Advances in Bioscience and Biotechnology
Vol.06 No.01(2015), Article ID:53638,11 pages
10.4236/abb.2015.61005
Thermodynamic Principle Revisited: Theory of Protein Folding
Yi Fang
Department of Mathematics, Nanchang University, Nanchang, China
Email: yifang@ncu.edu.cn, yi.fang3@gmail.com
Copyright © 2015 by author and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 10 January 2015; accepted 25 January 2015; published 29 January 2015
ABSTRACT
Anfinsen’s thermodynamic hypothesis is reviewed and misunderstandings are clarified. It really should be called the thermodynamic principle of protein folding. Energy landscape is really just the mathematical graph of the Gibbs free energy function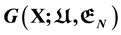 , a very high dimensional hyper surface. Without knowing it any picture of the Gibbs free energy landscape has no theoretical base, including the funnel shape claims. New insight given by newly obtained analytic Gibbs free energy function
, a very high dimensional hyper surface. Without knowing it any picture of the Gibbs free energy landscape has no theoretical base, including the funnel shape claims. New insight given by newly obtained analytic Gibbs free energy function 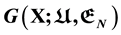 of protein folding derived via quantum statistical mechanics are discussed. Disputes such as target-based or cause-based; what is the folding force, hydrophobic effect or hydrophilic force? Single molecule or ensemble of molecules to be used for the statistical physics study of protein folding, are discussed. Classical observations of 1970’s and 1980’s about global geometric characteristics of native structures of globular proteins turn out to have grabbed the essence of protein folding, but unfortunately have been largely forgotten.
of protein folding derived via quantum statistical mechanics are discussed. Disputes such as target-based or cause-based; what is the folding force, hydrophobic effect or hydrophilic force? Single molecule or ensemble of molecules to be used for the statistical physics study of protein folding, are discussed. Classical observations of 1970’s and 1980’s about global geometric characteristics of native structures of globular proteins turn out to have grabbed the essence of protein folding, but unfortunately have been largely forgotten.
Keywords:
Protein Folding, Gibbs Free Energy, Quantum Statistic, Single Molecule

1. Introduction
1.1. The Second Law of Thermodynamics
The Second Law of Thermodynamics states that in an isolated system the entropy will increase. For a spontaneous process in a system of constant temperature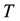 , pressure
, pressure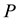 , and composition, the equivalent statement of the second law of thermodynamics states that the Gibbs free energy will be lowered, and at the new equilibrium state it will be at a minimum. Indeed, this applies to any process or any chemical reaction with constant temperatures and pressure [1] . The Gibbs free energy has the form
, and composition, the equivalent statement of the second law of thermodynamics states that the Gibbs free energy will be lowered, and at the new equilibrium state it will be at a minimum. Indeed, this applies to any process or any chemical reaction with constant temperatures and pressure [1] . The Gibbs free energy has the form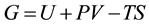 , where
, where 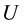 is the internal energy,
is the internal energy, 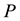 the pressure,
the pressure, 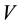 the volume,
the volume, 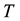 the temperature, and
the temperature, and 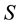 the entropy, of the system. Even
the entropy, of the system. Even 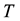 and
and 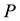 are not uniform in the system, moreover even they are not well defined in the system, the available free energy
are not uniform in the system, moreover even they are not well defined in the system, the available free energy 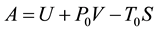 will be lowered and goes to minimum as long as the heat bath of the system has well defined temperature and pressure and on the boundary of the system they are constants
will be lowered and goes to minimum as long as the heat bath of the system has well defined temperature and pressure and on the boundary of the system they are constants 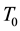 and
and 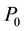 respectively [2] . This fundamental principle was known long before.
respectively [2] . This fundamental principle was known long before.
1.2. Anfinsen’s Thermodynamics Hypothesis of Protein Folding
A good example of applying this fundamental principle to lift experimental results to guiding theory of further research is Anfinsen’s Thermodynamic Hypothesis of protein folding [3] . After many years of experimental work proved that the refolding process of ribonuclease is spontaneous, Anfinsen summarised: “The studies on the renaturation of the fully denatured ribonuclease required many supporting investigations to establish, finally, the generality which we have occasionally called the ‘thermodynamic hypothesis’. This hypothesis states that the three-dimensional structure of a native protein in its normal physiological milieu (solvent, pH, ionic strength, presence of other components such as metal ions or prosthetic groups, temperature, and other) is the one in which the Gibbs free energy of the whole system is lowest; that is that the native conformation is determined by the totality of the inter atomic interactions, and hence by the amino acid sequence, in a given environment.” [3] .
Once we know that protein folding is a spontaneous process, the thermodynamic hypothesis should be upgraded to thermodynamic principle. The thermodynamic principle makes the protein folding problem a pure physics problem, all biological knowledge needed is how to specify the physiological environment for a particular protein and how to reasonably simplify the environment for the further study.
But this has not been recognised so far, instead, it is thought that in biological problems such as protein folding theoretical consideration is unpractical. Coincidently, in the early 1970’s, the same time when the thermodynamic principle of protein folding was established by Anfinsen, computer entered research and played more and more important role in protein folding research. Theory was neglected, simulations became essential as if they were experiments, but many cannot satisfy the essential requirement to experiments in experimental sciences, the reproducibility, see [4] . Furthermore, theoretical background justification of these simulations were rarely questioned. One wonders that if the increasing computer power were really guided by the thermodynamics principle, perhaps today the mystery of protein folding phenomenon would not be really a mystery anymore.
1.3. Reasons of the Thermodynamic Principle of Protein Folding Is Neglected
Why the thermodynamics principle were not actively and persistently pursued?
1.3.1. Do Not Believe It
First, some do not think that the thermodynamic principle is correct. For example, in [5] it is claimed that Anfinsen’s theory was disapproved for long time because “other complexities of biological systems for example solvents of different compositions may affect the folding/unfolding of proteins, the role of high dielectric constant of water, chaperone assisted folding of proteins and existence of stable folding intermediates.”
All the reasons listed above to “disapprove” the thermodynamic principle belong to neglecting that in the thermodynamic principle of protein folding environment plays the same important role as the peptide chain of a protein. In fact, Anfinsen never claimed “that the primary amino acid sequence of polypeptides contains all of the necessary information to direct their folding into functional native proteins” [6] . Instead, Anfinsen stressed that “in a given environment”. Solvents of different compositions, particular properties of water, chaperones to assist folding, etc., are constitutes of environment. For example, globular proteins have the simplest environment which can be simplified as only water molecules surrounding a protein molecule. For proteins needing chaperones to assist folding, chaperone molecules must appear in the environment. For membrane proteins, the environment must be described as including three layers, the middle one is hydrophobic, and the other two are mainly water molecules. Some proteins would not fold in environments not including certain constitutes, does not disapprove the thermodynamic principle, rather the proteins are not in their “normal physiological milieu”. This is just one example that the generality of the thermodynamic principle is often misunderstood and then thought as wrong.
1.3.2. Misunderstandings Caused by Energy Landscape “Theory”
Second, the main reason of the thermodynamic principle was not pursued enough is because of confusions caused by the energy landscape “theory”, both the EL (potential energy landscape) and GEL (Gibbs energy landscape) “theories”. Indeed to minimise the Gibbs free energy one should have a Gibbs free energy function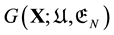 , where the variable
, where the variable 
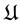


Without knowing
In fact, the GEL is just the graph of



Thus, if we know
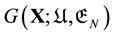
For example, in [1] it is claimed that pursuing of the thermodynamic principle (equivalented to GEL) leads to pitfalls, and the thermodynamic principle will not help to solve the protein folding problem [5] , [11] .
1.3.3. Lack of Mathematical Training
One of the main reasons of GEL, in fact, the thermodynamic principle, will not help solving the protein folding problem is that the second law of the thermodynamics cannot guarantee that the Gibbs free energy 



1.3.4. Should the Native Structure Be Only a Local Minimum?
Another main reason of GEL will not help solving the protein folding problem is that the native structure of a protein maybe is only at a local minimum, instead of the global minimum of Gibbs free energy. This is possible, but in circumstances not against the second law of thermodynamics, hence will not negate the thermodynamic principle. In this case, the initial conformation will determine which minimal points will be achieved by the native structure. We should know more of the conformation of newly synthesised poly peptide chain in a cell. Is it alway the same conformation or does it vary with each individual molecule? If it is the former, then starting with other initial conformations may lead to local or global minima different to the native structure. If it is the later, then perhaps the native structure is really the unique global minimum, since starting from all initial conformations lead to the same native structure. Judged from the experiment results of ribonuclease denaturation/renatu- ration, denatured ribonuclease still hold about 1% biological function. Since there are 105 patterns of fulfilling the 4 disulphide bonds, we may infer that perhaps each of the 105 patterns has the similar percentage in the denatured state. Yet, all these initial conformations refold to the same native structure in which the protein has 100% biological function. Thus we can infer that for ribonuclease the 
1.3.5. Mistaking Environment
A really legitimate concern about the thermodynamic principle is argued in [16] . It says that
“According to this hypothesis, if we define 



























The point is that the solvent composition 












After clarifying these suspicions on the thermodynamics principle, we will demonstrate what is the 
2. The Formula
We will not give the derivation of
Our understanding of the thermodynamic principle is that it emphasises holistic view, it requires a single molecule method and quantum statistics instead of classical statistics to derive the Gibbs free energy formula
2.1. The Function 
Unlike the potential energy function, the Gibbs free energy function, or, the GEL, is not pairwise additive as has been pointed in [6] . In fact, we cannot first consider local contributions and then sum them up to get the Gibbs free energy. This is emphasised by Anfinsen in the statement “that is that the native conformation is determined by the totality of the inter atomic interactions, and hence by the amino acid sequence, in a given environment.” [3] .
So that when trying to derive 









2.2. Single Molecule Treatment Is Necessary
Like any computer simulation of protein folding, we describe only one protein molecule in various conformations





Since one of the tasks of protein folding problem is to figure out the individual protein’s native structure, but in an ensemble of molecules, all available methods are actually neglecting the structures of individual molecule, we cannot use the ensemble method. Therefore, we have to take a single molecule






2.3. Classical or Quantum Statistics?
We have to figure out how to do statistics on this thermodynamic system
2.4. The Importance of Environment
Our tailor made thermodynamic system 


Since except for monomeric globular proteins, we have not figured out how to handle environment, our present function 
2.5. The Thermodynamic System 
Since only globular proteins allow us to simplify their physiological environment as consisting of only water molecules, we will only work on monomeric globular proteins here. A conformation of a polymeric globular protein is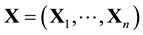




First, a conformation 


















To explain the formula

In general, any closed surface (connected, bounded, and has no boundary, for example, a sphere) 


where 



Rolling a sphere of radius 





















Let

be the first hydration shell surrounding


2.6. Hydrophobicity Levels
Any Gibbs free energy formula should not only have fairly general form for all proteins, or at least a class of proteins such as monomeric globular proteins, but also must be able to distinguish different proteins. That means that if 













Hence, we should find a way to distinguish proteins by their peptide chains. The hardest task is that given a peptide chain








To this purpose, we divide atoms in a protein according to their hydrophobicity levels. Atoms in a protein molecule are naturally existing in atom groups or moieties which have different physicochemical properties. One of these properties is the electronic charge distributions caused the tendency of forming hydrogen bonds either with other moieties (intra-molecular) or with other molecules in the environment (inter-molecular). Accordingly, we can divide these atom groups or moieties into different levlels of hydrophobicity, from the most hydrophobic (cannot form hydrogen bond) to the most hydrophilic, say there are 








For any compact (closed and bounded) set








where 



To each hydrophobic level













2.7. The Formula
Let 


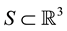

Theorem 1 Let 






In [12] - [15] , the quantum statistical derivation first get a intermediate formula, which is much familiar but with new meaning for

where 








We get formula (6) from (7).
3. New Insight
3.1. Structure Prediction
With theoretically established



Or, in case that 

As discussed before, in this situation, initial conformation 
To solve (9) there is no need of searching landscapes as seen so important in GEL “theory” [9] . Just following the 






When the native structure may take only a local minimum instead a global one, we have to try different initial conforms




Of course we can use another set of variables, i.e., the dihedral angles
In fact, the dihedral angles are the most efficient variables in solving (9) and (10). For the explicit derivative formulae of
3.2. Understanding the Folding Process
Theoretically derived 

path, the initial conformation 




Now consider an ensemble of 

and folding time, etc. Then indeed we should consider the distribution



where 




The rational is that any conformation 


thus less chance to appear in the full function state of the ensemble. Of course, this is only a conjecture and it is not so important to know, at least not as a claim made in [24] : “If one knew this distribution, then one could tell which conformations are more probable than the others under the given environment.” In fact, we now all know that in physiological situation the native structure is “more probable than the others under the given environment”, but, we still do not know the shape of the native structure. So to solve the protein folding problem, at least for the prediction of native structure from the knowledge of amino acid sequence, we have to know what is 
3.3. Force of Folding
In [6] , the folding force is claimed as

3.4. Understanding Denaturation and Refolding
If theoretically derived 















Furthermore, various thermodynamic functions, such as the entropy


3.5. Hydrophobic, Hydrophilic, Which Is the Folding Force?
There is a hot debate in [6] on which one is the main folding force, hydrophobic effect or hydrophilic force? Once we know










In terms of the force











To simplify the discussion of hydrophobic and hydrophilic effects (or force), we consider the simplest classification of hydrophobicity levels, i.e., 



Thus for fixed



Since






ing about secondary structure and hydrogen bond were considered. No calculation of the dihedral angles. No testing of positions of the donor and acceptor groups, let alone any intent to push them closer to form a hydrogen bond. Yet nevertheless, secondary structures and hydrogen bond appeared in statistical significance. Before this simulation, it was recognised that hydrogen bond must be explicitly modelled for helix formation and pairwise simulation without specifying hydrogen bonding cannot produce secondary structures [26] .
3.6. Continue a Classics Global View
If


4. Conclusion
The reasons that over four decades Anfinsen’s thermodynamic hypothesis has been dismissed as leading to pitfalls, as disapproved, as no importance at all, are analysed and clarified. They are due to misunderstanding and inability in deriving a Gibbs free energy formula of protein folding. The misunderstandings mainly come from neglecting environment’s role that Anfinsen so emphasised. The inability of deriving a Gibbs free energy function of protein folding comes from using ensemble of conformations that neglected individual conformation’s 3-dimensional shape. The no importance dismiss came coincidently with various of computer simulation without theoretical discuss of their theoretical bases. Newly derived Gibbs free energy function 



References
- Ben-Naim, A. (2011) Pitfalls in Anfinsen’s Thermodynamic Hypothesis. Chemical Physics Letters, 511, 126-128. http://dx.doi.org/10.1016/j.cplett.2011.05.049
- Müller, I. (2010) Miscellania about Entropy, Energy, and Available Free Energy. Symmetry, 2, 916-934. http://dx.doi.org/10.3390/sym2020916
- Anfinsen, C.B. (1973) Principles that Govern the Folding of Protein Chains. Science, 181, 223-230. http://dx.doi.org/10.1126/science.181.4096.223
- Wang, Y., Zhang, H., Li, W. and Scott, R.A. (1995) Discriminating Compact Nonnative Structure from the Native Structure of Globular Proteins. PNAS, 92, 709-713.
- Kaushik, A. and Gupta, E. (2013) Protein Folding grand Challenge: Hydrophobic vs. Hydrophilic Forces. Journal of Biomolecular Structure and Dynamics, 31, 1011-1012.
- Ben-Naim, A. (2012) Levinthal’s Question Revisited, and Answered. Journal of Biomolecular Structure and Dynamics, 30, 113-124. http://dx.doi.org/10.1080/07391102.2012.674286
- Lazaridis, T. and Karplus, M. (2003) Thermodynamics of Protein Folding: A Microscopic View. Biophysical Chemistry, 100, 367-395. http://dx.doi.org/10.1016/S0301-4622(02)00293-4
- Ben-Naim, A. (2013) Response to Comments on My Article: Liventhal’s Question Revisited and Answered. Ben-Naim A. (2012) Journal of Biomolecular Structure and Dynamics, 30, 113-124. Journal of Biomolecular Structure and Dynamics, 31, 1028-1033. http://dx.doi.org/10.1080/07391102.2012.748548
- Schafer, N.P., Kim, B.L., Zhang, Q. and Wolynes, P.G. (2013) Learning to Fold Protein Using Energy Landscape Theory. arXiv:1312.7283v1 [q-bio.BM] http://arxiv.org/pdf/1312.7283.pdf
- Liu, S.Q., Ji, X.L., Tao, Y., Tan, D.Y., Zhang, K.Q. and Fu, Y.X. (2012) Protein Folding, Binding and Energy Landscape: A Synthesis. In: Kaumaya, P.T.P., Ed., Protein Engineering, Intech, Rijeka, 207-253.
- Fang, Y. (2015) Why Ben-Naim’s Deepest Pitfall Does Not Exist. To Appear in Open Journal of Biophysics.
- Fang, Y. (2012) Gibbs Free Energy Formula for Protein Folding. In: Morales-Rodriguez, R., Ed., Thermodynamics― Fundamentals and Its Application in Science, Intech, Rijeka, 47-82. http://www.intechopen.com/books/thermodynamics-fundamentals-and-its-application-in-science
- Fang, Y. (2013) Ben-Naim’s Pitfalls: Don Quixote’s Windmill. Open Journal of Biophysics, 3, 13-21.
- Fang, Y. (2014) The Second Law, Gibbs Free Energy, Geometry, and Protein Folding. Journal of Advances in Physics, 3, 278-285.
- Fang, Y. (2014) A Gibbs Free Energy Formula for Protein Folding Derived from Quantum Statistics. Science China Physics, Mechanics & Astronomy, 57, 1547-1551. http://dx.doi.org/10.1007/s11433-013-5288-x
- Gillet, J. and Ghosh, I. (2013) Concepts on the Protein Folding Problem. Journal of Biomolecular Structure and Dynamics, 31, 1020-1023. http://dx.doi.org/10.1080/07391102.2012.748546
- Bader, R.F.W. (1990) Atoms in Molecules: A Quantum Theory. Clarendon Press, Oxford.
- Pippard, A.A. (1957) The Elements of Classical Thermodynamics. Cambridge University Press, Cambridge.
- Richards, F.M. (1977) Areas, Volumes, Packing, and Protein Structure. Annual Review of Biophysics and Bioengineering, 6, 151-176. http://dx.doi.org/10.1146/annurev.bb.06.060177.001055
- Tuñón, I., Silla, E. and Pascual-Ahuir, J.L. (1992) Molecular Surface Area and Hydrophobic Effect. Protein Engineering, Design and Selection, 5, 715-716. http://dx.doi.org/10.1093/protein/5.8.715
- Jackson, R.M. and Sternberg, M.J.E. (1993) Protein Surface Area Defined. Nature, 366, 638. http://dx.doi.org/10.1038/366638b0
- Eisenberg, D. and McLachlan, A.D. (1986) Solvation Energy in Protein Folding and Binding. Nature, 319, 199-203. http://dx.doi.org/10.1038/319199a0
- Fang, Y. and Jing, J. (2008) Implementation of a Mathematical Protein Folding Model. International Journal of Pure and Applied Mathematics, 42, 481-488.
- Ben-Naim, A. (2013) Comment on a Paper: “Ben-Naim’s ‘Pitfalls’: Don Quixote’s Windmill” by Y. Fang, Open Journal of Biophysics, 2013, 3, 13-21. Open Journal of Biophysics, 3, 275-276.
- Fang, Y. and Jing, J. (2010) Geometry, Thermodynamics, and Protein. Journal of Theoretical Biology, 262, 383-390. http://dx.doi.org/10.1016/j.jtbi.2009.09.013
- Hubner, I.A. and Shakhnovic, E.I. (2005) Geometric and Physical Considerations for Realistic Protein Models. Physical Review E, 72, Article ID: 022901. http://dx.doi.org/10.1103/PhysRevE.72.022901
- Lee, B. and Richards, F.M. (1971) The Interpretation of Protein Structures: Estimation of Static Accessibility. Journal of Molecular Biology, 55, 379-400. http://dx.doi.org/10.1016/0022-2836(71)90324-X
- Janin, J. (1976) Surface Area of Globular Proteins. Journal of Molecular Biology, 105, 13-14. http://dx.doi.org/10.1016/0022-2836(76)90192-3
- Richards, F.M. (1979) Packing Defects, Cavities, Volume Fluctuations, and Access to the Interior of Proteins. Including Some General Comments on Surface Area and Protein Structure. Carlsberg Research Communications, 44, 47-63. http://dx.doi.org/10.1007/BF02906521
- Novotny, J., Bruccoleri, R. and Karplus, M. (1984) An Analysis of Incorrectly Folded Protein Models: Implications for Structure Predictions. Journal of Molecular Biology, 177, 787-818. http://dx.doi.org/10.1016/0022-2836(84)90049-4
- Novotny, J., Rashin, A.A. and Bruccoleri, R. (1986) Criteria that Discriminate between Native Proteins and Incorrectly Folded Models. Proteins, 4, 19-30. http://dx.doi.org/10.1002/prot.340040105
- Fang, Y. (2005) Mathematical Protein Folding Problem. In: Hoffman, D., Ed., Global Theory of Minimal Surfaces. Proceedings of the Clay Mathematical Proceedings, Vol. 2, American Mathematical Society, Clay Mathematics Institute, 611-622.



