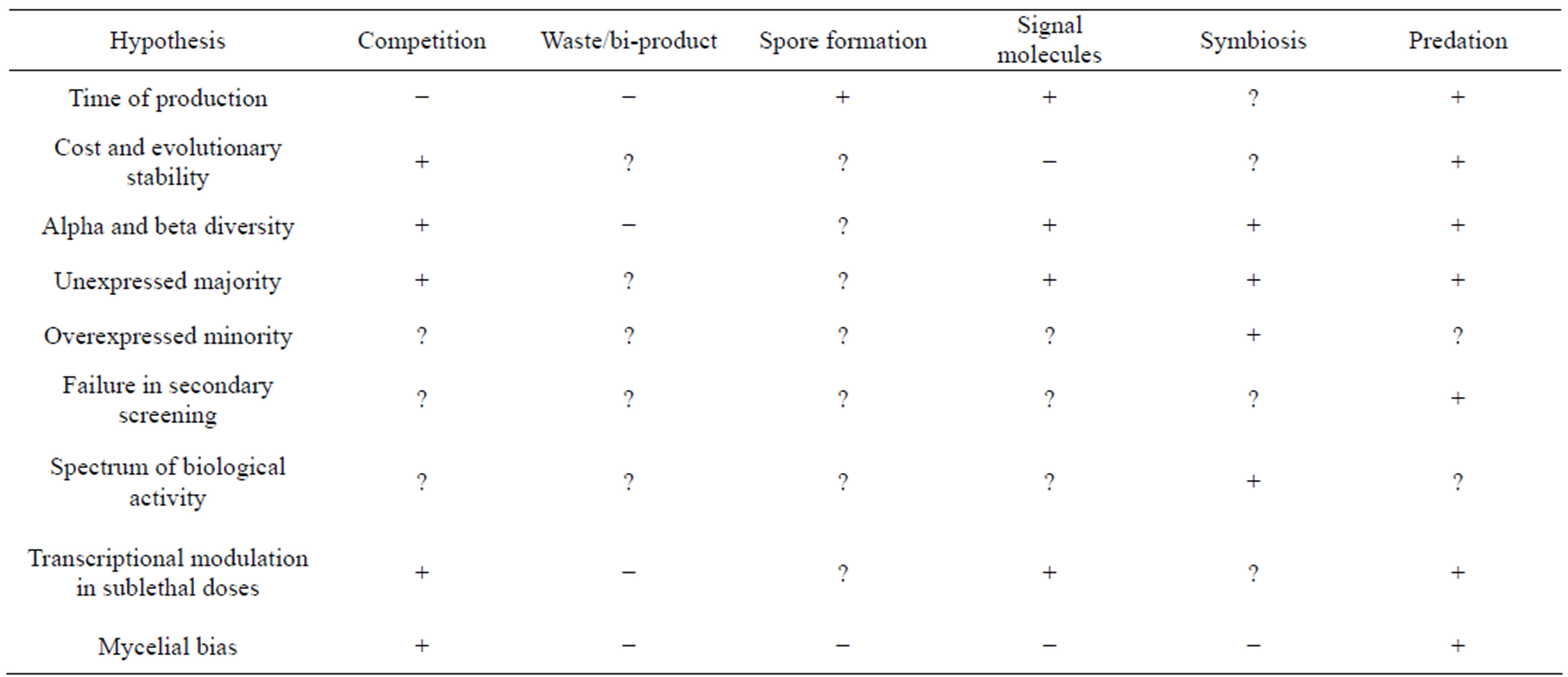
Table 2. Comparison of different hypotheses in explaining known patterns in antibiotic production. key: +, satisfactory explanation; ?, no clear explanation offered; –, a contradiction. See discussion for details.
predation hypotheses since conventional culture excludes all these interspecies interactions. It is also likely that the ecological role of these compounds is extremely local and therefore most of them are produced locally in small quantities under a specific set of conditions and therefore not detected in the overall culture. This is particularly applicable for predation which initiates under nutrient limiting conditions when energy economy is most critical. It is also likely that producing larger quantities will be maladaptive for predation since it will lyse prey cells at a greater distance from the producing tips releasing nutrients that would encourage the growth of other competitors. Therefore antibiotic release in large amounts is contrary to the predation hypothesis. The waste product and sporulation hypotheses offer no specific explanation to the apparently unexpressed gene clusters.
3.5. The Over-Expressed Minority
If energy economy is critical for secondary metabolite production, why a minority of them are overproduced in laboratory media? It appears that this phenomenon is distinct and may have evolved for a different function than the majority of apparently unexpressed ones. Predation would involve local and limited production whereas competition and symbiosis may need production of large quantities. Symbiosis is particularly promising here since the secondary metabolite needs to serve a host function that might be spatiotemporally away from the mycelium producing it. For example the host applies the antibiotic to keep a fungal garden free from contamination. For such purposes the compound needs to be produced in larger quantities. Also since the host provides nutrients in the symbiosis, production of large quantities becomes affordable for the actinomycete partner.
3.6. Difference in Expression on Solid Surface versus Liquid Environment
Predation by Myxobacteria as well as by Streptomyces is a surface phenomenon. Proponents of other hypotheses have not offered any explanation for the difference between antibiotic expression under solid versus liquid environments. It is interesting to note that the initial attempts for screening of Streptomyces antibiotics actually screened for predatory activity [74] which was soon replaced by screening for inhibition zones. The reason for this shift is not explicitly found in literature. It is possible that the rate of failure in secondary screening (typically shake flask) from predation screening (on solid medium) is higher than that from inhibition zone screening. Therefore the former was given up soon. The physical environment makes a radical difference for predation. The transcriptional modulation effects are also shown to be different on solid versus fluid media but so far no specific explanation has been offered for this difference [16]. The predation hypothesis can potentially explain this difference as the predatory pressure is substantially different in solid versus fluid environment and the defense strategies in solid and liquid environments also need to be different.
3.7. The Spectrum of Biological Activities
Secondary metabolites have a wide spectrum of activities apart from their antimicrobial action. Effects such as antitumor agents, immunosuppressive agents, hypocholesterolemic agents, enzyme inhibitors, antimigraine agents, and antiparasitic agents [78] are surprising on the background of most of the above hypotheses. Competition and predation hypotheses explain the antimicrobial activities but do not explain other bioactivities. The only possible natural role of other activities is in symbiotic associations. For example animal physiological action might have evolved as vertebrate toxicity that can be used in a symbiosis to protect the symbiotic partner from grazing or predation by animals. This is particularly important for organisms like sponges that have neither motility nor hard skeleton as a defense against predators.
3.8. Transcription Modulation in Sublethal Doses
The widely demonstrated transcriptional modulation by sublethal concentrations of antibiotics is the heart of the signaling hypothesis. Competition and predation can also potentially explain the transcriptional modulation as defense strategies of the susceptible organisms [17]. It is interesting in this regard that the activities boosted by low doses of antibiotics include stimulation of growth on solid but not in liquid environment (unpublished study from our lab), biofilm formation [79], Exo-polysaccharide synthesis [80], enhancement of motility or increased rate of mutations [18] all of which can be potentially protective [81]. However not all transcriptional modulation has a clear protective interpretation which leaves the possibility of other functions. We will have to wait until the functional relevance of most of the transcriptional modulations is clear. The waste product, sporulation and symbiosis hypotheses have no specific suggestion to explain transcriptional modulation activity in sensitive organisms.
3.9. The Mycelial Bias
A common sociobiological problem associated with any costly extracellular activity is that a cheater mutant that does not make the extracellular product but shares the benefit of its function can get a net advantage and therefore invade the wild type population. It is therefore difficult to explain the evolutionary stability of extracellular activity of unicellular forms of life. In yeast, the cheater or defector phenotypes which do not produce invertase and thereby give rise to a show-drift game are well known [82]. Cellulytic bacteria in a rumen environment can also accumulate cheaters and predatory protozoa offer a possible solution by selectively preying on cheater cells [83]. It is perhaps because of this sociobiological reason that efficiency of biodegradation of the rumen crucially depends upon predatory protozoa. The problem of cheaters appears to have been circumvented by the mycelial forms. In mycelial microorganisms the biomass is multinucleate with or without septa and growth occurs only at the tips. Since there is no individual cell identity except at the spore stage, a mutant “individual” is unlikely to get any specific advantage of faster replication. Therefore costly extracellular activity that is essential for growth could be more stable in mycelial forms than in cellular forms on an evolutionary time scale. This is likely to be the evolutionary reason why costly secondary metabolites are commonly produced by mycelial forms than by cellular forms among both prokaryotic and eukaryotic microorganisms.
It is well known that actinomycetes and fungi are the main sources of all antibiotics discovered so far. Among both prokaryotic and eukaryotic microorganisms it is the mycelial group that is the antibiotic producer. This may not be a coincidence. Further among the taxonomic group of actinobacteria, there is wide morphological diversity and unequal distribution of secondary metabolite diversity among the different morphological forms. We attempted a meta-analysis to see the association between mycelial morphology and the genomic repertoire for secondary metabolite production based on published reviews of genomic information on different members of actinobacteria [3,7,84]. The analysis showed a very tight association between mycelial morphology and the number of gene clusters in the genome for secondary metabolites. The mycelial forms have substantially greater number of gene clusters for secondary metabolites than the cellular forms (Figure 1). This is compatible with our hypothesis of mycelial forms being more resistant to cheating and therefore having greater evolutionary stability of extracellular products. Interestingly even among fungi, yeasts are not known to be antibiotic producers. Almost all fungal antibiotics are obtained from mycelial fungi.
This logic extends beyond antibiotic production and should apply equally to other extracellular functions such as biodegradation of complex polymers by extracellular enzymes. It may not be a coincidence therefore that both actinomycetes and fungi are efficient biodegraders of natural polymers where their mycelial morphology ensures evolutionary stability of extracellular functions.
Myxobacteria are non-mycelial predators and antibiotic producers and the sociobiological problem of cheating is known to be common among them. They seem to have evolved other mechanisms to maintain complex social behavior in spite of cheaters [85]. Our understandding of the sociobiology of antibiotic production is very elementary at this stage but we can appreciate that antibiotic production is a sociobiological phenomenon and mycelial morphology changes the norms of the game so that there is greater evolutionary stability.
4. EXPERIMENTAL EVIDENCE
Attempts to test one or more of the hypotheses are rare. Most of the evolutionary explanations are incidental and
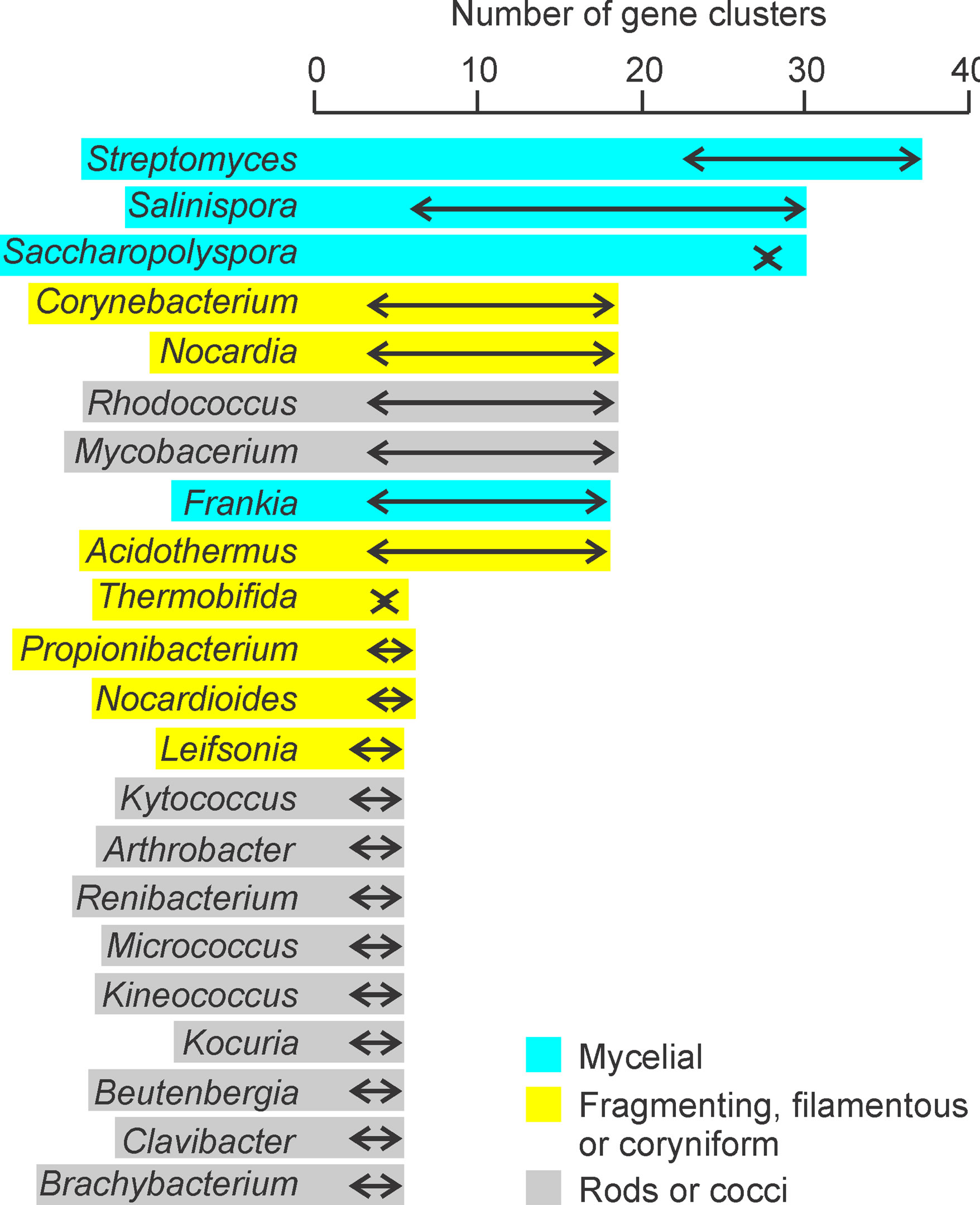
Figure 1. The gene clusters for secondary metabolites among different actinobacterial genera: From available genomic information it can be notices that the genera with mycelial morphology have greater diversity of secondary metabolites as compared to cellular morphologies.
attempts to make and test predictions are almost nonexistent. Nevertheless some fragmentary evidence for or against some of them can be found in literature. Evidence for competition is contradictory [29-31] and much of the positive evidence claimed to support competition hypothesis has other possible interpretations. Predation can account for the most of the observations interpreted as competition. No difference has been shown in the spore production as well survival of antibiotic producers and non-producers and therefore role of antibiotics in spore production is not experimentally supported. No specific benefit of signaling by antibiotics to the producers has been experimentally demonstrated so far. Although a number of cases of symbioses have been demonstrated so far, it is not know how common symbiotic association is among the entire group of mycelial actinomycetes and whether it is sufficient to account for the evolution of secondary metabolites. The association between antibiotic production and predatory abilities is strong across taxa and in at least one case, that of myxobacteria the role of antibiotics in predation has been directly demonstrated [70]. Currently the nature of evidence for involvement of secondary metabolites in predation by Streptomyces is indirect but in spite of that predation appears to be the most promising of all the hypotheses for the evolutionary origins and stability of antibiotics. The evidence for waste product hypothesis is necessarily negative by nature. This hypothesis was proposed whenever others were thought to be inconsistent. As evidence for any other hypothesis becomes strong, this hypothesis becomes increasingly irrelevant.
5. TOWARDS A SYNTHESIS
Compiling all the above arguments it can be seen (Table 2) that no single hypothesis explains all these observed phenomenon. It is likely therefore that secondary metabolites evolved by more than one selection pressures simultaneously or serially. Predation appears to be the most promising hypothesis of all which explains all the known phenomenon associated with antibiotics except the non antimicrobial actions of some of the secondary metabolites and over production of some of the antibiotics in nutrient rich environment. We feel that these two phenomena are better explained by the symbiotic role of actinomycetes. Symbiosis explains some important patterns that are not explained by other hypotheses. However the relatively smaller proportion of over-expressed compounds and compounds without antimicrobial activity makes it unlikely that secondary metabolites primarily evolved for symbiosis, although this has been suggested [86].
It is challenging to make a logically coherent sense out of a large number of different and diverse observations and experiments. However an attempt towards a synthesis can be largely rewarding and adds to our insights into a phenomenon (Figure 2). We will argue here that the predation hypothesis helps us join the wide array of facts
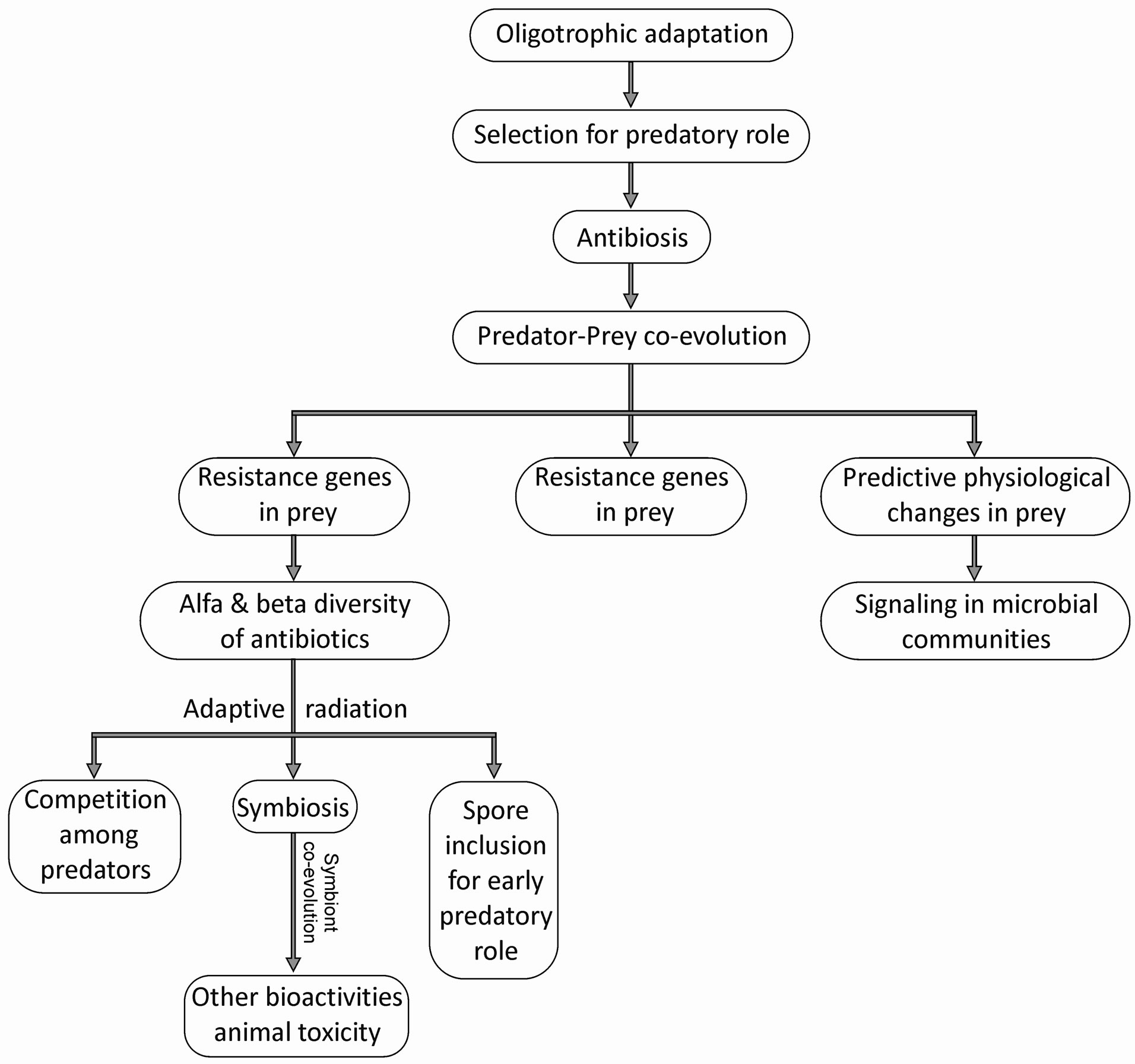
Figure 2. A possible evolutionary pathway for the secondary metabolites of Streptomyces: the ancestral characteristics are expected to be most widespread and the specialized once in the specifically evolved lines. The theme might be more generally applicable to all microbial secondary metabolites.
and observations and make a coherent and consistent theory. We propose that Streptomyces are primarily K selected oligophilic bacteria [87] that have a relatively slow growth rate and high growth yield and an ability to grow at extremely low nutrient concentrations and yet produce a surprisingly large amount of biomass. The concentration of soluble nutrients available in soil and other natural environments of Streptomyces is orders of magnitude smaller than what is generally offered in commonly used culture media. We have shown before that colony counts of bacteria from a variety of natural habitats are substantially greater in dilute but diverse media than those on the conventional rich nutrient media [87, 88]. A number of isolates that can grow on oligotrophic media fail to grow on conventional media on primary culture although they may adapt to it after one or more subcultures. This indicates that most bacteria in soil and other ambient environments are in an oligophilic phase of metabolism [87]. Our previous experience has suggested that actinomycetes are a predominant group among the isolates obtained on dilute media. (We use the terms oligophilic and copiophilic to indicate the characteristics of organisms and their metabolism and the terms oligotrophic and copiotrophic to indicate habitat characteristics and growth environments). As a specific adaptation to oligotrophic environments this genus has evolved the abilities to make use of every possible source of nutrient and as a result they have evolved remarkable biodegradation abilities, predatory abilities and mycelial morphology that is essential for the evolutionary stability of these functions.
It is likely therefore that secondary metabolites evolved primarily as weapons used in predation. This might be true across taxa as shown by the strong association (Table 1). Eventually some of the antibiotic producers entered non-obligate symbiotic associations with a variety of other organisms and the bioactivities diversified further to fulfill the new niches created. Symbiosis needs much larger amounts of the compound to be produced. Therefore overproduction of some of the compounds might have evolved in a symbiotic environment. Nevertheless symbiosis is only a small fraction of the diversity as well as life cycles of Streptomyces and therefore overproduction remained a minority activity. In a nutrient limited natural environment there can be competition between predators. Antibiotics may be used as weapons for this competition and the antibiotic response to nutrient depletion is not incompatible with this type of competition. Presence of a myxobacterial predator has been shown to increase antibiotic production by Streptomyces [89] which is in support of this possibility. Incorporation of antibiotics in spores may help adopting a predatory role immediately after germination in certain environments. Ultimately it is also likely that the transcription modulation response of potential prey organisms evolved primarily as a defense mechanism but eventually it diversified into other signals involved in interactions within multispecies community. Since the cost of production is compensated by the primary function, evolutionary stability will not be a problem for the secondarily acquired signaling function. This evolutionary synthesis is most plausible and makes logically coherent links with the fragments of prior observations and knowledge. The synthesis eliminates waste product hypothesis and logically links all other hypotheses with the predation function being the origin as well as the crucial link to the evolution of other functions.
6. TESTABLE PREDICTIONS AND RECOMMENDED LINES OF FURTHER RESEARCH
Some of the testable predictions that need carefully designed experiments include attempts to demonstrate the expression of secondary metabolite gene clusters locally at the tips of invading predatory mycelia and detection of novel antimicrobial compounds in the gel surrounding the invading tips. This would need extremely sensitive detection methods since the amounts are expected to be small.
The possibility of the role of antibiotics in competition between predators is certainly testable by relatively simple experiments and our synthesis predicts greater expression of antibiotics, both qualitatively and quantitatively during such a competition. There is at least one demonstrated example so far [89] but it is important to know how common this phenomenon is.
The proposed role of symbiosis in evolution of the diversity of bioactivity makes a testable prediction that non-antimicrobial bioactivities will be found more frequently in actinomycete species in symbiotic association with sponges and other species that have no other defense against grazing and predation. We already know a wide diversity of novel compounds in actinomycetes in marine symbioses [6,61,90,91] but quantitative ecological data are needed to test such a hypothesis. A counterpart of this prediction is that the free living mycelial actinomycetes should have a greater proportion of antimicrobial compounds. The main difficulty in testing these predictions is that since the symbiotic associations are non-obligate, the symbiotic species will also be found free living. Nevertheless quantitatively rigorous experimental designs should be able to detect the difference if real. If true, such an association can be useful in directing screening attempts for the drug discovery industry. Another possible prediction of the synthesis is that if the so far “unexpressed” secondary metabolites have a role in predation in an oligotrophic environment, they could be much more potent in their antimicrobial action since they have to work at extremely low concentrations. This is a possible new hope for the drug discovery sector.
REFERENCES
- Watve, M., Tickoo, R., Jog, M. and Bhole, B. (2001) How many antibiotics are produced by the genus Streptomyces ? Archives of Microbiology, 176, 386-390. doi:10.1007/s002030100345
- Challis, G.L. and Hopwood, D.A. (2003) Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proceedings of the National Academy of Sciences of the USA, 100, 14555-14561. doi:10.1073/pnas.1934677100
- Adegboye, M.F. and Babalola, O.O. (2012) Taxonomy and ecology of antibiotic producing actinomycetes. African Journal of Agricultural Research, 7, 2255-2261.
- Bentley, S.D., Chater, K.F., Cerdeno-Tarraga, A.-M., Challis, G.L., Thomson, N.R., James, K.D., Harris, D.E., Quail, M.A., Kieser, H., Harper, D., Bateman, A., Brown, S., Chandra, G., Chen, C.W., Collins, M., Cronin, A., Fraser, A., Goble, A., Hidalgo, J., Hornsby, T., Howarth, S., Huang, C.-H., Kieser, T., Larke, L., Murphy, L., Oliver, K., O’Neil, S., Rabbinowitsch, E., Rajandream, M.-A., Rutherford, K., Rutter, S., Seeger, K., Saunders, D., Sharp, S., Squares, R., Squares, S., Taylor, K., Warren, T., Wietzorrek, A., Woodward, J., Barrell, B.G., Parkhill, J. and Hopwood, D.A. (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature, 417, 141-147. doi:10.1038/417141a
- Jayapal, K.P., Lian, W., Glod, F., Sherman, D.H. and Hu, W.-S. (2007) Comparative genomic hybridizations reveal absence of large Streptomyces coelicolor genomic islands in Streptomyces lividans. BMC Genomics, 8, 229. doi:10.1186/1471-2164-8-229
- Udwary, D.W., Zeigler, L., Asolkar, R.N., Singan, V., Lapidus, A., Fenical, W., Jensen, P.R. and Moore, B.S. (2007) Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proceedings of the National Academy of Sciences of the USA, 104, 10376-10381. doi:10.1073/pnas.0700962104
- Nett, M., Ikeda, H. and Moore, B.S. (2009) Genomic basis for natural product biosynthetic diversity in the actinomycetes. Natural product reports, 26, 1362-1384. doi:10.1039/b817069j
- Rosamond, J. and Allsop, A. (2000) Harnessing the power of the genome in the search for new antibiotics. Science, 287, 1973-1976. doi:10.1126/science.287.5460.1973
- Baltz, R. (2007) Antimicrobials from Actinomycetes: Back to the Future. Microbe, 2, 125-131.
- Jenke-Kodama, H., Müller, R. and Dittmann, E. (2008) Evolutionary mechanisms underlying secondary metabolite diversity. Progress in Drug Research, 65, 121-140.
- Hesseltine, C.W. (1972) Biotechnology report: Solid state fermentations. Biotechnology and Bioengineering, 14, 517-532. doi:10.1002/bit.260140402
- Raimbault, M. (1998) General and microbiological aspects of solid substrate fermentation. Electronic Journal of Biotechnology, 1, 26-27. doi:10.2225/vol1-issue3-fulltext-9
- Pérez-Guerra, N., Torrado-Agrasar, A., López-Macias, C. and Pastrana, L. (2003) Main characteristics and applications of solid substrate fermentation. Electronic Journal of Environmental, Agricultural and Food Chemistry, 2, 1579-4377.
- Krishna, C. (2005) Solid-state fermentation systems-an overview. Critical Reviews in Biotechnology, 25, 1-30. doi:10.1080/07388550590925383
- Soares, V.F., Castilho, L.R., Bon, E.P.S. and Freire, D.M.G. (2005) High-yield Bacillus subtilis protease production by solid-state fermentation. Applied Biochemistry and Biotechnology, 121, 311-319. doi:10.1385/ABAB:121:1-3:0311
- Goh, E.-B. Yim, G., Tsui, W., McClure, J., Surette, M.G. and Davies, J. (2002) Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proceedings of the National Academy of Sciences of the USA, 99, 17025-17030. doi:10.1073/pnas.252607699
- Blázquez, J., Gómez-Gómez, J.-M., Oliver, A., Juan, C., Kapur, V. and Martín, S. (2006) PBP3 inhibition elicits adaptive responses in Pseudomonas aeruginosa. Molecular Microbiology, 62, 84-99. doi:10.1111/j.1365-2958.2006.05366.x
- Linares, J.F., Gustafsson, I., Baquero, F. and Martinez, J.L. (2006) Antibiotics as intermicrobial signaling agents instead of weapons. Proceedings of the National Academy of Sciences of the USA, 103, 19484-19489. doi:10.1073/pnas.0608949103
- Waksman, S. (1961) The Actinomycetes classification, identification and description of genera and species. Literary Licensing, LLC.
- Gottlieb, D. (1976) The production and role of antibiotics in soil. The Journal of Antibiotics, 29, 987-1000. doi:10.7164/antibiotics.29.987
- Katz, E. and Demain, A.L. (1977) The peptide antibiotics of Bacillus: Chemistry, biogenesis, and possible functions. Bacteriological Reviews, 41, 449-474.
- Malik, V.S. (1980) Microbial secondary metabolism. Trends in Biochemical Sciences, 5, 68-72. doi:10.1016/0968-0004(80)90071-7
- Martin, J.F. and Demain, A.L. (1980) Control of antibiotic biosynthesis. Microbiological Reviews, 44, 230-251.
- Fredrickson, A. and Stephanopoulos, G. (1981) Microbial competition. Science, 213, 972-979. doi:10.1126/science.7268409
- Williams, S. and Vickers, J. (1986) The ecology of antibiotic production. Microbial Ecology, 12, 43-52. doi:10.1007/BF02153221
- Williams, D.H., Stone, M.J., Hauck, P.R. and Rahman, S.K. (1989) Why are secondary metabolites (natural products) biosynthesized? Journal of Natural Products, 52, 1189-1208. doi:10.1021/np50066a001
- Kolter, R. and Moreno, F. (1992) Genetics of ribosomally synthesized peptide antibiotics. Annual Review of Microbiology, 46, 141-161. doi:10.1146/annurev.mi.46.100192.001041
- Baba, T. and Schneewind, O. (1998) Instruments of microbial warfare: Bacteriocin synthesis, toxicity and immunity. Trends in Microbiology, 6, 66-71. doi:10.1016/S0966-842X(97)01196-7
- Rasool, K.A. and Wimpenny, J.W.T. (1982) Mixed continuous culture experiments with an antibiotic-producing Streptomycete and Escherichia coli. Microbial Ecology, 8, 267-277. doi:10.1007/BF02011430
- Turpin, P.E., Dhir, V.K., Maycroft, K.A., Rowlands, C. and Wellington, E.M.H. (1992) The effect of Streptomyces species on the survival of Salmonella in soil. FEMS Microbiology Letters, 101, 271-280. doi:10.1111/j.1574-6968.1992.tb05784.x
- Wiener, P. (1996) Experimental studies on the ecological role of antibiotic production in bacteria. Evolutionary Ecology, 10, 405-421. doi:10.1007/BF01237726
- De Lorenzo, V., Martínez, J.L. and Asensio, C. (1984) Microcin-mediated interactions between Klebsiella pneumoniae and Escherichia coli strains. Journal of General Microbiology, 130, 391-400.
- Manske, R.H.F., Holmes, H.L. and James, W. (1950) The alkaloids: Chemistry and physiology. Academic press, New York.
- Mothes, K. (1955) Physiology of alkaloids. Annual Review of Plant Physiology, 6, 393-432. doi:10.1146/annurev.pp.06.060155.002141
- Turner, W.B. (1971) Fungal metabolites. Academic Press, London.
- Luckner, M. (1972) Secondary metabolism in plants and animals. Academic Press, London.
- Robinson, T. (1974) Metabolism and function of alkaloids in plants. Science, 184, 430-435. doi:10.1126/science.184.4135.430
- Swain, T. (1977) Secondary compounds as protective agents. Annual Review of Plant Physiology, 28, 479-501. doi:10.1146/annurev.pp.28.060177.002403
- Haslam, E. (1986) Secondary metabolism-fact and fiction. Natural Product Reports, 3, 217-249. doi:10.1039/np9860300217
- Neidleman, S.L. (1989) Advances in applied microbialogy. Academic Press, London.
- Vining, L.C. (1990) Functions of Secondary Metabolites. Annual Review of Microbiology, 44, 395-427. doi:10.1146/annurev.mi.44.100190.002143
- Seigler, D.S. (1998) Plant secondary metabolism. Springer, US.
- Hartmann, T. (2007) From waste products to ecochemicals: Fifty years research of plant secondary metabolism. Phytochemistry, 68, 2831-2846. doi:10.1016/j.phytochem.2007.09.017
- Dworkin, M.M., Rosenberg, E. and Schleifer, K.-H. (2006) The prokaryotes, a handbook on the biology of bacteria: Eco-physiology. Springer, US.
- Adamidis, T., Riggle, P. and Champness, W. (1990) Mutations in a new Streptomyces coelicolor locus which globally block antibiotic biosynthesis but not sporulation. Journal of Bacteriology, 172, 2962-2969.
- Ueda, K., Kawai, S., Ogawa, H., Kiyama, A., Kubota, T., Kawanobe, H. and Beppu, T. (2000) Wide distribution of interspecific stimulatory events on antibiotic production and sporulation among Streptomyces species. The Journal of Antibiotics, 53, 979-982. doi:10.7164/antibiotics.53.979
- Kaltenpoth, M., Göttler, W., Herzner, G. and Strohm, E. (2005) Symbiotic bacteria protect wasp larvae from fungal infestation. Current Biology, 15, 475-479. doi:10.1016/j.cub.2004.12.084
- Kroiss, J., Kaltenpoth, M., Schneider, B., Schwinger, M.-G., Hertweck, C., Maddula, R.K., Strohm, E. and Svatoš, A. (2010) Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nature Chemical Biology, 6, 261-263. doi:10.1038/nchembio.331
- Becking, J.H. (1970) Plant-endophyte symbiosis in nonleguminous plants. Plant and Soil, 32, 611-654. doi:10.1007/BF01372898
- Klemmedson, J.O. (1979) Ecological Importance of Actinomycete-Nodulated Plants in the Western United States. Botanical Gazette, 140, S91-S96. doi:10.1086/337042
- Goodfellow, M. and Williams, S.T. (1983) Ecology of actinomycetes. Annual Review of Microbiology, 37, 189- 216. doi:10.1146/annurev.mi.37.100183.001201
- Currie, C.R. (2001) A community of ants, fungi, and bacteria: A multilateral approach to studying symbiosis. Annual Review of Microbiology, 55, 357-380. doi:10.1146/annurev.micro.55.1.357
- Currie, C.R., Bot, A.N.M. and Boomsma, J.J. (2003) Experimental evidence of a tripartite mutualism: Bacteria protect ant fungus gardens from specialized parasites. Oikos, 101, 91-102. doi:10.1034/j.1600-0706.2003.12036.x
- Currie, C.R., Scott, J.A., Summerbell, R.C. and Malloch, D. (2003) Corrigendum: Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature, 423, 461. doi:10.1038/nature01563
- Cafaro, M.J. and Currie, C.R. (2005) Phylogenetic analysis of mutualistic filamentous bacteria associated with fungus-growing ants. Canadian Journal of Microbiology, 51, 441-446. doi:10.1139/w05-023
- Currie, C.R., Poulsen, M., Mendenhall, J., Boomsma, J.J. and Billen, J. (2006) Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science, 311, 81-83. doi:10.1126/science.1119744
- Mueller, U.G., Dash, D., Rabeling, C. and Rodrigues, A. (2008) Coevolution between attine ants and actinomycete bacteria: A reevaluation. Evolution, 62, 2894-2912. doi:10.1111/j.1558-5646.2008.00501.x
- Poulsen, M., Bot, A.N.M., Currie, C.R., Nielsen, M.G. and Boomsma, J.J. (2003) Within-colony transmission and the cost of a mutualistic bacterium in the leaf-cutting ant acromyrmex octospinosus. Functional Ecology, 17, 260-269. doi:10.1046/j.1365-2435.2003.00726.x
- Poulsen, M., Cafaro, M., Boomsma, J.J. and Currie, C.R. (2005) Specificity of the mutualistic association between actinomycete bacteria and two sympatric species of acromyrmex leaf-cutting ants. Molecular Ecology, 14, 3597-3604. doi:10.1111/j.1365-294X.2005.02695.x
- Lee, J.H. and Lee, H.K. (2001) Microbial symbiosis in marine sponges. Society, 39, 254-264.
- Gandhimathi, R., Arunkumar, M., Selvin, J., Thangavelu, T., Sivaramakrishnan, S., Kiran, G.S., Shanmughapriya, S. and Natarajaseenivasan, K. (2008) Antimicrobial potential of sponge associated marine actinomycetes. Journal of Medical Mycology, 18, 16-22.
- Sabarathnam, B., Manilal, A., Sujith, S., Kiran, G.S., Selvin, J., Thomas, A. and Ravji, R. (2010) Role of sponge associated actinomycetes in the marine phosphorrous biogeochemical cycles. American-Eurasian Journal of Agriculture and Environmental Sciences, 8, 253-256.
- McLain, E.E. (2006) Actinomycetes and antibiotics Host defense through symbiosis. Ph.D. Thesis, University of Kansas.
- Davies, J. (1990) What are antibiotics? Archaic functions for modern activities. Molecular Microbiology, 4, 1227- 1232. doi:10.1111/j.1365-2958.1990.tb00701.x
- Davies, J. (2006) Are antibiotics naturally antibiotics? Journal of Industrial Microbiology and Biotechnology, 33, 496-499. doi:10.1007/s10295-006-0112-5
- Davies, J., Spiegelman, G.B. and Yim, G. (2006) The world of subinhibitory antibiotic concentrations. Current Opinion in Microbiology, 9, 445-453. doi:10.1016/j.mib.2006.08.006
- Diggle, S.P., Gardner, A., West, S.A. and Griffin, A.S. (2007) Evolutionary theory of bacterial quorum sensing: When is a signal not a signal? Philosophical Transactions of the Royal Society B: Biological Sciences, 362, 1241- 1249. doi:10.1098/rstb.2007.2049
- Yim, G., Wang, H.H. and Davies, J. (2006) The truth about antibiotics. International Journal of Medical Microbiology, 296, 163-170. doi:10.1016/j.ijmm.2006.01.039
- Yim, G., Wang, H.H. and Davies F.R.S.J. (2007) Antibiotics as signalling molecules. Philosophical Transactions of the Royal Society B: Biological Sciences, 362, 1195 -1200. doi:10.1098/rstb.2007.2044
- Xiao, Y., Wei, X., Ebright, R. and Wall, D. (2011) Antibiotic production by myxobacteria plays a role in predation. Journal of Bacteriology, 193, 4626-4633. doi:10.1128/JB.05052-11
- Casida Jr., L.E. (1988) Minireview: Nonobligate bacterial predation of bacteria in soil. Microbial Ecology, 15, 1-8. doi:10.1007/BF02012948
- Casida Jr., L.E. (1982) Ensifer adhaerens gen. nov., sp. nov.: A bacterial predator of bacteria in soil. International Journal of Systematic Bacteriology, 32, 339-345. doi:10.1099/00207713-32-3-339
- Watve, M. and Kumbhar, C. (2007) Streptomyces sp. as predators of bacteria. Nature Proceedings. http://precedings.nature.com/documents/1263/version/2
- Waksman, S.A. and Woodruff, H.B. (1941) Actinomyces antibioticus, a new soil organism antagonistic to pathogenic and non-pathogenic bacteria 1. Journal of Bacteriology, 42, 231-249.
- Rigali, S., Titgemeyer, F., Barends, S., Mulder, S., Thomae, A.W., Hopwood, D.A. and Van Wezel, G.P. (2008) Feast or famine: The global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Reports, 9, 670-675. doi:10.1038/embor.2008.83
- Corno, G., Caravati, E., Callieri, C. and Bertoni, R. (2008) Effects of predation pressure on bacterial abundance, diversity, and size-structure distribution in an oligotrophic system. Journal of Limnology, 67, 107-119. doi:10.4081/jlimnol.2008.107
- Morgan, A.D., MacLean, R.C., Hillesland, K.L. and Velicer, G.J. (2010) Comparative analysis of myxococcus predation on soil bacteria. Applied and Environmental Microbiology, 76, 6920-6927. doi:10.1128/AEM.00414-10
- Demain, A.L. (1999) Pharmaceutically active secondary metabolites of microorganisms. Applied Microbiology and Biotechnology, 52, 455-463. doi:10.1007/s002530051546
- Hoffman, L.R., D’Argenio, D.A., MacCoss, M.J., Zhang, Z., Jones, R.A. and Miller, S.I. (2005) Aminoglycoside antibiotics induce bacterial biofilm formation. Nature, 436, 1171-1175. doi:10.1038/nature03912
- Bader, M.W., Navarre, W.W., Shiau, W., Nikaido, H., Frye, J.G., McClelland, M., Fang, F.C. and Miller, S.I. (2003) Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Molecular Microbiology, 50, 219-230. doi:10.1046/j.1365-2958.2003.03675.x
- Allison, D.G. and Gilbert, P. (1995) Modification by surface association of antimicrobial susceptibility of bacterial populations. Journal of Industrial Microbiology and Biotechnology, 15, 311-317. doi:10.1007/BF01569985
- Gore, J., Youk, H. and Van Oudenaarden, A. (2009) Snowdrift game dynamics and facultative cheating in yeast. Nature, 459, 253-256.
- Modak, T., Pradhan, S. and Watve, M. (2007) Sociobiology of biodegradation and the role of predatory protozoa in biodegrading communities. Journal of Biosciences, 32, 775-780. doi:10.1007/s12038-007-0078-0
- Oliynyk, M., Samborskyy, M., Lester, J.B., Mironenko, T., Scott, N., Dickens, S., Haydock, S.F. and Leadlay, P.F. (2007) Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nature Biotechnology, 25, 447-453. doi:10.1038/nbt1297
- Velicer, G.J. and Vos, M. (2009) Sociobiology of the myxobacteria. Annual Review of Microbiology, 63, 599- 623. doi:10.1146/annurev.micro.091208.073158
- Clardy, J., Fischbach, M. and Currie, C. (2009) The natural history of antibiotic. Current Biology, 19, R437-R441. doi:10.1016/j.cub.2009.04.001
- Watve, M., Shejval, V., Sonawane, C., Rahalkar, M., Matapurkar, A., Shouche, Y., Patole, M., Phadnis, N., Champhenkar, A., Damle, K., Karandikar, S., Kshirsagar V. and Jog, M. (2000) The “K” selected oligophilic bacteria: A key to uncultured diversity? Current Science, 78, 1535-1542.
- Nagarkar, P.P., Ravetkar, S.D. and Watve, M.G. (2001) Oligophilic bacteria as tools to monitor aseptic pharmaceutical production units. Applied and Environmental Microbiology, 67, 1371-1374. doi:10.1128/AEM.67.3.1371-1374.2001
- Pérez, J., Muñoz-Dorado, J., Braña, A.F., Shimkets, L.J., Sevillano, L. and Santamaría, R.I. (2011) Myxococcus xanthus induces actinorhodin overproduction and aerial mycelium formation by Streptomyces coelicolor. Microbial Biotechnology, 4, 175-183.
- Fiedler, H.-P., Bruntner, C., Bull, A.T., Ward, A.C., Goodfellow, M., Potterat, O., Puder, C. and Mihm, G. (2005) Marine actinomycetes as a source of novel secondary metabolites. Antonie Van Leeuwenhoek, 87, 37-42. doi:10.1007/s10482-004-6538-8
- Imamura, N., Nishijima, M., Adachi, K. and Sano, H. (1993) Novel antimycin antibiotics, urauchimycins A and B, produced by marine actinomycete. The Journal of Antibiotics, 46, 241-246. doi:10.7164/antibiotics.46.241
- Manage, P.M., Kawabata, Z. and Nakano, S. (2000) Algicidal effect of the bacterium Alcaligenes denitrificans on Microcystis spp. Aquatic Microbial Ecology, 22, 111-117. doi:10.3354/ame022111
- Sayyed, R. and Chincholkar, S. (2009) SiderophoreProducing Alcaligenes feacalis Exhibited More Biocontrol Potential Vis-à-Vis Chemical Fungicide. Current Microbiology, 58, 47-51. doi:10.1007/s00284-008-9264-z
- Sayyed, R.Z. and Chincholkar, S.B. (2006) Purification of siderophores of Alcaligenes faecalis on Amberlite XAD. Bioresource Technology, 97, 1026-1029. doi:10.1016/j.biortech.2005.04.045
- Yutani, M., Taniguchi, H., Borjihan, H., Ogita, A., Fujita, K.-I. and Tanaka, T. (2011) Alliinase from Ensifer adhaerens and its use for generation of fungicidal activity. AMB Express, 1, 2. doi:10.1186/2191-0855-1-2
- Ogita, A., Fujita, K.-I. and Tanaka, T. (2012) Enhancing effects on vacuole-targeting fungicidal activity of amphotericin B. Frontiers in Microbiology, 3, 100.
- Germida, J.J. and Casida, L.E. (1983) Ensifer adhaerens predatory activity against other bacteria in soil, as monitored by indirect phage analysis. Applied and Environmental Microbiology, 45, 1380-1388.
- Drugă, B., Suteu, D., Roşca-Casian, O., Pârvu, M. and Dragos, N. (2011) Two novel Alliin lyase (alliinase) genes from twisted-leaf garlic (Allium obliquum) and mountain garlic (Allium senescens ssp. montanum). Notulae Botanicae Horti Agrobotanici, Cluj-Napoca, 39, 293-298.
- Davidov, Y., Huchon, D., Koval, S.F. and Jurkevitch, E. (2006) A new α-proteobacterial clade of bdellovibrio-like predators: Implications for the mitochondrial endosymbiotic theory. Environmental Microbiology, 8, 2179-2188. doi:10.1111/j.1462-2920.2006.01101.x
- Wang, Z., Kadouri, D.E. and Wu, M. (2011) Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics, 12, 453. doi:10.1186/1471-2164-12-453
- Lambina, V.A., Afinogenova, A.V., Romay Penobad, Z., Konovalova, S.M. and Andreev, L.V. (1983) New species of exoparasitic bacteria of the genus Micavibrio infecting gram-positive bacteria. Mikrobiologiia, 52, 777-780.
- Dashiff, A., Junka, R. A., Libera, M. and Kadouri, D. E. (2011) Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. Journal of Applied Microbiology, 110, 431-444. doi:10.1111/j.1365-2672.2010.04900.x
- Kadouri, D., Venzon, N.C. and O’Toole, G.A. (2007) Vulnerability of Pathogenic Biofilms to Micavibrio aeruginosavorus. Applied and Environmental Microbiology, 73, 605-614. doi:10.1128/AEM.01893-06
- Makkar, N.S. and Casida, L.E. (1987) Cupriavidus necator gen. nov., sp. nov: A nonobligate bacterial predator of bacteria in soil. International Journal of Systematic Bacteriology, 37, 323-326. doi:10.1099/00207713-37-4-323
- Casida, L.E. (1988) Response in soil of Cupriavidus necator and other copper-resistant bacterial predators of bacteria to addition of water, soluble nutrients, various bacterial species, or Bacillus thuringiensis spores and crystals. Applied and Environmental Microbiology, 54, 2161- 2166.
- Kreutzer, M.F., Kage, H. and Nett, M. (2012) Structure and biosynthetic assembly of cupriachelin, a photoreactive siderophore from the bioplastic producer Cupriavidus necator H16. Journal of the American Chemical Society, 134, 5415-5422. doi:10.1021/ja300620z
- Casida, L.E. (1992) Competitive ability and survival in soil of Pseudomonas strain 679-2, a dominant, nonobligate bacterial predator of bacteria. Applied and Environmental Microbiology, 58, 32-37.
- Cain, C.C., Lee, D., Waldo 3rd, R.H., Henry, A.T., Casida Jr., E.J., Wani, M.C., Wall, M.E., Oberlies, N.H. and Falkinham 3rd, J.O. (2003) Synergistic antimicrobial activity of metabolites produced by a nonobligate bacterial predator. Antimicrobial Agents Chemotherapy, 47, 2113- 2117. doi:10.1128/AAC.47.7.2113-2117.2003
- Li, S., Jochum, C.C., Yu, F., Zaleta-Rivera, K., Du, L., Harris, S.D. and Yuen, G.Y. (2008) An antibiotic complex from Lysobacter enzymogenes strain C3: Antimicrobial activity and role in plant disease control. Phytopathology, 98, 695-701. doi:10.1094/PHYTO-98-6-0695
- Ji, G.-H., Wei, L.-F., He, Y.-Q., Wu, Y.-P. and Bai, X.-H. (2008) Biological control of rice bacterial blight by Lysobacter antibioticus strain 13-1. Biological Control, 45, 288-296. doi:10.1016/j.biocontrol.2008.01.004
- Ensign, J.C. and Wolfe, R.S. (1966) Characterization of a Small proteolytic enzyme which lyses bacterial cell walls. Journal of Bacteriology, 91, 524-534.
- Daft, M.J., McCord, S.B. and Stewart, W.D.P. (1975) Ecological studies on algal-lysing bacteria in fresh waters. Freshwater Biology, 5, 577-596. doi:10.1111/j.1365-2427.1975.tb00157.x
- Zhang, W., Li, Y., Qian, G., Wang, Y., Chen, H., Li, Y.-Z., Liu, F., Shen, Y. and Du, L. (2011) Identification and characterization of the anti-mrsa WAP-8294A2 biosynthetic gene cluster from Lysobacter enzymogenes OH11. antimicrob. Agents Chemother. http://aac.asm.org/content/early/2011/09/19/AAC.05370-11
- Ensign, J.C. and Wolfe, R.S. (1965) Lysis of bacterial cell walls by an enzyme isolated from a myxobacter. Journal of Bacteriology, 90, 395-402.
- Park, J.H., Kim, R., Aslam, Z., Jeon, C.O. and Chung, Y.R. (2008) Lysobacter capsici sp. nov., with antimicrobial activity, isolated from the rhizosphere of pepper, and emended description of the genus Lysobacter. International Journal of Systematic and Evolutionary Microbiology, 58, 387-392. doi:10.1099/ijs.0.65290-0
- Bonner, D.P., O’Sullivan, J., Tanaka, S.K., Clark, J.M. and Whitney, R.R. (1988) Lysobactin, a novel antibacterial agent produced by Lysobacter sp. II. Biological properties. The Journal of Antibiotics, 41, 1745.
- Hashizume, H., Hirosawa, S., Sawa, R., Muraoka, Y., Ikeda, D., Naganawa, H. and Igarashi, M. (2004) Tripropeptins, novel antimicrobial agents produced by lysobacter sp. Part 2. Structure elucidation. ChemInform, 57, 52- 58.
- Nakayama, T., Homma, Y., Hashidoko, Y., Mizutani, J. and Tahara, S. (1999) Possible role of xanthobaccins produced by Stenotrophomonas sp. strain SB-K88 in suppression of sugar beet damping-off disease. Applied and Environmental Microbiology, 65, 4334-4339.
- Alonso, A., Sanchez, P. and Martínez, J.L. (2000) Stenotrophomonas maltophilia D457R contains a cluster of genes from gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrobial Agents Chemotherapy, 44, 1778-1782. doi:10.1128/AAC.44.7.1778-1782.2000
- Ryan, R.P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M.B., Berg, G., Lelie van de, D. and Dow, J.M. (2009) The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nature Reviews Microbiology, 7, 514-525.
- Shoji, J., Hinoo, H., Wakisaka, Y., Koizumi, K., Mayama, M. and Matsuura, S. (1977) Isolation of two new polymyxin group antibiotics. (Studies on antibiotics from the genus Bacillus. XX). The Journal of Antibiotics, 30, 1029-1034. doi:10.7164/antibiotics.30.1029
- Lim, H.-S., Kim, Y.-S. and Kim, S.-D. (1991) Pseudomonas stutzeri YPL-1 genetic transformation and antifungal mechanism against Fusarium solani, an agent of plant root rot. Antimicrobial Agents Chemotherapy, 57, 510-516.
- Leisinger, T. and Margraff, R. (1979) Secondary metabolites of the fluorescent Pseudomonads. Microbiological Reviews, 43, 422-442.
- Gumbo, J.R. (2010). The isolation and identification of predatory bacteria from a Microcystis algal bloom. African Journal of Biotechnology, 9, 663-671.
- Vining, L.C. (1990) Functions of secondary metabolites. Annual Review of Microbiology, 44, 395-427. doi:10.1146/annurev.mi.44.100190.002143
- Jurkevitch, E. (2007) Predatory behaviors in bacteria—Diversity and transitions. American Society for Microbiology, 2, 67-72.
- Chen, H., Young, S., Berhane, T.-K. and Williams, H.N. (2012) Predatory Bacteriovorax communities ordered by various prey species. PLoS ONE, 7, 34174. doi:10.1371/journal.pone.0034174
- McBride, M.J. and Zusman, D.R. (1996) Behavioral analysis of single cells of Myxococcus xanthus in response to prey cells of Escherichia coli. FEMS Microbiology Letters, 137, 227-231. doi:10.1111/j.1574-6968.1996.tb08110.x
- Erol, O., Schäberle, T.F., Schmitz, A., Rachid, S., Gurgui, C., El Omari, M., Lohr, F., Kehraus, S., Piel, J., Müller, R. and König, G.M. (2010) Biosynthesis of the myxobacterial antibiotic corallopyronin A. ChemBioChem, 11, 1253-1265.
- Suzuki, Y., Ojika, M. and Sakagami, Y. (2004) Biotransformation of cystothiazole A, a myxobacterial antibiotic, into novel derivatives by the mother producer, Cystobacter fuscus. Bioscience, Biotechnology, Biochemistry, 68, 390-396. doi:10.1271/bbb.68.390
- Schäberle, T.F., Goralski, E., Neu, E., Erol, Ö., Hölzl, G., Dörmann, P., Bierbaum, G. and König, G.M. (2010) Marine Myxobacteria as a source of antibiotics—comparison of physiology, polyketide-type genes and antibiotic production of three new isolates of Enhygromyxa salina. Marine Drugs, 8, 2466-2479. doi:10.3390/md8092466
- Singh, B.N. (1947) Myxobacteria in soils and composts; their distribution, number and lytic action on bacteria. Microbiology, 1, 1-10.
- Gaspari, F., Paitan, Y., Mainini, M., Losi, D., Ron, E. Z. and Marinelli, F. (2005) Myxobacteria isolated in Israel as potential source of new anti-infectives. Journal of Applied Microbiology, 98, 429-439. doi:10.1111/j.1365-2672.2004.02477.x
- Beebe, J.M. (1941) The Morphology and cytology of Myxococcus xanthus, N. Sp. 1. Journal of Bacteriology, 42, 193-223.
- Shimkets, L., Dworkin, M. and Reichenbach, H. (2006) The Myxobacteria. In: Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H. and Stackebrandt, E., Eds. The Prokaryotes, Springer, New York, 31-115. http://www.springerlink.com/content/q6033452111246hh/abstract/
- Trick, I. and Lingens, F. (1984) Characterization of Herpetosiphon spec.—A gliding filamentous bacterium from bulking sludge. Applied Microbiology and Biotechnology, 19, 191-198. doi:10.1007/BF00256453
- Quinn, G. and Skerman, V. (1980) Herpetosiphon—Nature’s scavenger? Current Microbiology, 4, 57-62. doi:10.1007/BF02602893
- Nett, M., Erol, Ö., Kehraus, S., Köck, M., Krick, A., Eguereva, E., Neu, E. and König, G.M. (2006) Siphonazole, an unusual metabolite from Herpetosiphon sp.. Angewandte Chemie International Edition, 45, 3863- 3867.
- Furusawa, G., Yoshikawa, T., Yasuda, A. and Sakata, T. (2003) Algicidal activity and gliding motility of Saprospira sp. SS98-5. Canadian Journal of Microbiology, 49, 92-100. doi:10.1139/w03-017
- Saw, J., Yuryev, A., Kanbe, M., Hou, S., Young, A.G., Aizawa, S.-I. and Alam, M. (2012) Complete genome sequencing and analysis of Saprospira grandis str. Lewin, a predatory marine bacterium. Standards in Genomic Sciences, 6, 84-93.
- Mincer, T.J., Spyere, A., Jensen, P.R. and Fenical, W. (2004) Phylogenetic analyses and diterpenoid production by marine bacteria of the genus Saprospira. Current Microbiology, 49, 300-307. doi:10.1007/s00284-004-4358-8
- Lewin, R.A. (1997) Saprospira grandis, a flexibacterium that can catch bacterial prey by “ixotrophy”. Microbial Ecology, 34, 232-236. doi:10.1007/s002489900052
- Sangkhobol, V. and Skerman, V.B.D. (1981) Saprospira species—Natural predators. Current Microbiology, 5, 169-174. doi:10.1007/BF01578523
- Casida, L.E. (1983) Interaction of Agromyces ramosus with other bacteria in soil. Applied Microbiology and Biotechnology, 46, 881-888.
- Cherniakovskaia, T.F., Dobrovol’skaia, T.G. and Bab’eva, I.P. (2004) The ability of saprotrophic bacteria isolated from natural habitats to lyse yeasts. Mikrobiologiia, 73, 567-570.
- Arcamone, F., Cassinelli, G., Fantini, G., Grein, A., Orezzi, P., Pol, C. and Spalla, C. (1969) Adriamycin, 14- hydroxydaunomycin, a new antitumor antibiotic from S. Peucetius var. caesius. Biotechnology and Bioengineering, 11, 1101-1110. doi:10.1002/bit.260110607
- Casida, L.E. (1980) Bacterial predators of micrococcus luteus in soil. Applied Microbiology and Biotechnology, 39, 1035-1041.
- Cassinelli, G., Grein, A., Orezzi, P., Pennella, P. and Sanfilippo, A. (1967) New antibiotics produced by Streptoverticillium orinoci, n. sp. Arch Mikrobiol, 55, 358-368. doi:10.1007/BF00406442
- Konev, I.E., Efimova, V.M., Etingov, E.D. and Zaval’- naia, N.M. (1978) Streptoverticillium griseoviridum n. sp., a producer of the candidin-amphotericin B group, antifungal heptaene nonaromatic antibiotic 0185. Antibiotiki, 23, 143-148.
- Evans, J.R., Napier, E.J. and Fletton, R.A. (1978) G1499- 2, a new quinoline compound isolated from the fermentation broth of Cytophaga johnsonii. The Journal of Antibiotics, 31, 952-958. doi:10.7164/antibiotics.31.952
- Shaaban, M., Maskey, R.P., Wagner-Döbler, I. and Laatsch, H. (2002) Pharacine, a natural p-cyclophane and other indole derivatives from Cytophaga sp. strain AM13.1. Journal of Natural Products, 65, 1660-1663. doi:10.1021/np020019a
- Rashidan, K.K. and Bird, D.F. (2001) Role of predatory bacteria in the termination of a Cyanobacterial bloom. Microbial Ecology, 41, 97-105.
- Ainsworth, G.C., Brown, A.M. and Brownlee. G. “Aerosporin”, an Antibiotic Produced by Bacillus aerosporus Greer. http://www.nature.com/nature/journal/v160/n4060/abs/160263a0.html
- Gonzalez-Pastor, J.E., Hobbs, E.C. and Losick, R. (2003) Cannibalism by sporulating bacteria. Science, 301, 510- 513. doi:10.1126/science.1086462
- Pichard, B., Larue, J.P. and Thouvenot, D. (1995) Gavaserin and saltavalin, new peptide antibiotics produced by Bacillus polymyxa. FEMS Microbiology Letters, 133, 215-218. doi:10.1111/j.1574-6968.1995.tb07887.x
- Al-Janabi, A.A.H.S. (2006) Identification of bacitracin produced by local isolate of Bacillus licheniformis. Journal of Biotechnology, 5, 1600-1601.
- Bie, X.-M., Lü, F.-X., Lu, Z.-X., Huang, X.-Q. and Shen, J. (2006) Isolation and identification of lipopeptides produced by Bacillus subtilis fmbJ. Shengwu Gongcheng Xuebao/Chinese Journal of Biotechnology, 22, 644-649.
- Awais, M., Shah, A.A.L.I. and Hameed, A. (2007) Isolation, Identification and optimization of bacitracin produced by bacillus sp. Time, 39, 1303-1312.
- Nandy, S.K., Bapat, P.M. and Venkatesh, K.V. (2007) Sporulating bacteria prefers predation to cannibalism in mixed cultures. FEBS Letters, 581, 151-156. doi:10.1016/j.febslet.2006.12.011
- Stansly, P.G. and Schlosser, M.E. (1947) Studies on polymyxin: Isolation and identification of bacillus polymyxa and differentiation of polymyxin from certain known antibiotics. Journal of Bacteriology, 54, 549-556.
- Be’er, A., Zhang, H.P., Florin, E.-L., Payne, S.M., Ben-Jacob, E. and Swinney, H.L. (2009) Deadly competetion between sibling bacterial colonies. Proceedings of the National Academy of Sciences of the USA, 106, 428- 433. doi:10.1073/pnas.0811816106
- Selim, S., Negrel, J., Govaerts, C., Gianinazzi, S. and Van Tuinen, D. (2005) Isolation and partial characterization of antagonistic peptides produced by Paenibacillus sp. strain B2 Isolated from the sorghum mycorrhizosphere. Applied Microbiology and Biotechnology, 71, 6501-6507. doi:10.1128/AEM.71.11.6501-6507.2005
- Beatty, P.H. and Jensen, S.E. (2002) Paenibacillus polymyxa produces fusaricidin-type antifungal antibiotics active against Leptosphaeria maculans, the causative agent of blackleg disease of canola. Canadian Journal of Microbiology, 48, 159-169. doi:10.1139/w02-002
- Raza, W., Yang, W. and Shen, Q.R. (2008) Paenibacillus polymyxa: Antibiotics, hydrolytic enzymes and hazard assessment. Journal of Plant Pathology, 90, 419-430.
- McBride, M.J., Xie, G., Martens, E.C., Lapidus, A., Henrissat, B., Rhodes, R.G., Goltsman, E., Wang, W., Xu, J., Hunnicutt, D.W., Staroscik, A.M., Hoover, T.R., Cheng, Y.-Q. and Stein, J.L. (2009) Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Applied Microbiology and Biotechnology, 75, 6864-6875. doi:10.1128/AEM.01495-09
- Banning, E.C. and Erin, C. (2010) Biology and potential biogeochemical impacts of novel predatory flavobacteria. http://18.7.29.232/handle/1721.1/57797

