Advances in Bioscience and Biotechnology
Vol.3 No.1(2012), Article ID:17188,10 pages DOI:10.4236/abb.2012.31005
Disorder structural predictions of the native EWS and its oncogenic fusion proteins in rapport with the function*
![]()
IBFBMI-BAS, Sofia, Bulgaria
Email: todorova@bio21.bas.bg
Received 8 October 2011; revised 15 November 2011; accepted 28 December 2011
Keywords: Intrinsicaly Disordered Proteins; Predictors; Relationship Sequence-Disorder-Function; EWS Oncogenic Fusion Proteins
ABSTRACT
The Intrinsic structural disorder (ISD) of native EWS and its fusion oncogenic proteins, including EWS/FliI, EWS/ATF1 and EWS/ZSG, was estimated by different Predictors. The ISD difference between the wild type and the oncogenic fusions found in the CTD is due to the fusion partner, usually a transcription factor (TF). A disordered region was found in the sequence (AA 132 - 156) of the NTD (EAD) of EWS, consisting of the longest region free of Y motifs. The IQ domain (AA 258 - 280), a Y-free region, flanked by two Y-boxes, is also disordered by all used Predictors. The EWS functional regions RGG1, RGG2 and RGG3 are predominantly disordered. A strong dependence was found between the structure of EWS protein and its oncogenic fusions, and their estimated ISD. The oncogenic function of the fusions is related to a decreased ISD in the CTD, due to the fused TF. The Predictors shown that the different isoforms have similar profiles, shifted with some amino acids, due to the translocations. On the bases of the prediction results, an analysis was made of the EWS sequence and its functional regions with increased ISD to make a relationship sequence-disorder-function that could be helpful in the design of antitumor agents against the corresponding malignances.
1. INTRODUCTION
Ewing’s Sarcoma Oncogene (EWS) on chromosome 22q12 is encoding a RNA binding protein that is target of tumor-specific chromosomal translocations in Ewing sarcoma tumor, Myxoid liposarcoma, Malignant melanoma of soft parts, Desmoplastic small round cell tumor, Peripheral neurectodermal tumour, Angiomatoid fibrous histiocytoma, Extra-skeletal myxoid chondrosarcomas, Rhabdomyosarcoma, Locally destructive tumour, Myoepithelioma tumours of soft tissue, Hidradenoma or eccrine acrospiroma, Mucoepidermoid carcinoma, Neuroblastoma, Olfactory neuroblastoma, Solid pseudopapillary tumour of the pancreas and Acute myeloid leukemia. Around 85% of Ewing tumours carry the EWSR1/FLI-1 fusion. The cellular function, the mechanism of participation in the multiple levels of gene expression and the role of EWS in pathogenesis of the resulting cancers are not well defined. The interaction mechanism of EWS in self-association and oligomerization involves N-terminal and centrally localized amino acids, while for optimal association are required full-length EWS molecules [1]. The EWS activation domain (EAD) is located within its N-terminal 286 amino acids, and like many chromatin organizing proteins is an intrinsically disordered (ID) protein [2].
Here is made an attempt to estimate the Intrinsic structural disorder (ISD) of EWS and its reported fusion oncogenic proteins by different methods of prediction. On the bases of the prediction results, an analysis was made of the EWS sequence and its functional regions with increased ISD to make a relationship sequence-disorderfunction that could be used to design antitumor agents against the corresponding malignances.
2. MATERIALS AND METHODS
Methods and Sequences
The protein intrinsic disorder (ID) was predicted by IUPred [3], GlobPlot2 [4], DisEMBL [5], FoldIndex [6], RONN [7], PONDR [8], VL3H [9]. The Predictors were used to estimate the protein disorder of native EWS and of the known oncogenic fusions EWS/FliI, EWS/ATF1, EWS/ZSG, and others with known sequence around the break-point, such as EWS/ERG, EWS/WT1, EWS/CHOP and EWS-CHN. All estimations are made with the RNA-binding protein EWS isoform 2 (656 Amino acids (AA)) [Homo sapiens], NP_005234.1. Estimates were realized by the same method to compare the IPD of all fusions by their isomers, including also the globular domains. The used isoforms of EWS oncogenic proteins in the predictions were fusions EWS/FLI1 (1)-(3), EWS/ ATF1 (4), (5) and EWS/ZSG (6)-(8):
1) EWS-Fli1 (476 AA); AAK11227.1.
2) EWS-Fli1 (526 AA); ADX41460.1.
3) EWS-Fli1 (504 AA); ADX41459.1.
4) EWS/ATF1 (432 AA); ADX41458.1.
5) EWS/ATF1 (537 AA); ADX41457.1.
6) EWS/ZSG long A isoform (713 AA); AAG09036.1.
7) EWS/ZSG long B isoform (609 AA); AAG09037.1.
8) EWS/ZSG short isoform (609 AA); AAG09035.1.
The results from different Predictors were compared for the first time on the example of native EWS protein and its oncogenic fusions. A new approach is the analysis on the bases of the prediction results of the functional regions of EWS and its oncogenic fusions to make a relationship sequence-disorder-function.
3. RESULTS AND DISCUSSIONS
3.1. Feature of the Protein Intrinsic Disorder
Intrinsicaly disordered proteins (IDPs) lack stable tertiary and/or secondary structure and participate in both one-to-many and many-to-one signaling, important for transient protein-protein and protein-nucleic acid interactions. The alternative splicing and posttranslational modifications of IDPs are linked to their functions in cellular regulation, recognition and signal transduction. Human diseases associated proteins, including cancer, are enriched in ID and enter in high-specificity-low-affinity interactions by one-to-many binding mode via plasticity [10]. The ID Transcription factors (TFs) can provide significant advantages in response to molecular targets, allowing multiple partners interactions and fine control over binding affinity, thus representing targets for therapeutic drugs. Some types of DNA-binding domains (DBDs) in TFs are highly unstructured in isolation, and undergo a disorder-to-order transition upon binding to specific DNA [11]. Modular DBDs, such as zinc finger domains, recognize a DNA sequence in a regulatory region of a targeted area. Chromosomal translocations represent major genetic aberration leading to cancer. The high level of ISD is enabling fusion proteins to evade cellular surveillance mechanisms that eliminate misfolded proteins. Predictions of translocation-related human proteins show 43.3% disorder vs 20.7% in all human proteins, fewer Pfam domains and translocation breakpoints tending to avoid domain splitting [12]. The vicinity of the breakpoint is significantly more disordered than the rest of fusion proteins. The ISD is enabling the long-range structural communication in the oncogenic function. The fusion of a DBD to a transactivator domain results in an aberrant TF, such as EWS/Fli1, EWS/ATF, EWS/ZSG, EWS/ERG, EWS/WT1, EWS/ CHOP and EWS-CHN.
3.2. Predictions of the Intrinsic Disorder
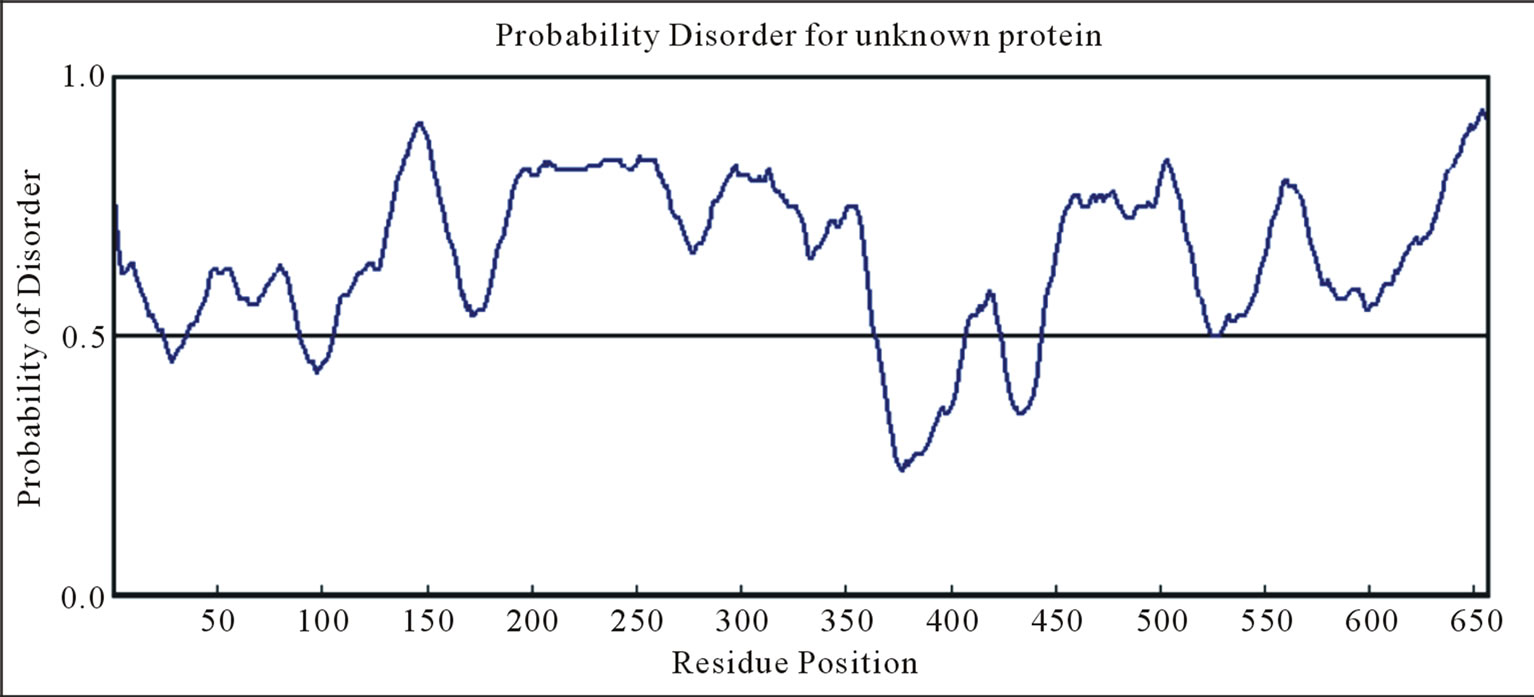
The plots of the IPD predictions for the different isoforms of fusions EWS/FLI1 (1)-(3), EWS/ATF1 (4), (5) and EWS/ZSG (6)-(8), estimated by IUPRED (long disorder), were shown on Figure 1. The results show identical distribution with high ID in the N-terminal domain (NTD) of EWS (EAD, transactivation domain). The ID difference is in the C-terminal domain (CTD) that is RNA binding for EWS native and DNA binding for the EWS fusions, originated from different TFs. The CTD of EWS shown full disorder, while the CTDs in the fusions shown also globular domains, characteristic for the fused TFs. The DisEMBLs predictions of the IPD were shown on Figure 3.
3.2.1. EWS Native (AA 1 - 656)
EWSR1 protein is composed from 656 AA. The EAD (AA 1 - 285) contains multiple DHRs (consensus SYGQQS) where Tyr is mandatory, a site interacting with SF1 (AA 228 - 264), and an IQ domain binding calmodulin (AA 256 - 285). The CTD contains three RGG regions (AA 300 - 340, AA 454 - 513 (RGP rich), AA 559 - 640), a RNA recognition motif (RRM or RNAbinding domain (RBD)) at (AA 361 - 447), and a RanBP2 type Zinc finger (518 - 549). Four EWS isoforms are known: isoform 1 (661 AA), isoform 2 (656 AA), isoform 3 (655 AA) and isoform 4 (600 AA). The NTD of EWS is involved in self-association and dimerization. An RNA binding component is involved in EWS oligomerization [1].
The ID of EWS was estimated by Predictors: IUPred (long disorder) (Figure 1(a)), RONN (Figure 2(a)), GlobPlot2, DisEMBL (Figure 2(b)), FoldIndex, PONDR, VL3H. A similar distribution with high ID was found in the NTD (EAD, Trans-activation domain (TAD)), but also in the CTD (RBD) of EWS. The prediction of the NTD (AA 1 - 240) by Porter, PaleAle, BrownAle, XStout, XXStout, 3Distill, Porter+, SCLpred shown an animal nuclear localization, found also experimentally in vivo and in vitro. Thus, the NTD of EWS consists of large disordered regions and very small amount of short ordered regions, while the CTD is almost completely disordered:
1) A common disordered region was found by all Predictors, containing the AA 132 - 156 in the NTD, consisting of the longest region free of Y motifs, and functionally responsible for multiple interactions with different factors.
2) The IQ domain (AA 258 - 280, binding calmodulin)
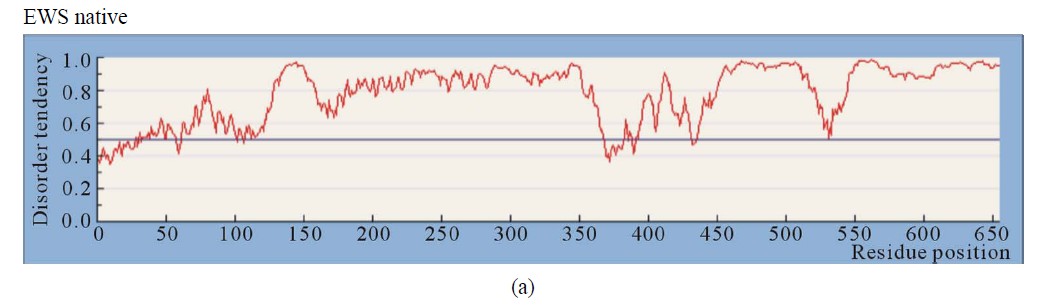
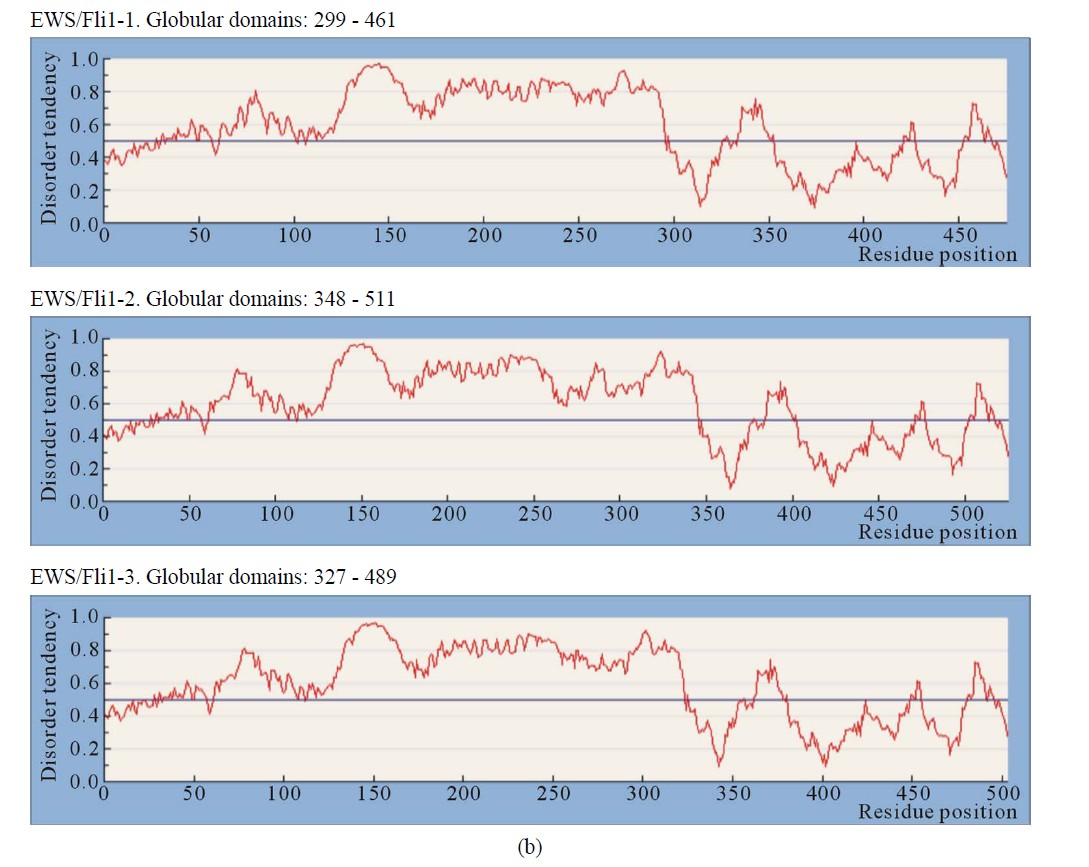
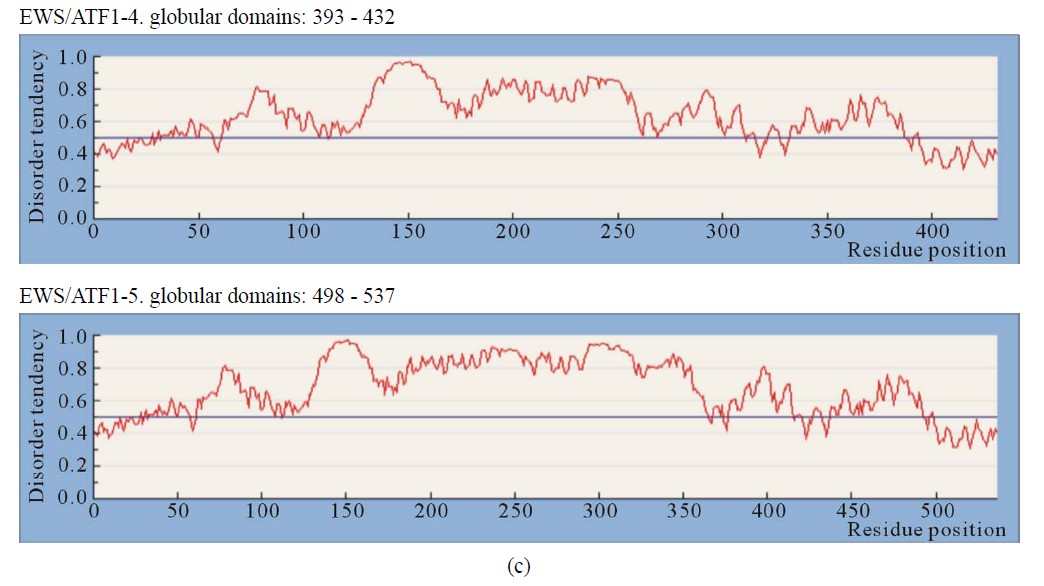
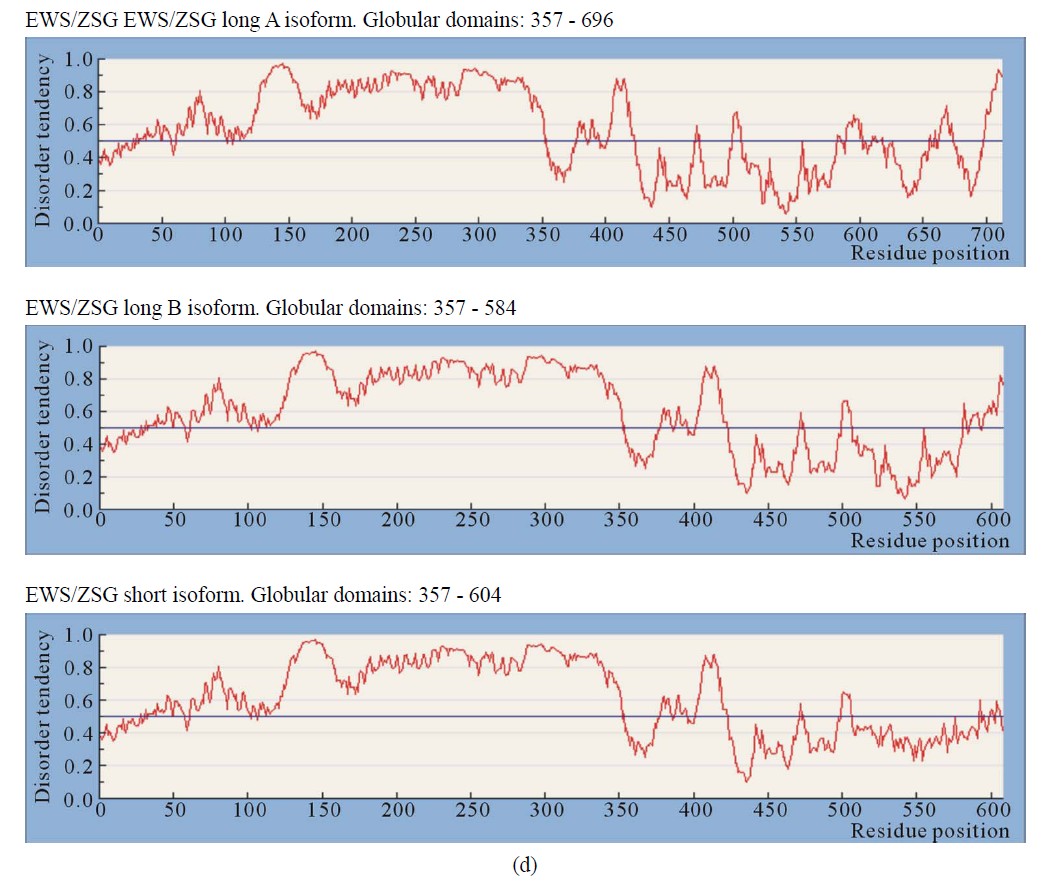
Figure 1. IUPRED predictions (Long disorder) of intrinsic disorder (ID) for EWS and fusion proteins EWS/ATF1, EWS/Fli1 and EWS/ZSG. Values above 0.5 are considered disordered. (a) Predictions of ID for native EWS. There are not Globular domains; (b) Predictions of ID for the fusion protein EWS/Fli1 (3 isoforms). Globular domains: AA 299 - 461 (isoform1), AA 348 - 511 (isoform2), AA 327 - 489 (isoform 3); (c) Predictions of ID for the fusion protein EWS/ATF1 (2 isoforms). Globular domains: AA 393 - 432 (isoform 1), AA 498 - 537 (isoform 2); (d) Predictions of ID for the fusion protein EWS/ZSG (3 isoforms). Globular domains: AA 357 - 696 (isoform (a)), AA 357 - 584 (isoform (b)), AA 357 - 604 (short isoform).
contains the NTD longest region, free from repetitive structures, flanked by two Y-containing boxes, and disordered by RONN, IUPred, FoldIndex, PONDR, while by GlobPlot the connecting region has a break in the disorder.
3) The regions, containing RGG1 (5 motifs), RGG2 (4 motifs) and RGG3 (12 motifs) are predominantly disordered by all used Predictors.
4) The NTD (AA 1 - 264) is almost completely disordered, and especially the regions of AA (80 - 95), 130 - 160, 195 - 205, 240 - 250. The region, surrounding the AA 100 (95 - 120) is folded by Foldindex, Disembl, Iupred.
5) The IQ domain (AA 258 - 280), containing the second long Y-free region, flanked by two Y-boxes, is disordered by all used Predictors.
6) The RRM region (RNA recognition motif), consisting of about 100 conserved residues, is ordered by Globplot2.
7) The ZF (Cys2 - Cys2) Zing finger domain, containing AA (518 - 549) is partially ordered by RONN, and flanked by highly disordered regions. The C2H2-type finger domains consist of folded modules, connected to highly conserved linker sequences [11].
3.2.2. EWS/ATF1
The EWS/ATF1 is a fusion between EWS and the TF ATF1. The bZIP domain (AA 214 - 271), consisting of a basic region that directly contacts DNA and a leucine zipper (ZIP) that allows dimerization, is necessary and sufficient for dimerization and DNA-binding [13]. Transactivation by EWS/ATF1 does not require dimerization [13]. The inhibition of B-ZIP TFs could be therapeutically used in cancer cells (clear cell sarcoma) [14].
IUPRED predicted a globular domain in the last 39 AA (bZIP) at (AA 498 - 537) and (AA 393 - 432) for isoforms 5) and 4) of EWS/ATF1. The disorder profile is identical in the NTD (AA 1 - 264, EAD) and similar in the
 (a)
(a)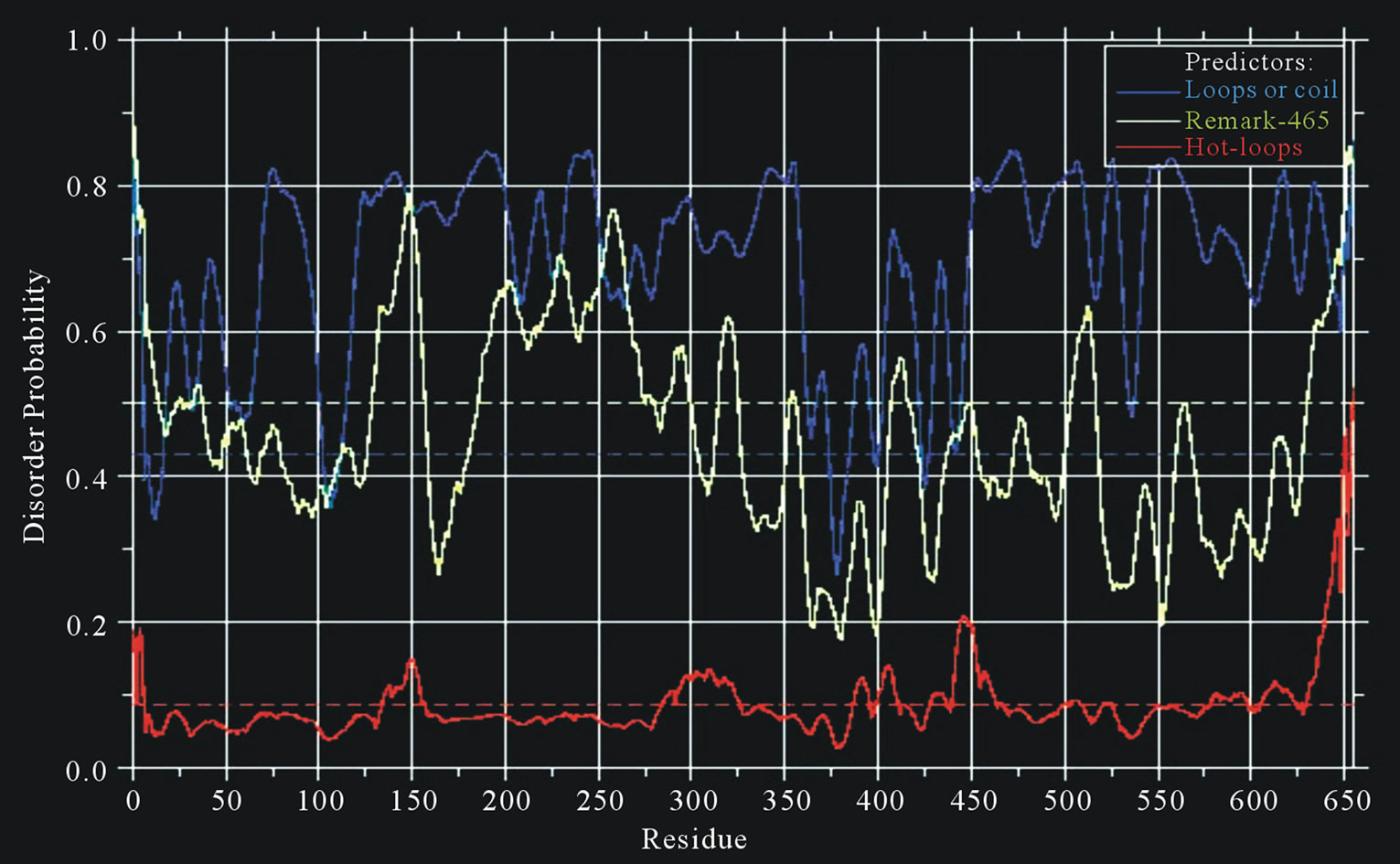 (b)
(b)
Figure 2. Predictions of ISD for native EWS protein: (a) Predictions of ID for native EWS by RONN; (b) Predictions of ID for native EWS by DisEMBLs.
CTD, where the isoforms differ (Figure 1(c)). The EWS/ ATF1 isoforms have increased ID of regions flanking bZIP (AA 457 - 550). The CTD is almost ordered, including the bZIP domain that is folded and linked by highly conserved sequences, mobile and unstructured. The results, obtained by different methods are similar, where the critical elements (particularly the breakpoint), are connected by long segments of structural disorder. The calculated distance/disorder between the oncogenic elements TAD and b-ZIP was of 280/265 AA [12]. The region (AA 340 - 353) is structured (Disembl) (Figure 3(b)). Potential globular domains (GlobDoms) by Globplot2 in EWS/ATF1 are (AA 294 - 432 in (4)) and (AA 399 - 537 in (5)). The disordered region AA 136 - 152 was used to generate Ab against the EWS protein [15].
3.2.3. EWS/Fli1
The EWS/Fli1 (ETS type, AA 281 - 361) is a fusion between the NTD of EWSR1 protein (1 - 265) and the DBD of the human FLI1 protein (452 AA) at AA 260. Self-association of EWS and EWS-FLI1 (but not FLI1) and interaction of EWS-FLI1 with EWS and FLI1, was observed in vivo. The NTD (EAD) contributed to homo and heterotypic interactions. EWS-FLI1 self-associates and binds to FLI1 via its CTD DBD [1].
IUPRED shown globular domains in EWS/Fli1 close to, but not at the C-terminal end of the isoform, where disordered are the last 13 AA. Globular sequences included about 60 AA situated in the CTD of EWS/Fli11: (AA 299 - 459) in 1), (AA 348 - 509) in 2), and (AA 327 - 489) in 3). The disorder profile is the same in the NTD EWS/Fli1-1
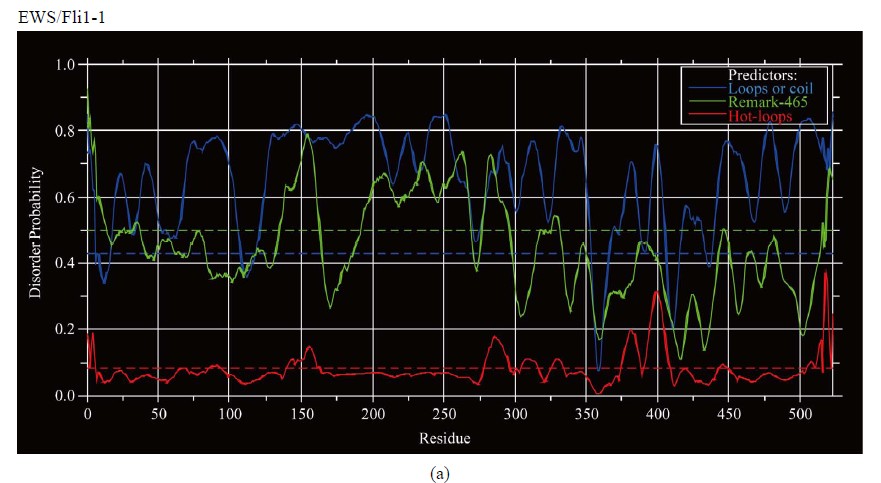
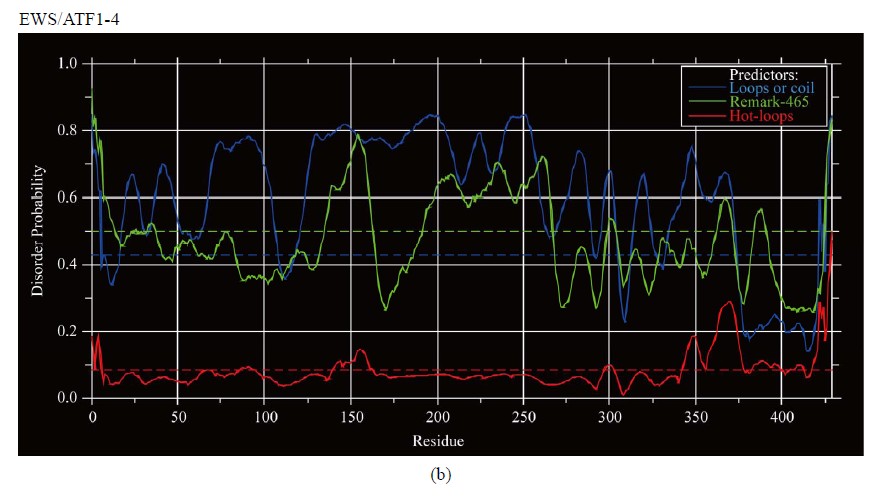
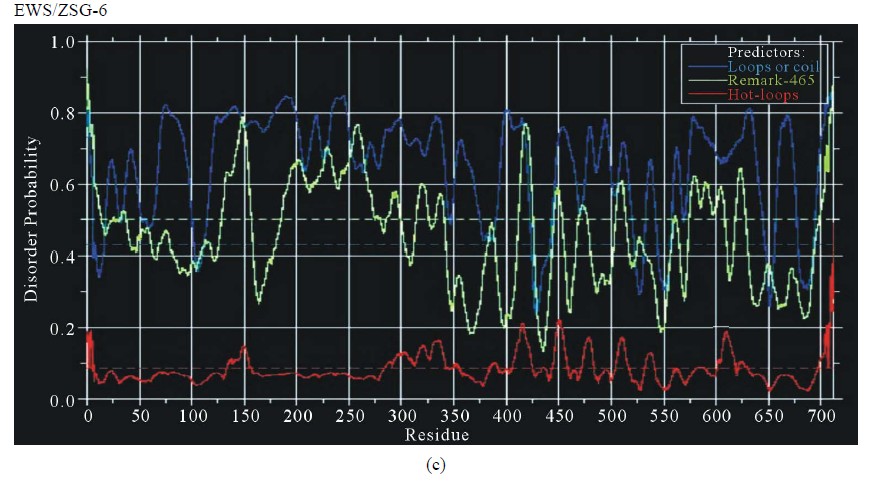
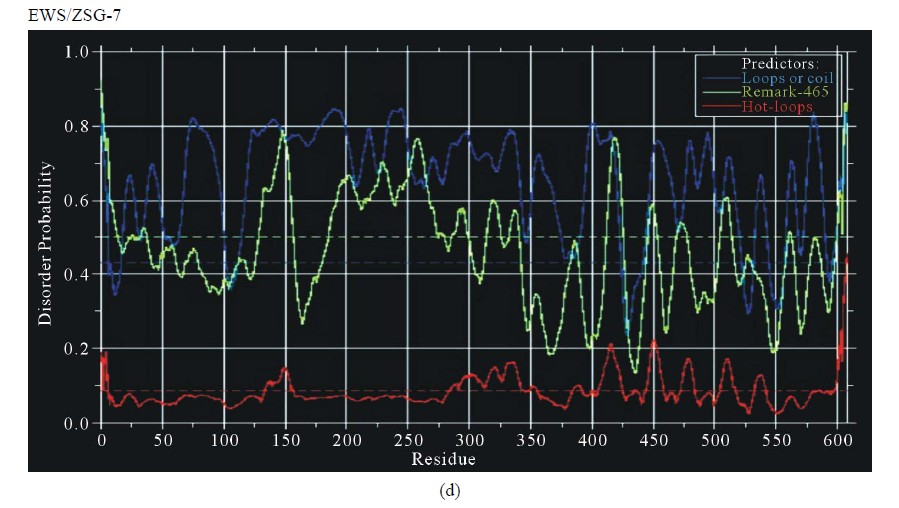
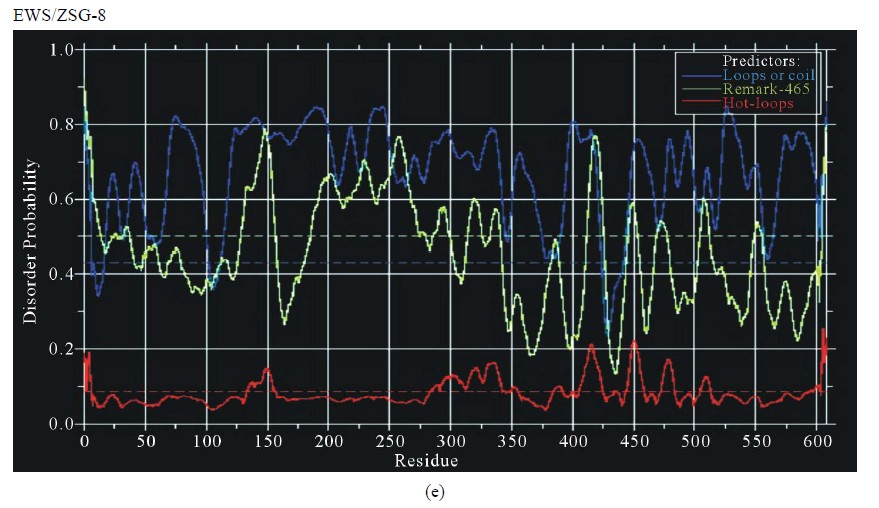
Figure 3. Predictions of ISD for EWS fusion proteins by DisEMBLs: (a) Predictions of ID for EWS/Fli1-1; (b) Predictions of ID for EWS/ATF1-4; (c) Predictions of ID for EWS/ZSG-6; (d) Predictions of ID for EWS/ZSG-7; (e) Predictions of ID for EWS/ZSG-8.
(EAD) and similar in the CTD, where the isoforms differ (Figure 1(b)). Native EWS-Fli1 approaches a largely unfolded conformation [16]. The CTD originated from Fli1 is almost ordered with low propensity for disorder by all Predictors. The region (АА 278 - 297) is structured by Disembl in some isoforms (Figure 3(a)). Potential globular domains by Globplot2 are 1) (AA 302 - 421), 2) (AA 352 - 471) and 3) (AA 330 - 449).
3.2.4. EWS/ZSG
EWS/ZSG is a Zing-finger type oncogenic protein (EFT) with rare frequency in tumors. The IPD of EWS/ZSG, estimated by IUPRED, shown a long disordered region, originated from EWS, followed by a globular domain, comprising residues 357 - 584 (long B isoform), 357 - 696 (long A isoform) and 357 - 604 (protein short isoform), followed by a short disordered region at the Cterminal end of the molecule (Figure 1(d)). The ID of the EAD in EWS/ZSG is analogous to EWS/Fli1 and EWS/ATF1. The profile is the same in the NTD (EAD) and similar in the CTD, where the isoforms differ. The IPD of EWS/ZSG differs from the other EWS oncogenic fusions by the total disorder in the CTD, similar to the native EWS. Predictions of ISD by DisEMBLs shown similar profiles for all isoforms (Figures 3(c)-(e)). Calculated Potential globular domains (GlobDoms) by Russell/Linding definition (Globplot 2) lacked in all isoforms of EWS/ZSG.
3.2.5. EWS/WT1
EWS/WT1 is a zing-finger type oncogenic protein, frequent in DSR tumors. EWS/WT1 self-association maps to the fusion junction (negatively influenced by phosphorylation), where DBD and self-association domains overlap, but the DNA-binding does not depend on selfassociation. The binding of several EWS/WT1 molecules leads to homotypic associations that translate into transcriptional effects [17]. In EWS/WT1 the NTD (EAD) was disordered, while the rest of the protein, containing the zing fingers of WT1, are globular (AA 15 - 89) estimated by IUPRED.
3.2.6. EWS/ERG
The ERG is an ETS type of TF, similar to Fli1. In EWS/ ERG (type 1e) the NTD (EAD) residues are disordered, and the rest of the protein, containing ERG (AA 84 - 247) forms a globular domain (IUPRED). The breakpoint is around AA 80. In the EWS/ERG (type 9e) the globular domain is composed from AA 23 - 189. The complete sequence is not available, but the profile of the different isoforms is similar. Erg/Ets-2 dimer formation may prevent Ets-2 from acting as TF [18,19]. The monomer is functionally active, while the heterodimeric complexes are inactive.
3.2.7. EWS/CHOP
CHOP is a bZIP type of TF, similar to ATF1. EWS/ CHOP is composed from two highly disordered regions, connected with a globular linker, comprising AA 91 - 137, estimated by IUPRED. The complete sequence is not available.
3.3. Estimation of the Different Methods of Protein Disorder Prediction
Comparing, the EWS oncogenic fusions show similar ID in the NTD (AA 1 - 264, EAD). The CTD disorder of fusions differs from that of native protein and between the fusion proteins. A strong relationship was found between the structure and estimated ID of EWS and its oncogenic fusions, by all used Predictors. Finally, the oncogenic function is related to a decreased IPD in the CTD, due to the fused partner, a TF. The different isoforms shown similar profiles, shifted with some amino acids, due to translocations. All methods follow the same shape with small differences that does not influence the complete profile. Thus, Predictors could be used to study the relationship structure-function-protein disorder of native EWS and its oncogenic fusions. A relation structure/disorder/function was found in some regions of the proteins. The disordered region found in AA 132 - 156 of EAD consisted of the longest region, free of Y motifs. The IQ domain (AA 258 - 280), a Y-free region, flanked by two Y-boxes, is also disordered by all used Predictors. The EWS functional regions RGG1, RGG2 and RGG3 are predominantly disordered. This is consistent with the finding that the particular AA composition of the EAD creates an enabling structure with several critical Tyr residues, dispersed in a polar/neutral environment, favoring hydrogen bonding interactions and flexibility [20].
3.4. Relationship between IPD and Multimerization in EWS and Its Oncogenic Fusions
The common structural features limited to TET family members suggest that they bind RNA and/or ssDNA in a unique way [15]. TBP dimerization inhibits DNA-binding thus regulating TBP-DNA interaction [21]. The formation of inactive homo or heterodimers could be a general mode of regulating transcription factors activity in vivo [22]. Multimerization is characteristic for EWS and many EWS fusion oncoproteins. The self-association may be important for the function of EWS and its oncogenic fusions, and realized in different ways. The mechanism may be related to the function of EWS in the normal cells and EWS fusions in cancer cells. The homoand hetero-association may have impact in TF regulation, as well as trans-activation in vivo. The capability of these molecules to associate is closely related to their high level of IPD, flexibility and accessibility to interact with regions inside the same molecule, and with other molecules with same or different nature, thus allowing structural adaptation and multi-partner interactions in their regulative and oncogenic functions. The phenomenon of ISD possibly is linked to the functional regions and the specific interactions undertaken by them. Such is the interaction found in vitro between EAD (AA 1 - 57) and hsRPB7 [23].
Disorder Predictors can help identify some local domains within longer regions of disorder. Combining prediction of ID with other techniques provides an alternative strategy for protein structural characterization and drug target identification, small molecule design and assay development. Disordered proteins are potential targets of small molecule therapeutics. Small molecules that disable EWS-FLI1 function with minimal toxicity (sparing hematopoietic stem cells) could potentially provide a therapy for patients with ESFT and other related sarcomas [24].
4. CONCLUSION
Summarizing, the data from the ID predictions, using several Predictors, have shown a relationship between the functional characteristics of proteins, their structure and amino acid composition. This relationship function-structure-disorder could be used in the design of potential antitumor agents against the EWS fusions related tumors. A contribution of this work is that the fusions oncogenic potential could be studied in the relation functional regions-intrinsic disorder.
REFERENCES
- Spahn, L., Siligan, C., Bachmaier, R., Schmid, J.A., Aryee, D.A.T. and Kovar, H. (2003) Homotypic and heterotypic interactions of EWS, Fli-1 and their oncogenic fusion proteins. Oncogene, 22, 6819-6829. doi:10.1038/sj.onc.1206810
- Ng, K.P., Potikyan, G., Savene, R.O.V., Denny, C.T., Uversky, V.N. and Lee, K.A.W. (2007) Multiple aromatic side chains within a disordere structure are critical for transcription and transforming activity of EWS family oncoproteins. PNAS, 104, 479-484. doi:10.1073/pnas.0607007104
- Dosztanyi, Z., Csizmok, V., Tompa, P. and Simon, I. (2005) IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics, 21, 3433-3434. doi:10.1093/bioinformatics/bti541
- Linding, R., Russell, R.B., Neduva, V. and Gibson, T.J. (2003) GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Research, 31, 3701- 3708. doi:10.1093/nar/gkg519
- Linding, R., Jensen, L.J., Diella, F., Bork, P., Gibson, T.J. and Russell, R.B. (2003) Protein disorder prediction: Implications for structural proteomics. Structure, 11, 1453- 1459. doi:10.1016/j.str.2003.10.002
- Prilusky, J., Felder, C.E., Zeev-Ben-Mordehai, T., Rydberg, E.H., Man, O., Beckmann, J.S., Silman, I., Sussman, J.L. (2005) FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics, 21, 3435-3438. doi:10.1093/bioinformatics/bti537
- Yang, Z.R., Thomson, R., McNeil, P. and Esnouf, R.M. (2005) RONN: The bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics, 21, 3369-3376. doi:10.1093/bioinformatics/bti534
- Xue, B., Dunbrack, R.L., Williams, R.W., Dunker, A.K. and Uversky, V.N. (2010) PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta, 1804, 996-1010.
- Obradovic, Z., Peng, K., Vucetic, S., Radivojac, P., Brown, C.J. and Dunker, A.K. (2003) Predicting intrinsic disorder from amino acid sequence. Proteins, 53, 566-572. doi:10.1002/prot.10532
- Uversky, V.N., Oldfield, C.J., Midic, U., Xie, H., Xue, B., Vucetic, S., Iakoucheva, L.M., Obradovic, Z. and Dunker, A.K. (2009) Unfoldomics of human diseases: Linking protein intrinsic disorder with diseases. BMC Genomics, 10, S7. doi:10.1186/1471-2164-10-S1-S7
- Liu, J., Perumal, N.B., Oldfield, C.J., Su, E.W., Uversky, V.N. and Dunker, A.K. (2006) Intrinsic disorder in transcription factors. Biochemistry, 45, 6873-6888. doi:10.1021/bi0602718
- Hegyi, H., Buday, L. and Tompa, P. (2009) Intrinsic Structural Disorder Confers Cellular Viability on Oncogenic Fusion Proteins. PLoS Computational Biology, 5, e1000552. doi:10.1021/bi0602718
- Pan, S., Ming, K.Y., Dunn, T.A., Li, K.K. and Lee, K.A. (1998) The EWS/ATF1 fusion protein contains a dispersed activation domain that functions directly. Oncogene, 16, 1625-1631. doi:10.1038/sj.onc.1201671
- Heyerdahl, S.L., Rozenberg, J., Jamtgaard, L., Rishi, V., Varticovski, L., Akah, K., Scudiero, D., Shoemaker, R.H., Karpova, T.S., Day, R.N., McNally, J.G. and Vinson, C. (2010) The arylstibonic acid compound NSC13746 disrupts B-ZIP binding to DNA in living cells. European Journal of Cell Biology, 89, 564-573. doi:10.1016/j.ejcb.2009.11.029
- Bertolotti, A., Melot, T., Acker, J., Vigneron, M., Dellatre, O. and Tora, L. (1998) EWS, but not EWS-Fli-1, is associated with both TFIID and RNA-polymerase II: Interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA Polymerase II complexes. Molecular Cell Biology, 18, 1489-1497.
- Uren, A., Tcherkasskaya, O. and Toretsky, J.A. (2004) Recombinant EWS-FLI1 oncoprotein activates transcription. Biochemistry, 42, 13579-13589. doi:10.1021/bi048776q
- Kim, J., Lee, J.M., Branton, P.E. and Pelletier, J. (1999) Modification of EWS/WT1 functional properties by phosphorylation. Proceedings of the National Academy of Sciences of the United States of America, 96, 14300- 14305. doi.org/10.1073/pnas.96.25.14300
- Butticè, G., Duterque-Coquillaud, M., Basuyaux, J.P., Carrère, S., Kurkinen, M. and Stéhelin, D. (1996) Erg, an Ets-family member, differentially regulates human collagenase1 (MMP1) and stromelysin1 (MMP3) gene expression by physically interacting with the Fos/Jun complex. Oncogene, 13, 2297-2306.
- Basuyaux, J.P., Ferreira, E., Stéhelin, D. and Butticè, G. (1997) The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner. Journal of Biological Chemistry, 272, 26188- 26195. doi.org/10.1074/jbc.272.42.26188
- Lee, K.A. (2007) Ewings family oncoproteins: Drunk, disorderly and in search of partners. Cell Research, 17, 286-288. doi:10.1038/cr.2007.22
- Taggart, A.K. and Pugh, B.F. (1996) Dimerization of TFIID when not bound to DNA. Science, 272, 1331-1333. doi:10.1126/science.272.5266.1331
- Carrère, S., Verger, A., Flourens, A., Stehelin, D. and Duterque-Coquillaud, M. (1998) Erg proteins, transcription factors of the Ets family, form homo, heterodimers and ternary complexes via two distinct domains. Oncogene, 16, 3261-3268. doi:10.1038/sj.onc.1201868
- Todorova, R. (2009) In vitro interaction between the N-terminus of the Ewing’s sarcoma protein and the subunit of RNA polymerase II hsRPB7. Molecular Biology Reports, 36, 1269-1274. doi:10.1007/s11033-008-9308-2
- Erkizan, H.V., Kong, Y., Merchant, M., Schlottmann, S., Barber-Rotenberg, J.S., Yuan, L., Abaan, O.D., Chou, T.-H., Dakshanamurthy, S., Brown, M.L., Uren, A. and Toretsky, J.A. (2009) A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing’s sarcoma. Nature Medicine, 15, 750-757. doi:10.1038/nm.1983
NOTES
*Intrinsic disorder of EWS and its oncogenic fusions in rapport to function.

