Journal of Biomaterials and Nanobiotechnology
Vol.4 No.2A(2013), Article ID:29852,10 pages DOI:10.4236/jbnb.2013.42A005
Thiocyanate Oxidation by Coculture from a Coke Wastewater Treatment Plant
![]()
1The Key Lab of Pollution Control and Ecosystem Restoration in Industry Clusters, Ministry of Education, College of Environment and Energy, South China University of Technology, Guangzhou, China; 2Shenzhen Water (Group) Co. Ltd., Shenzhen, China; 3Jingtian Environmental Protection Technology Co. Ltd., Guangzhou, China; 4College of Bioscience and Bioengineering, South China University of Technology, Guangzhou, China.
Email: *cechwei@scut.edu.cn
Copyright © 2013 Huijing Huang et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received January 23rd, 2013; revised February 27th, 2013; accepted April 6th, 2013
Keywords: Thiocyanate Biodegradation; Carbon Source; Coke Wastewater; Biokinetics; Andrew-Haldane Model
ABSTRACT
Bacterial strains in an activated sludge aerobic reactor from a coke wastewater were found to be able to utilize thiocyanate as carbon source when the thiocyanate-containing wastewater was deprived of carbon source. This study showed that three thiocyanate-oxidizing bacterial strains, Burkholderia sp., Chryseobacterium sp., and Ralstonia sp. were isolated from the activated sludge of a coke wastewater treatment plant as evidenced by the fact that complete decomposition of thiocyanate was achieved either by coculture or individual pure culture. The thiocyanate biodegradation by the coculture occurred with an optimal pH range between 6.5 and 8.5 and an optimal temperature range between 30˚C and 40˚C. The biodegradation kinetics of thiocyanate was well fitted with the Andrew-Haldane model, which demonstrated a distinct substrate concentration-inhibited bacterial growth pattern. The effects of different types of additional carbon, nitrogen or sulfur sources on thiocyanate biodegradation were also investigated. Analysis of the end-products indicated that thiocyanate degradation by these strains should proceed via two pathways.
1. Introduction
Coke wastewater is a typical industrial wastewater primarily present in most of steel production facilities, as coke produced by heating coal in the absence of air is an indispensable material in the metallurgical industry. The liquid effluent of coking process contains considerable amounts of toxic compounds such as phenol, benzene, pyridine, quinoline, ammonium, thiocyanate and cyanide [1]. Current laboratory and field studies [2,3] have demonstrated that activated sludge systems are effective in removing these compounds particularly under aerobic conditions. In this manner, the degradation of phenols and other readily biodegradable substrates always proceeds much faster than thiocyanate degradation [4]. Nevertheless, the aerobic reactor has the capability of removing thiocyanate as evidenced from our field data (obtained from a real coke wastewater treatment plant located in Shaoguan, Guangdong, China). We observed distinct removal of thiocyanate when phenol concentrations were less than about 5 mg·L−1. Thus, the thiocyanate-oxidizing bacteria available in the activated sludge were believed to be responsible for thiocyanate biodegradation due to the consumption of readily biodegradable carbon sources by heterotrophic bacteria.
It is well documented that the aerobic microbial attacked on thiocyanate occurred as follows [5,6]. That is, thiocyanate was hydrolyzed to cyanate (OCN−) and sulfide (S2−), followed by further hydrolyzation of cyanate to ammonium (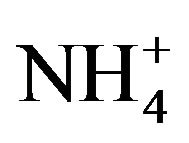 ) and oxidation of sulfide to sulfate (
) and oxidation of sulfide to sulfate (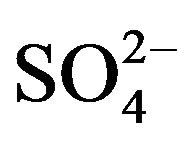 ). The overall process is summarized by the simplified reaction:
). The overall process is summarized by the simplified reaction:
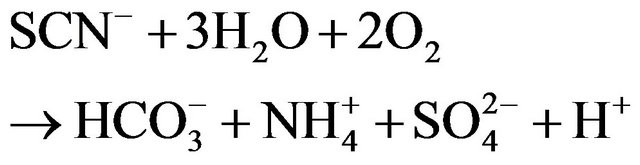 (1)
(1)
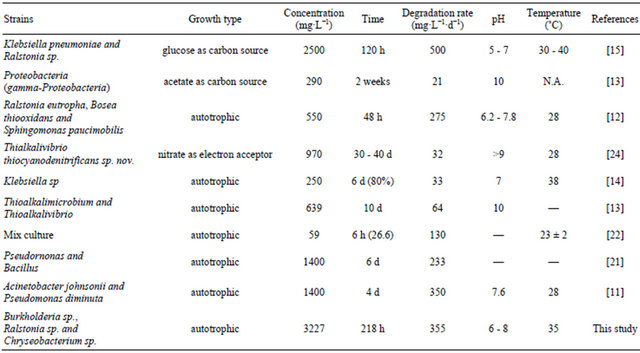
This unique ability makes thiocyanate-oxidizing strains attractive for applications in treating real industrial wastewater containing thiocyanate since this toxic compound is thought to be recalcitrant to biodegradation by many heterotrophs [7,8]. To date, several microorganisms able to grow using thiocyanate as sole carbon, nitrogen, and sulfur sources have been reported. These include Klebsiella [9] isolated from a gold mine soil, Thiobacillus [10] isolated from the activated sludge used for the treatment of wastewater from a coke-oven factory, Acinetobacter and Pseudomonas [11] isolated from a sludge suspension of an urban sewage treatment plant, Ralstonia eutropha, Bosea thiooxidans and Sphingomonas paucimobilis [12] isolated from a gold mine tailings reservoir, and Thioalkalimicrobium and Thioalkalivibrio [13] isolated from a soda solonchak soil.
In the present work, efforts were devoted to investigating the thiocyanate-oxidizing strains from the activated sludge of a coke wastewater treatment plant (Shaoguan, China) which has undergone long-term stable operation. Pure culture or coculture of three bacterial strains including Burkholderia sp., Chryseobacterium sp., and Ralstonia sp. were obtained to be able to discompose thiocyanate in the absence of external carbon sources. Among which, Burkholderia sp. and Chryseobacterium sp. were the newly reported strains capable of growing with thiocyanate as sole carbon source. The determination of optimal bacterial growth conditions and the estimation of biokinetic parameters such as bacterial growth rate and yield were performed. The biokinetics of the thiocyanate biodegradation process were studied by fitting the experimental data of the specific bacterial growth rates with the Andrew-Haldane model [14]. Moreover, the end-products of thiocyanate degradation were analyzed.
2. Materials and Methods
2.1. Enrichment and Isolation of Thiocyanate-Oxidizing Bacteria
The mineral base growth medium for enrichment and isolation of thiocyanate-oxidizing bacteria contained (per liter) 7.0 g of K2HPO4, 56 g of NaH2PO4, 4.13 g of MgCl2·6H2O, 0.5 g of FeCl3·6H2O, 1 g of CaCl2·2H2O and KSCN (its concentration was varied depending on the degradation tests) as sole carbon, nitrogen and sulfur sources. Two mL of a trace element solution consisting of 0.2 g·L−1 CuCl2, 0.5 g·L−1 KI, 2 g·L−1 MnCl2·H2O, 2 g·L−1 ZnCl2·7H2O, 2.5 g·L−1 HBO3, 1 g·L−1 Na2MoO4·2H2O was also added to the growth medium (1 L). The medium pH was adjusted by adding diluted NaOH or HCl solution. The Luria-Bertani (LB) culture medium was composed of 0.3% beef extract, 1% peptone, 0.5% NaCl with a controlled pH of 7.6.
Samples of the activated sludge were obtained from the aerobic tank of the Shaogang Coke Wastewater Treatment Plant. The sludge was starved of solute organic carbon after being strongly agitated for three days. Then 10 mL of suspension was inoculated in a 250 mL Erlenmeyer flask containing 100 mL mineral medium and 100 mg·L−1 SCN−. The flasks were placed on an orbital shaker at 32˚C, 200 rpm for bacterial inoculation. The enrichment of culture was carried out by stepwise increase in thiocyanate concentration of 100, 300, 500, 700 and 1000 mg·L−1 when thiocyanate was completely degraded. Samples of the acclimatized cultures were plated on LB medium solidified with 2.5% agar, in which individual colonies were purified and tested for aseptic growth in the mineral medium. The plates were incubated at 32˚C for 3 - 5 days. Three morphologically different bacterial isolates denoted as SCN-1, SCN-2 and SCN-3 were isolated and purified.
2.2. DNA Extraction, PCR Amplification and Sequencing
Isolates were then subjected to identification using 16S rRNA analysis followed by comparison with database sequences. To sequence the 16S rRNA gene of the isolates, polymerase chain reaction (PCR) amplification of 16S rRNA gene sequences was carried out using the primers F27 (5’-AGAGTTTGATCCTGGCTCAG-3’) and R1522 (5’-AAGGAGGTGATCCAGCCGCA-3’). The bacteria were placed in 20 μL sterilized deionized water and then boiled for 5 min to prepare the PCR template for 16S rRNA fragment sequencing. PCR amplification was conducted in an automated thermal cycler (PTC-200, MJ Research, USA) using the following protocol: initial denaturation for 4 min at 94˚C and 30 cycles of denaturetion for 1 min at 94˚C, annealing for 1 min at 55˚C, extension for 1.5 min at 72˚C, followed by a final extension for 7 min at 72˚C. PCR mixtures (50 μL) contained 41.2 μL of deionized H2O, 5.0 μL of 10 × PCR buffer, 1.0 μL of dNTPs (10 mM), 1.0 μL of primer F27 (10 μM), 1.0 μL of primer R1522 (10 μM), 0.3 μL of DNA template, and 0.5 μL of Taq DNA Polymerase (5 U(μL)−1). The 1.4 kb PCR fragments of each strain was sequenced at the Shanghai Sangon Biological Engineering Technology & Services Co., Ltd. and the sequences were logged in GenBank to compare with registered/submitted sequences.
The 16S rRNA sequences of the isolates were compiled and concatenated in FASTA format. The concatenated sequences were first aligned using CLUSTALX (version 1.83). Each output alignment was then converted to FASTA format using BIOEDIT. The alignment was then converted to MEGA format for phylogenetic analyses. Distance analyses were performed with the MEGA3 (version 3.0) package using the Neighbor-Joining (NJ) method with Kimura 2-model. The reliability of the phylogenetic analysis was subjected to a bootstrap test with 1000 replicates.
2.3. Effects of Different Operational Parameters on Thiocyanate Biodegradation
The coculture obtained was tested for thiocyanate biodegradation at different pH, temperature and initial thiocyanate concentrations. Thiocyanate degradation rate was defined as
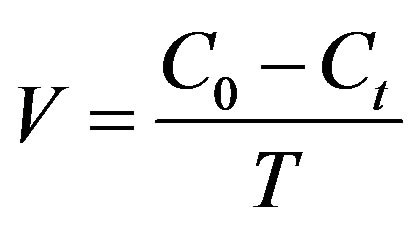 (2)
(2)
where V (mg·L−1·h−1) is thiocyanate degradation rate, C0, Ct (mg·L−1) are initial thiocyanate concentration and concentration at time T (h), respectively. To optimize the operational parameters, batch tests of thiocyanate degradation were performed by varying pH in the range of 3.5 - 9.5 and temperature in the range of 20˚C - 40˚C. The tolerance thiocyanate concentration and the inhibition of the bacterial growth were evaluated by changing its concentration from 50 - 3200 mg·L−1. The effect of additional carbon, nitrogen or sulfur nutrient on thiocyanate biodegradation efficiency was examined by supplementing different types of materials including carbon source like glucose, sucrose, starch and sodium acetate (150 mg·L−1 for each), nitrogen source like KNO3, NH4Cl, urea and peptone (150 mg·L−1 for each), or sulfur source like Na2SO4, Na2SO3, sulfur, Na2S, and Na2S2O3 (150 mg·L−1 for each) to the mineral medium, respectively. All the experiments were conducted in a 250 mL flask filled with the mineral medium and at shaking conditions (200 rpm).
2.4. The End-Products Analysis
Shake flask (150 ml medium in 250 ml flask) experiments were carried out for biodegradation of thiocyanate. During this process, ammonia and sulfate concentrations in the medium were measured for the identification of final products.
2.5. Analytical Methods
The supernatant (1 mL) was taken at different time intervals from each flask and filtered with a 0.45 µm filter paper. The residual thiocyanate concentration was analyzed colorimetrically as ferric thiocyanate with triplicates. Control experiments were conducted with an unionculated flask. The dry weight of bacterial cell was determined by centrifugation (5000 g for 15 min), washing (0.15 mol·L−1 phosphate buffers), and drying (105˚C) of the cell to constant weight. Ammonia concentrations were analyzed by Nessler reagent colorimetric methods. Sulfate concentrations were analyzed by using an ionchromatograph (Dionex ICS-2000, USA) with an Ionpac AG4A-SC/AS4A-SC column pair and eluted isocraticcally with 1.0 mM NaHCO3/3.5 mM Na2CO3 at 1 ml·min−1.
3. Results and Discussion
3.1. Isolation and Identification of Thiocyanate-Oxidizing Bacteria
Starting with the activated sludge from the aerobic tank of the coke wastewater treatment plant, the microbial population was acclimated to a mineral medium with thiocyanate as sole carbon, nitrogen and sulfur sources. After three-month selective enrichment, thiocyanate-degrading microorganisms were cultured by successive passages in the mineral medium. Figure 1 shows the decomposing of 500 mg·L−1 thiocyanate by the enrichment cultures at initial pH 7.0. After a lag period of 20 h for the microbial growth in relation to a slow rate of thiocyanate removal, concentrations of thiocyanate rapidly decreased over time during which concentrations of cell dramatically increased and eventually reached a relatively steady value. A complete disappearance of thiocyanate was detected within 90 h. These results clearly indicate the growth and proliferation of strains enriched from the active sludge since no other organic compounds as a carbon source were added to the liquid medium.
Similar to that observed in a previous report [13], the enrichment cultures were not able to form colonies on the agar medium containing thiocyanate. Instead, a LB agar medium was chosen to obtain microbial colonies from the enrichment cultures. Three morphologically distinctive colonies named SCN-1, SCN-2 and SCN-3 were isolated after 3-day incubation. The phylogenetic analysis of the three strains on the based of 16S rRNA sequences demonstrated that SCN-1, SCN-2 and SCN-3 belonged to the genus Burkholderia sp., Chryseobacterium sp. and Ralstonia sp. (with 99% 16S rRNA sequence similarity for each), respectively (Figure 2). The three strains were deposited in National Center for
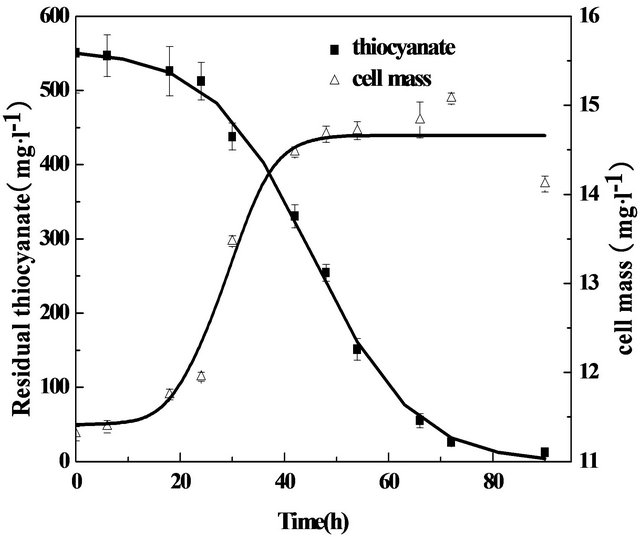
Figure 1. Growth curve of thiocyanate-degrading bacteria at 32˚C and pH 7.0 and under agitation (200 rpm).
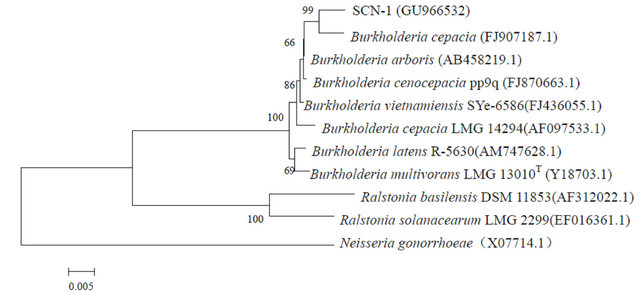
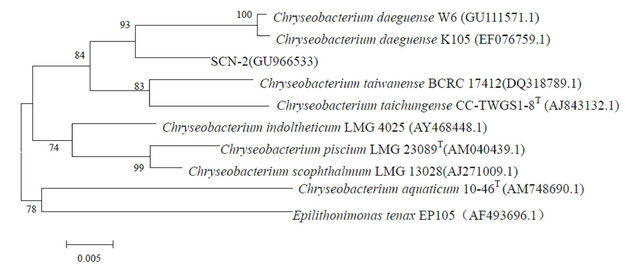
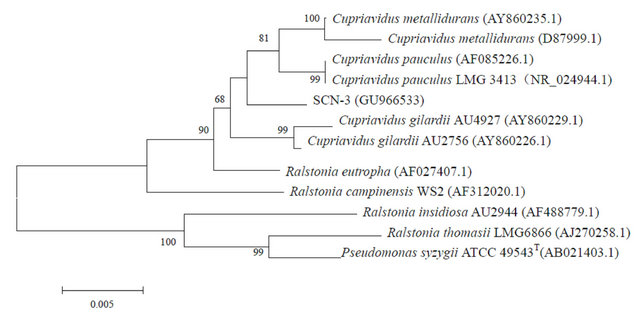
Figure 2. Neighbor-Joining trees of SCN-1, SCN-2 and SCN-3 and phylgenetically related bacteria constructed on the basis of their 16S rRNA sequences. GenBank accession numbers are given beside the strain numbers and bootstrap probability values (based on 1000 replications) are indicated the major branch points.
Bio-technology Information (NCBI), USA with the accession numbers of GU966532, GU966533 and GU- 966534, respectively.
The abilities of three pure strains for thiocyanate degradation were also examined. Plots of thiocyanate concentration decreased as a function of incubation time exhibited distinctly similar trends (Figure 3) for all the pure strains, indicating that each of them can work as an efficient thiocyanate-oxidizing strain. It was evident from Figure 3 that Burkholderia sp. has the relatively strongest ability to decompose thiocyanate, as the time required for the complete disappearance of thiocyanate by Burkholderia sp. (70 h) was shorter than that associated with Chryseobacterium sp. (120 h) or Ralstonia sp. (80 h). A previous study [15] showed that Ralstonia sp. in combination with Klebsiella pneumoniae were able to utilize
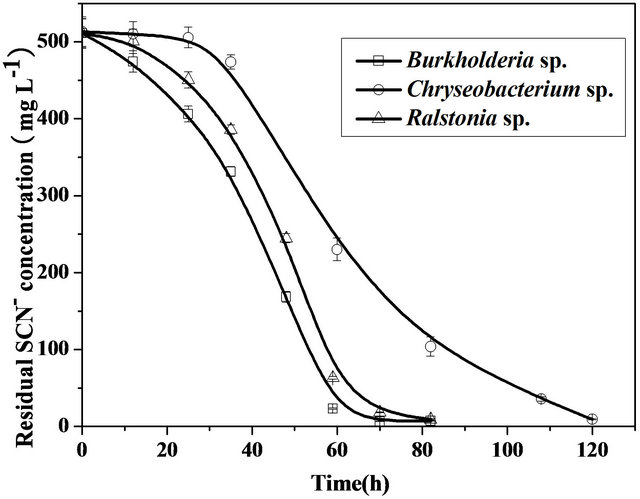
Figure 3. Comparison of thiocyanate biodegradation by Burkholderia sp., Chryseobacterium sp., and Ralstonia sp. in the mineral medium containing 500 mg·L−1 thiocyanate at 32˚C, pH 7.0 and under agitation (200 rpm).
thiocyanate as nitrogen source, but the individual contribution of this strain to the biodegradation of thiocyanate was not investigated, which indicated that they were cometabolic degradation. Burkholderia sp. were reported to be able utilize thiocyanate only when glucose were present. No reports concerning the biodegradation of thiocyanate as a carbon source of Chryseobacterium sp. were found in the literature.
3.2. Effects of Initial pH, Temperature and Initial Thiocyanate Concentration on Thiocyanate Degradation
Biodegradation of thiocyanate by strains has been reported to be sensitive to the initial pH of medium, because the variations in pH values may influence threedimensional shape of the enzymes in the microorganisms. Figure 4 shows the pH-dependent thiocyanate degradation rate in the presence of coculture isolated from the coke wastewater. The coculture was able to degrade thiocyanate effectively in the pH ranging from 6.5 to 8.5, a relatively wider pH range that it can tolerate as compared to many organisms reported elsewhere [9,16-18]. For example, the degradation of thiocyanate was inhibited even by a small change in pH (0.2 - 0.5 units) for Thiobacillus thioparus THI115 [10] and Acremonium strictum [19]. The fastest degradation was observed at pH 7.7 with a degradation rate of 7.5 mg·L−1·h−1. Degradation of thiocyanate was significantly decreased when pH was modulated below 6.0 or above 9.0. These observations imply that the coculture of Burkholderia sp., Chryseobacterium sp., Ralstonia sp. can be considered to be neutrophilic bacteria, similar to Klebsiella sp. [9], Thiobacillus sp. [17], and Pseudomonas putida [16] which were found to be well grown at an optimum pH ranging from 7.0 to 8.0.
Temperature of the growth medium is another important factor affecting thiocyanate degradation. Figure 5 illustrates that the coculture used in the present study was able to effectively degrade thiocyanate at temperature range between 30˚C - 40˚C. The highest degradation rate of 7.2 mg SCN− L−1·h−1 was obtained when the temperature was controlled at 35˚C. This result was in agreement with Klebsiella pneumonia and Ralstonia sp. which enabled thiocyanate removal potential at optimal temperature ranging from 30˚C to 40˚C [16]. In contrast, several researchers demonstrated that Klebsiella [9], Pseudomonas diminuta, Acinetobacter johnsonii [11] degraded thiocyanate efficiently at 25˚C to 30˚C.
To determine the effect of the initial thiocyanate concentration on its degradation rate, a series of batch experiments were conducted at optimized pH (7.7) and
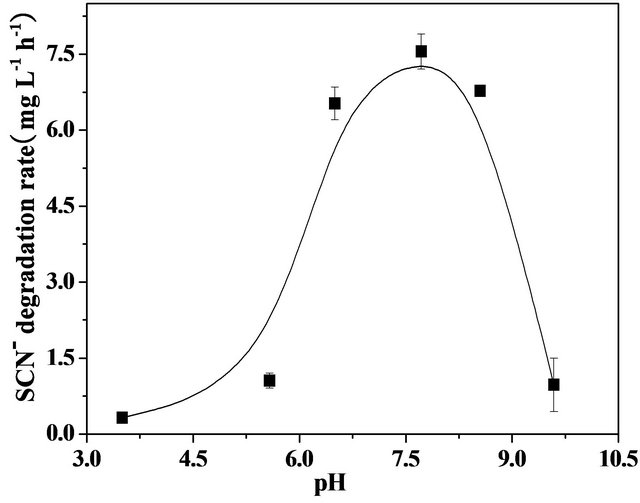
Figure 4. Effect of pH on thiocyanate biodegradation by the co-culture in the mineral medium containing 300 mg·L−1 thiocyanate under agitation (200 rpm).
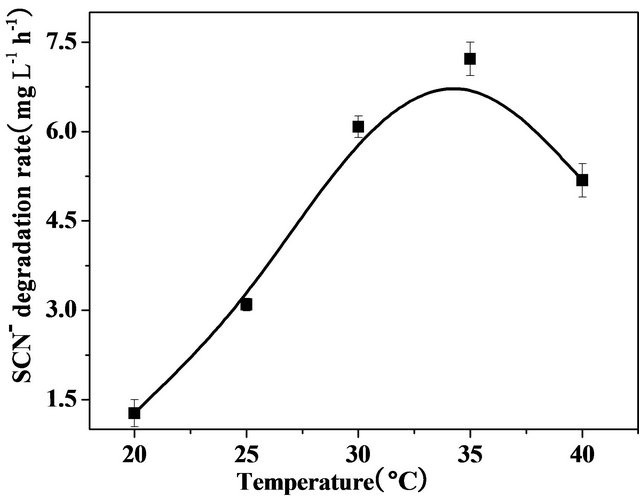
Figure 5. Effect of temperature on thiocyanate biodegradation by the coculture in the mineral medium containing 300 mg·L−1 thiocyanate under agitation (200 rpm).
temperature (35˚C) with the thiocyanate concentration varying in the range of 50 to 3200 mg·L−1. Figure 6 presents the removal of thiocyanate over incubation time with the various thiocyanate concentrations. Of particular note was the apparent zero-order degradation kinetics observed in all the samples. The apparent degradation rates were thus calculated from the zero-order kinetics law equation. Increasing the initial thiocyanate concentration from 250 to 3200 mg·L−1 resulted in an increase in the degradation rate. However, the plot did not exhibit a linear relationship between them, but indicated a substrate inhibition pattern. Similar observations of such an inhibition effect by thiocyanate were also reported in other microorganisms for thiocyanate degradation [14,15,20].
The coculture had maximum tolerance concentration of 3200 mg·L−1 at which the degradation rate of thiocyanate was 350 mg·L−1·d−1. Comparisons of these values with those associated with other bacteria were carried out as shown in Table 1. All these degradation tests were conducted under the batch conditions. It can be seen that the value of 3200 mg·L−1 represents the highest thiocyanate tolerance concentration among all the bacteria listed in Table 1. For example, it was more than one order of magnitude larger than the tolerance concentration (250 mg·L−1·d−1) in relation to Klebsiella sp. [14]. Moreover, the degradation rate obtained at the tolerance concentration in this study was found to be larger than those obtained in most of the reported bacteria [11-14,21-24] . The heterotrophic growth of Klebsiella pneumoniae and Ralstonia sp. [15] with the help of additional glucose utilizing thiocyanate exhibited a larger degradation rate than the growth of Burkholderia sp., Chryseobacterium sp., Ralstonia sp.
3.3. Biokinetics of Thiocyanate Degradation
Non-linear regression analysis was performed to estimate the biokinetic parameters of thiocyanate degradation using the Andrew-Haldane model [14] as described by the following equations.
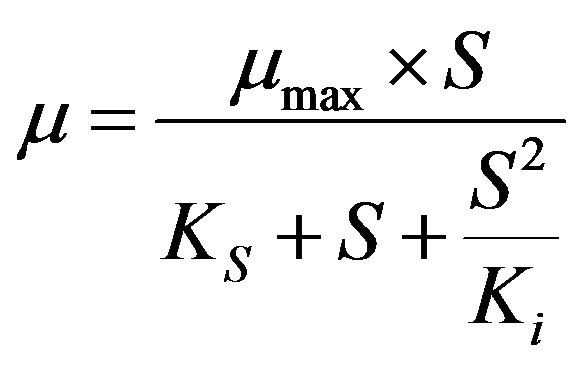 (3)
(3)
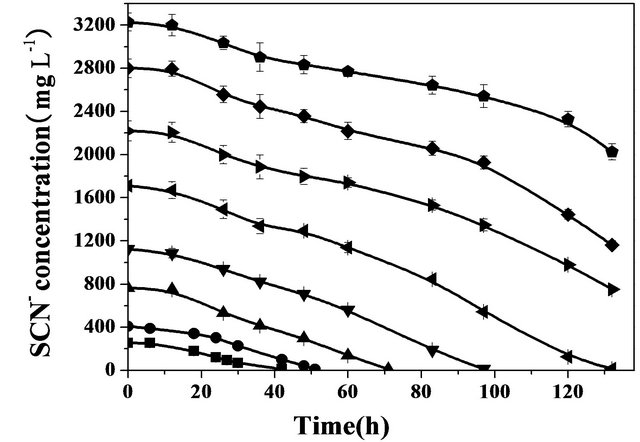
Figure 6. Effect of initial concentration of thiocyanate on thiocyanate biodegradation by the coculture at pH 7.7, 35˚C and under agitation (200 rpm).
Table 1. Comparisons of thiocyanate biodegradation by different bacterial strains with batch tests.
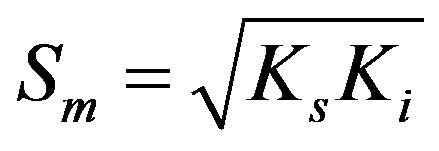 (4)
(4)
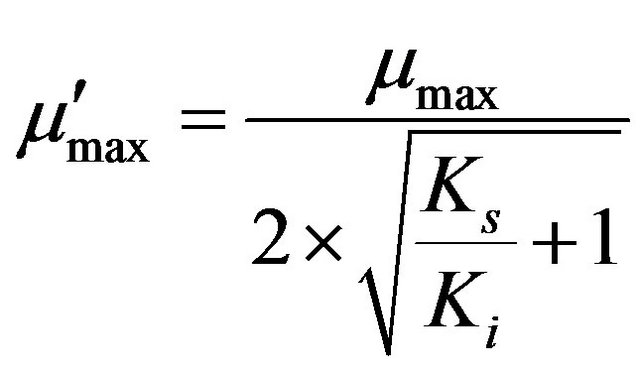 (5)
(5)
where µmax (d−1) is the maximum specific growth rate (the rate may approach µmax but will never reach it, d−1); S (mg·L−1) is the substrate concentration; Ks (mg·L−1) is the half saturation coefficient, numerically equal to the substrate concentration at which the specific growth rate is half of the maximum rate; Ki (mg·L−1) is the inhibition coefficient; Sm (mg·L−1) is the maximal substrate concentration resulting in the apparent maximum growth rate; and 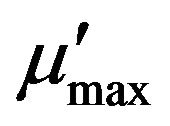 (d−1) is the apparent maximum specific growth rate if the substrate is inhibitory.
(d−1) is the apparent maximum specific growth rate if the substrate is inhibitory.
Figure 7 shows the plots of obtained and simulated specific growth rates against the initial thiocyanate concentration in the batch tests at pH 7.7 and 35˚C. The high determination efficient (0.91) approximate to 1 suggests this substrate-inhibition model is adequately fitted with the experimental data. The derived values of μmax, Ks, Ki, Sm and 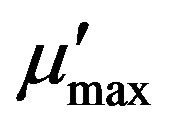 were 0.75 d−1, 332.75 mg SCN−·L−1, 881.05 mg SCN−·L−1, 541.45 mg SCN−·L−1 and 0.32 d−1, respectively.
were 0.75 d−1, 332.75 mg SCN−·L−1, 881.05 mg SCN−·L−1, 541.45 mg SCN−·L−1 and 0.32 d−1, respectively.
3.4. Growth Stimulants of Thiocyanate-Oxidizing Bacteria
Different external carbon, nitrogen and sulfur nutrients were added to the thiocyanate-containing medium in order to examine whether these additives can stimulate or inhibit thiocyanate biodegradation by the coculture. Table 2 compared the influences of various additives on biomass production and thiocyanate biodegradation. Clear visible was that a significant increase in biomass production was obtained when glucose or acetate was used an additional carbon source. In contrast, these two substrates caused a significant decrease in thiocyanate biodegradation rate, suggesting that the coculture prefers to utilize glucose or acetate rather than thiocyanate as the carbon source. The amendment of sucrose or starch gave
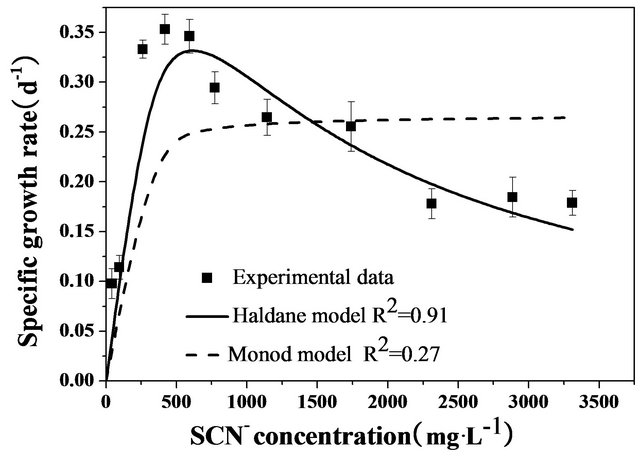
Figure 7. Evolution of specific growth rate of the coculture as a function of initial thiocyanate concentration in the batch tests at pH 7.7, 35˚C and under agitation (200 rpm).
Table 2. Effects of additional carbon, nitrogen or sulfur sources on thiocyanate biodegradation by the coculture.
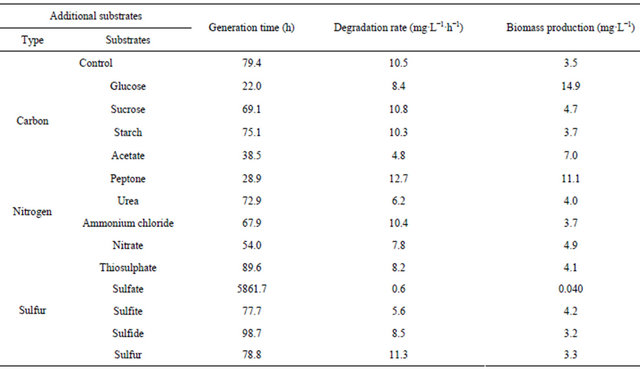
rise to a negligible effect on the growth of the coculture. The presence of urea, nitrate or ammonium chloride as a nitrogen source did not significantly improve the bacterial proliferation, but the former two sources caused a distinct decrease in the degradation rate. It was noted that the addition of peptone was effective in enhancing biomass production and thiocyanate biodegradation. The presence of thiosulphate, sulfate, sulfite or sulfide as a sulfur source also caused a decrease in the degradation rate. Especially when sulfate was used, the production of bacteria was almost completely prevented. The addition of sulfur to the growth medium was beneficial for thiocyanate biodegradation.
3.5. The End-Products of Thiocyanate Degradation
The products generated in the medium during thiocyanate degradation were analyzed. Thiocyanate depletion with concomitant accumulation of ammonium nitrogen and sulfate confirmed that the end oxidation compounds of thiocyanate degradation were ammonium nitrogen and sulfate. Equation (1) represents the thiocyanate transformation to various end-products.
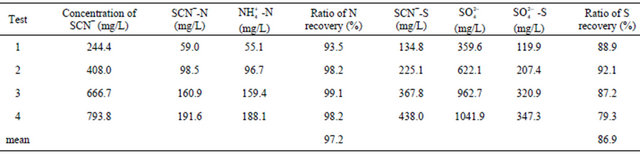
Table 3 shows the mass balance during thiocyanate degradation. The overall mass balance recoveries in conversion of nitrogen and sulfur of thiocyanate into ammonium and sulfate were 97.2% and 86.9%, respectively. Different from the final ratio of nitrogen recovery, the conversion of SCN−-N to 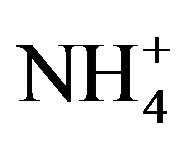 -N was about 90% when thiocyanate was completely degraded prior to the decay phase. As no other nitrogen compounds (i.e., nitrite and nitrate) was detected, the diminution of these ratios could be explained by bacterial cell synthesis. Continuous increase in
-N was about 90% when thiocyanate was completely degraded prior to the decay phase. As no other nitrogen compounds (i.e., nitrite and nitrate) was detected, the diminution of these ratios could be explained by bacterial cell synthesis. Continuous increase in 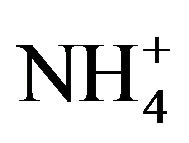 -N concentration after the complete degradation of thiocyanate was likely due to endogenous decay of the cell mass. Similar conversion ratios near 100% have been reported in previous studies using Klebsiella sp. [14], Acremonium strictum [19] and consortium of bacteria [11].
-N concentration after the complete degradation of thiocyanate was likely due to endogenous decay of the cell mass. Similar conversion ratios near 100% have been reported in previous studies using Klebsiella sp. [14], Acremonium strictum [19] and consortium of bacteria [11].
As illustrated above, the production of ammonium was in good balance with consumed thiocyanate; howeverthe sulfur balance was not achieved. The sulfur balance performed in Table 3 shows that the weight ratio of 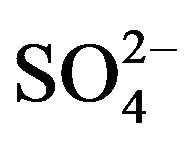 -S/SCN−-S is 0.8, compared with a theoretical value of 1.0 in reaction (1). After the complete degradation of thiocyanate, sulfate concentration increased slowly. The conversion ratio was relatively lower than previous works [11,14,21]. It is known that sulfur contributes a minor effect to bacterial cell synthesis because bacterial cell mass usually contains sulfur less than 1%. As a result, microbial sulfur uptake was indeed insignificant. Hung et al. [25] have reported that elemental sulfur (S0) was present during the aerobic degradation of thiocyanate by activated sludge-derived culture. Therefore, the yellow particles visible during incubation in these experiments were supposed to be elemental sulfur. The following equation may occur during thiocyanate degradation by the isolated bacteria:
-S/SCN−-S is 0.8, compared with a theoretical value of 1.0 in reaction (1). After the complete degradation of thiocyanate, sulfate concentration increased slowly. The conversion ratio was relatively lower than previous works [11,14,21]. It is known that sulfur contributes a minor effect to bacterial cell synthesis because bacterial cell mass usually contains sulfur less than 1%. As a result, microbial sulfur uptake was indeed insignificant. Hung et al. [25] have reported that elemental sulfur (S0) was present during the aerobic degradation of thiocyanate by activated sludge-derived culture. Therefore, the yellow particles visible during incubation in these experiments were supposed to be elemental sulfur. The following equation may occur during thiocyanate degradation by the isolated bacteria:
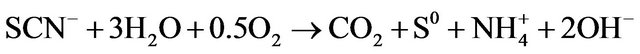 (6)
(6)
According to the previous study [25], elemental sulfur was thought to play an important role in energy reserve for sulfur-oxidation bacteria. When the supply of other sulfur species was depleted, additional energy could be obtained from oxidation of the sulfur deposits [26]:
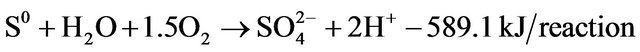 (7)
(7)
In the light of above discussion, elemental sulfur should be another sulfur product of thiocyanate degradation, which indicated that three strains may follow different kinds of degradation pathways.
4. Conclusion
Thiocyanate is an important component in coke wastewater, accounting for 15% of the total COD. Thiocyanate autotrophs were believed to be responsible for thiocyanate decomposition when the biodegradable carbon sources are consumed by heterotrophic bacteria. Here, we showed that three bacterial strains including Burkholderia sp., Chryseobacterium sp., and Ralstonia sp., isolated from a local coke wastewater treatment plant, can utilize thiocyanate as sole carbon source. At pH 7.7
Table 3. Recoveries of thiocyanate to sulphate and ammonia during different assays for increasing initial thiocyanate concentrations.
and 35˚C, the thiocyanate biodegradation proceeded with the highest rate, following the substrate inhibition pattern. This study may provide useful insights into the design and operation of a large-scale aerobic bioreactor using the bacteria obtained from this study for the treatment of thiocyanate in the coke wastewater.
5. Acknowledgements
The work was financially supported by the National Natural Science Foundation of China (No. 21037001), National Key Project for Basic Research of China (No. 2008BAC32B06-1), and National High Technology Research and Development Program of China (No. 2009- AA06Z319).
REFERENCES
- C. Wei, M. He and Y. Ren, “Pollution Characteristics of Coking Wastewater and Control Stragies: Biological Treatment Process and Technology,” Acta Scientiae Circumstantiae, Vol. 27, No. 7, 2007, pp. 1083-1093. (in Chinese)
- Y. S. Jeong and J. S. Chung, “Biodegradation of Thiocyanate in Biofilm Reactor Using Fluidized-Carriers,” Process Biochemistry, Vol. 41, No. 3, 2006, pp. 701-707. doi:10.1016/j.procbio.2005.09.004
- I. Vazquez, J. Rodriguez, E. Maranon, L. Castrillon and Y. Fernandez, “Study of the Aerobic Biodegradation of Coke Wastewater in a Two and Three-Step Activated Sludge Process,” Journal of Hazardous Materials, Vol. 137, No. 3, 2006, pp. 1681-1688. doi:10.1016/j.jhazmat.2006.05.007
- C. Staib and P. Lant, “Thiocyanate Degradation during Activated Sludge Treatment of Coke-Ovens Wastewater,” Biochemical Engineering Journal, Vol. 34, No. 2, 2007, pp. 122-130. doi:10.1016/j.bej.2006.11.029
- J. Stratford, A. E. X. O Dias and C. J. Knowles, “The Utilization of Thiocyanate as a Nitrogen Source by a Heterotrophic Bacterium: The Degradative Pathway Involves Formation of Ammonia and Tetrathionate,” Microbiology, Vol. 140, No. 10, 1994, pp. 2657-2662. doi:10.1099/00221287-140-10-2657
- N. V. Grigor’eva, T. F. Kondrat’eva, E. N. Krasil’nikova and G. I. Karavaiko, “Mechanism of Cyanide and Thiocyanate Decomposition by an Association of Pseudomonas putida and Pseudomonas stutzeti Strains,” Microbiology, Vol. 75, No. 3, 2006, pp. 266-273. doi:10.1134/S0026261706030040
- Y. M. Kim, D. Park, D. S. Lee and J. M. Park, “Inhibitory Effects of Toxic Compounds on Nitrification Process for Cokes Wastewater Treatment,” Journal of Hazardous Materials, Vol. 152, No. 3, 2008, pp. 915-921. doi:10.1016/j.jhazmat.2007.07.065
- X. Pan, Y. Li, H. Huang, Y. Ren and C. Wei, “Biodegradation of Thiocyanate and Inhibitory Interaction with Phenol, Ammonia in Coking Wastewater,” CIESC Journal, Vol. 60, No. 12, 2009, pp. 3089-3096. (in Chinese)
- C. Lee, J. Kim, J. Chang and S. Hwang, “Isolation and Identification of Thiocyanate Utilizing Chemolithotrophs from Gold Mine Soils,” Biodegradation, Vol. 14, No. 3, 2003, pp. 183-188. doi:10.1023/A:1024256932414
- S. J. Kim and Y. Katayama, “Effect of Growth Conditions on Thiocyanate Degradation and Emission of Carbonyl Sulfide by Thiobacillus thioparus THI115,” Water Research, Vol. 34, No. 11, 2000, pp. 2887-2894. doi:10.1016/S0043-1354(00)00046-4
- C. Boucabeille, A. Bories and P. Ollivier, “Degradation of Thiocyanate by a Bacterial Coculture,” Biotechnology Letters, Vol. 16, No. 4, 1994, pp. 425-430. doi:10.1007/BF00245064
- C. A. Du Plessis, P. Barnard, R. M. Muhlbauer and K. Naldrett, “Empirical Model for the Autotrophic Biodegradation of Thiocyanate in an Activated Sludge Reactor,” Letters in Applied Microbiology, Vol. 32, No. 2, 2001, pp. 103-107. doi:10.1046/j.1472-765x.2001.00859.x
- D. Y. Sorokin, T. P. Tourova, A. M. Lysenko and J. G. Kuenen, “Microbial Thiocyanate Utilization under Highly Alkaline Conditions,” Applied and Environmental Microbiology, Vol. 67, No. 2, 2001, pp. 528-538. doi:10.1128/AEM.67.2.528-538.2001
- J. H. Ahn, J. Kim, J. Lim and S. H. Hwang, “Biokinetic Evaluation and Modeling of Continuous Thiocyanate Biodegradation by Klebsiella sp.,” Biotechnology Progress, Vol. 20, No. 4, 2004, pp. 1069-1075. doi:10.1021/bp049967n
- A. U. Chaudhari and K. M. Kodam, “Biodegradation of Thiocyanate Using Co-Culture of Klebsiella pneumoniae and Ralstonia sp.,” Applied Microbiology and Biotechnology, Vol. 85, No. 4, 2010, pp. 1167-1174. doi:10.1007/s00253-009-2299-7
- K. D. Chapatwala, G. Babu, O. K. Vijaya, K. P. Kumar, and J. H. Wolfram, “Biodegradation of Cyanides, Cyanates and Thiocyanates to Ammonia and Carbon Dioxide by Immobilized Cells of Pseudomonas putida,” Journal of Industrial Microbiology and Biotechnology, Vol. 20, No. 1, 1998, pp. 28-33. doi:10.1038/sj.jim.2900469
- M. B. Stott, P. D. Franzmann, L. R. Zappia, H. R. Watling, L. P. Quan and B. J. Clark, et al., “Thiocyanate Removal from Saline CIP Process Water by a Rotating Biological Contactor, with Reuse of the Water for Bioleaching,” Hydrometallurgy, Vol. 62, No. 2, 2001, pp. 93-105. doi:10.1016/S0304-386X(01)00185-2
- N. V. Grigor’eva, Y. V. Smirnova and L. E. Dulov, “Thiocyanate Decomposition under Aerobic and OxygenFree Conditions by the Aboriginal Bacterial Community Isolated from the Wastewater of a Metallurgical Works,” Microbiology, Vol. 78, No. 4, 2009, pp. 402-406. doi:10.1134/S002626170904002X
- H. K. Kwon, S. H. Woo and J. M. Park, “Thiocyanate Degradation by Acremonium strictum and Inhibition by Secondary Toxicants,” Biotechnology Letters, Vol. 24, No. 16, 2002, pp. 1347-1351. doi:10.1023/A:1019825404825
- Y. S. Jeong and J. S. Chung, “Simultaneous Removal of COD, Thiocyanate, Cyanide and Nitrogen from Coal Process Wastewater Using Fluidized Biofilm Process,” Process Biochemistry, Vol. 41, No. 5, 2006, pp. 1141- 1147. doi:10.1016/j.procbio.2005.12.010
- Y. L. Paruchuri, N. Shivaraman and P. Kumaran, “Microbial Transformation of Thiocyanate,” Environmental Pollution, Vol. 68, No. 1-2, 1990, pp. 15-28. doi:10.1016/0269-7491(90)90011-Z
- C. H. Hung and S. G. Pavlostathis, “Kinetics and Modeling of Autotrophic Thiocyanate Biodegradation,” Biotechnology and Bioengineering, Vol. 62, No. 1, 1999, pp. 1-11. doi:10.1002/(SICI)1097-0290(19990105)62:1<1::AID-BIT1>3.0.CO;2-Q
- D. Y. Sorokin, T. P. Tourova, A. M. Lysenko, L. L. Mityushina and J. G. Kuenen, “Thioalkalivibrio thiocyanoxidans sp. nov. and Thioalkalivibrio paradoxus sp. nov., Novel Alkaliphilic, Obligately Autotrophic, Sulfur-Oxidizing Bacteria Capable of Growth on Thiocyanate, from Soda Lakes,” International Journal of Systematic and Evolutionary Microbiology, Vol. 52, 2002, pp. 657-664.
- D. Y. Sorokin, T. P. Tourova, A. N. Antipov, G. Muyzer and J. G. Kuenen, “Anaerobic Growth of the Haloalkaliphilic Denitrifying Sulfur-oxidizing Bacterium Thialkalivibrio thiocyanodenitrificans sp. nov. with Thiocyanate,” Microbiology, Vol. 150, No. 7, 2004, p. 2435. doi:10.1099/mic.0.27015-0
- C. H. Hung and S. G. Pavlostathis, “Aerobic Biodegradation of Thiocyanate,” Water Research, Vol. 31, No. 11, 1997, pp. 2761-2770. doi:10.1016/S0043-1354(97)00141-3
- T. D. Brock, M. T. Madigan, J. M. Martinko and J. Parker, “Biology of Microorgannism,” 7th Edition, Prentice Hall, Englewood Cliffs, 1994.
NOTES
*Corresponding author.

