Agricultural Sciences
Vol.5 No.5(2014), Article ID:44635,10 pages DOI:10.4236/as.2014.55044
Low-Cost Strategies for Development of Molecular Markers Linked to Agronomic Traits in Prunus
Juan A. Salazar1*, Mousa Rasouli1,2*, Reza Fatahi Moghaddam3, Zabihollah Zamani3, Ali Imani4, Pedro Martínez-Gómez1#
1Department of Plant Breeding, CEBAS-CSIC, Murcia, Spain
2Department of Landscape Engineering, Faculty of Agriculture, Malayer University, Malayer, Iran
3Horticulture Science Department, Faculty of Agriculture, University of Tehran, Karaj, Iran
4Department of Horticulture, Seed and Plant Improvement Institute, Karaj, Iran
Email: #pmartinez@cebas.csic.es
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 13 January 2014; revised 21 March 2014; accepted 4 April 2014
ABSTRACT
Evaluation of agronomic traits in Prunus breeding programs is a tedious process because of the long juvenile period of trees, the influence of juvenility and the existence of climatic factors affecting the expression of the trait. For these reasons, marker-assisted selection (MAS) strategies are particularly useful in these cases. The objective of this work is the analysis of alternative lowcost strategies for development of molecular markers linked to agronomic traits in Prunus including the application of modified Bulked segregant analysis (BSA) using Simple sequence repeat (SSRs) markers and the application of Random amplified polymorphism microsatellite (RAMP) markers. First BSA results showed that two SSR loci were found to be tightly linked to flowering time in almond. On the other hand, RAMP analysis has been demonstrated to be a potentially valuable molecular marker for the study of genetic relationships in Prunus. Results showed the dominant nature of these markers with a great abundance and transferability although with a reduced polymorphism. In addition, RAMP application in F1 progenies showed its suitability for molecular characterization and mapping, and later Quantitative trait loci (QTL) or BSA analysis.
Keywords:Prunus, Breeding, Flowering, Molecular Markers, SSR, RAMP, BSA, Marker-Assisted Selection

1. Introduction
The genus Prunus (Rosaceae family) include more than 200 species widely grown around the world [1] [2] . The annual worldwide production of the most important Prunus species cultivated were around 41 million metric tons in 2011, including 21.52 million tons of peach and nectarine fruits [P. persica (L.) Batsch]; 11.35 million tons of prune (P. domestica L.), plum (P. salicina Lindl), sloe (P. spinosa L.), and cherry plum fruits (P. cerasifera Ehrh.); 3.84 million tons of apricot fruits (P. armeniaca L.); 2.24 million tons of sweet (P. avium L.), sour (P. cerasus L.) and ground (P. fruticosa Pall.) cherry fruits; and 2.01 million tons of almond kernels [P. amygdalus (Batsch) syn. P. dulcis (Miller) Webb] (http://faostat.fao.org). In Prunus breeding programs, evaluation of agronomic traits in Prunus species is a tedious process because of the long juvenile period of trees, the influence of the juvenility on the expression of the trait, and the existence of climatic factors affecting this evaluation. For these reasons, marker-assisted selection (MAS) is particularly useful in these cases [3] [4] .
Studies of development of molecular markers linked to agronomic traits in Prunus were initially performed in almond using isoenzyme analysis. Nevertheless, this first approach was very limited because of the reduced polymorphism and the low variation [3] . Restriction Fragment Length Polymorphisms (RFLPs) provided a more efficient method because of its codominant nature and unlimited number of markers. However, the application of these markers has been limited due to their complexity and time-consuming. More recently, the utilization of PCR-based markers less laborious and time consuming has increased the possibilities of mapping. Random Amplified Polymorphic DNAs (RAPDs) were the first PCR marker assayed although its dominant nature and low repeatability limited drastically its utilization [3] . For these reasons, simple sequence repeat sequences (SSRs) are being become the markers of choice for molecular characterization and mapping in Prunus because of their high polymorphism, abundance, codominant inheritance and transportability across Prunus species [5] [6] .
To compensate for the weakness of these two approaches (SSR and RAPD); limited repeatability of RAPDs and cost of SSRs, a new low-cost marker called Random amplified polymorphism microsatellite (RAMP) was developed [7] . RAMP markers involved a SSR primer which is used to amplify genomic DNA in the presence or absence of RAPD primers (Figure 1). The resulting products can be resolved using submarine agarose electrophoresis. The amplification products derived from the anchored primer are only detected. Advantages of RAMP include high polymorphism, widely distributed throughout the genome, with an easily and low cost application. However, mixture interpretation is more difficult and repeatability could be reduced [7] [8] . RAMP has been successfully employed in diversity studies in different fruit species including peach [9] and pomegranate (Punica granatum L.) [10] . This new low-cost marker has been adopted in recent research for evaluation of DNA polymorphisms at different levels [11] .
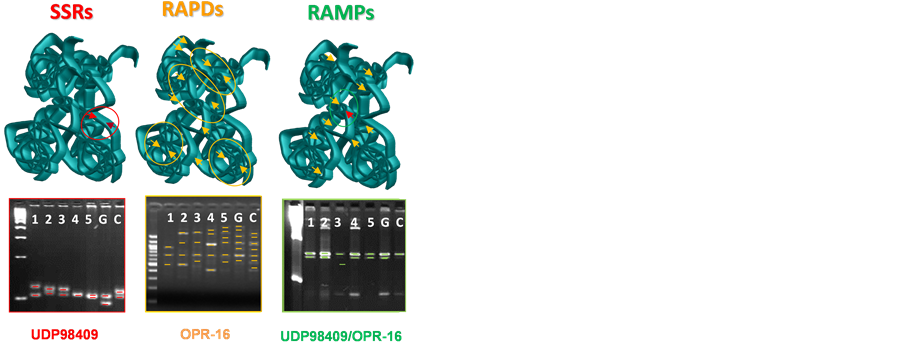
Figure 1. Schematic representation of Simple sequence repeat (SSR), Random amplified polymorphic DNA (RAPD) and Random amplified polymorphism microsatellite (RAMP) markers and application of UDP98409 SSR, OPR-16 RAPD and UDP98409/ OPR-16 RAMP application in the F1 apricot progeny “Goldrich” (G) × “Currot”.
The main approach for the development of molecular markers for MAS strategies in Prunus continue being the use of segregating progenies and the identification of quantitative trait loci (QTLs) linked to these traits [3] [4] [12] . However, bulk segregant analysis (BSA), where two pooled DNA samples are formed from plant sources that have similar genetic backgrounds but differ in one particular trait, is another powerful approach for the analysis of molecular marker-horticultural trait association [13] with lower costs in comparison with QTL analysis [12] . A strategy combining different markers with bulk segregant analysis was used to identify markers linked to loci of specific fruit characters in peach × almond crosses [14] . In addition, Ballester et al. [15] using this methodology identified RAPD markers associated with self-incompatibility and flowering time in almond.
The objective of this work was the development of alternative low-cost strategies for molecular markers development in Prunus by the application of modified bulked segregant analysis using SSR markers and the application of RAMP markers for the molecular characterization of several almond and apricot F1 progenies to evaluate their use for molecular characterization and mapping.
2. Material and Methods
2.1. Plant Material
The plant material assayed included a F1 almond progeny of seventy eight seedlings from the cross between the intermediate flowering Italian cultivar “Tuono” and the extra-late flowering Iranian cultivar “Shahrood-12” (T × S). In addition, to check the transferability of the molecular markers assayed across Prunus genus, a F1 apricot progeny of one hundred seedlings from the cross between the North American cultivar “Goldrich” and the Spanish “Currot” (G × C) was assayed.
2.2. Flowering Time Evaluation and Bulk Segregant Analysis in Almond
During two consecutive years (2008 and 2009) flowering time was evaluated in the progenitors and the F1 almond population T × S every 2 days and expressed in as extra-early, early, late, and extralate. In this population, four bulks (extra-early, early, late and extra-late) consisting in a DNA pool from several descendants selected from the almond progeny were selected for the future study using SSR markers (Figure 2).
2.3. Molecular Characterization of the Almond Progeny Using SSRs
Total genomic DNA was isolated using the procedure described by Doyle and Doyle [16] . Extracted almond genomic DNA was PCR-amplified using 71 pairs of primers flanking microsatellite sequences previously cloned and sequenced in different Prunus species including almond, peach, cherry and apricot (Table 1). Microsatellite amplifications and evaluation were performed as described in Sánchez-Pérez et al. [17] . Amplified PCR products were separated using Metaphor® agarose (Cambrex, East Rutherford, NJ, USA) and stained with GelRedTM (Biotium, Hatwad, CA, USA). These SSR markers were well distributed across the Prunus genome. Fifty six of this SSR makers were previously used in the first linked map of the population “R1000” × “Desmayo Largueta” [18] completing this map with the assay of 15 new SSRs according to previous information of Dondini et al. [19] and Sánchez-Pérez et al. [20] (Figure 3).
2.4. Molecular Characterization of Almond and Apricot Progenies Using RAMPs
Extracted DNA was PCR-amplified using a combination of two pair of primers (forward and reverse) flanking nuclear SSR sequences cloned in peach (UDP96003) and apricot (UDAp473) and 2 selected RAPD primers (OPA8, OPB11 and OPR-16) purchased from Operon Technologies (Huntsville, USA). PCR reactions were performed according to the protocol optimized by Sánchez-Pérez et al. [16] to SSR markers assaying different annealing temperatures (from 35˚C to 52˚C). Amplified PCR products were separated using regular LD-2® agarose (Conda, Madrid, Spain) and stained with GelRedTM (Biotium, Hatwad, CA, USA).
3. Results and Discussion
3.1. Flowering Time Evaluation in Almond
Figure 2 showed the distribution of the seventy almond seedlings of the F1 progeny “Tuono” × “Shahrood” for
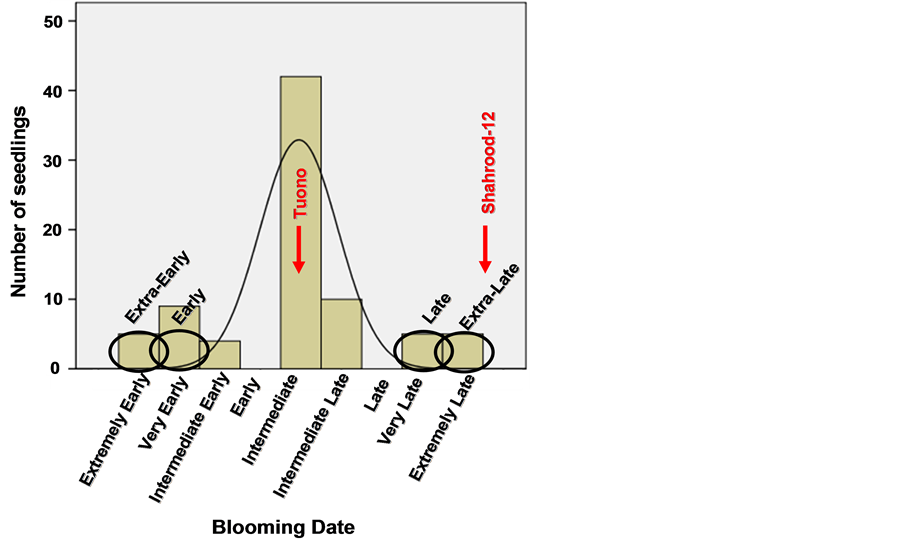
Figure 2. Flowering date evaluation in the almond F1 progeny of 78 seedlings from the cross between the “Tuono” and “Shahrood-12” during the years 2008 and 2009 and selection of DNA pools (extraearly, early, late and extra-late) from different seedlings.
Table 1. Origin of simple microsatellite (SSR) markers asayed.

Figure 3. Location in the almond linkage map of the seventy one SSR markers assayed in the molecular characterization of the almond and apricot plant material assayed.
flowering time according to the mean values of the two years.
Results showed a quantitative inheritance of flowering date in the almond progeny studied. The seedlings evaluated showed a wide range of flowering dates and in mots of cases a range between both progenitors. However, some of these descendants were earlier than the early progenitor “Tuono”. This can show the effect of genetic background on the expression of this trait.
Flowering time showed a quantitative inheritance in the family assayed with a clear modal distribution in agreement with previous results in other almond progenies [32] -[36] . However, our results are not in accordance with previous results studying some descendants of “Tardy Nonpareil” which a bimodal distribution was observed for this trait. This bimodal distribution was explained by the presence of a late flowering major gene (Lb), quantitatively modified by other minor genes [32] [33] . In our progeny where the late flowering donor is a Iranian almond cultivars it is not present this Lb gene.
Flowering date was considered an agronomical trait with a high heritability [36] [37] . In this sense, Dicenta et al. [38] established that the best strategy to obtain late-flowering descendants is to cross progenitors as lateflowering as possible. When the offspring showed a bimodal distribution, we must select the latest-flowering, probably carrying the late-flowering allele (in the case of descendants from “Tardy Nonpareil”) which could be transmitted to its descendants.
3.2. Bulked Segregant Analysis for Flowering Time in Almond Using SSR Markers
In the analysis of co-segregation of SSR markers and the T × S almond population, two microsatellite loci (CPPCT008 and EPDCU2584) were found to be tightly linked to this important agronomic trait (Figure 4).
Results corroborated the suitability of the use of SSR markers for the assessment of molecular genetic variability in almond and the high degree of transportability between peach SSR in almond previously reported by Martínez-Gómez et al. [5] . SSR detection using Metaphor® agarose gel electrophoresis was an efficient and would be able to resolve most of allelic variation. In this sense, we can indicate that the use of MetaPhor® agarose and Gel Red Nucleic Acid Gel Sating® appears good indicated for molecular characterization of mapping of population, due to its good resolution in comparison with the rest of agaroses, less toxicity in comparison with the use of ethidium bromide, and lower cost and easier routine application in comparison with the automatic capillary sequencing.
Modified Bulk Segregant Analysis
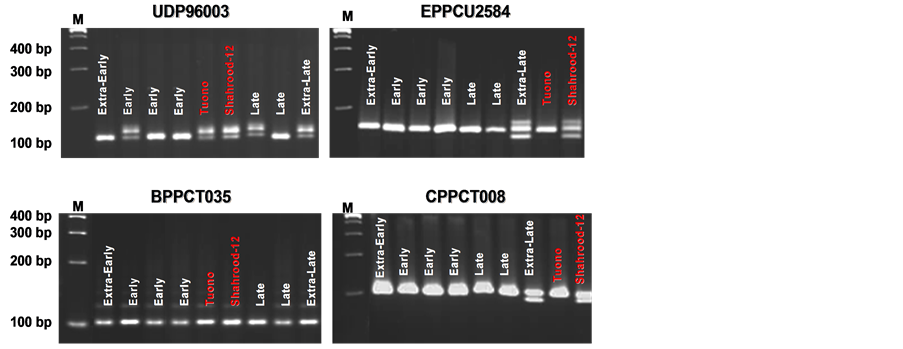
F1.almond (‘Tuono’ x ‘Shahrood-12’) population.
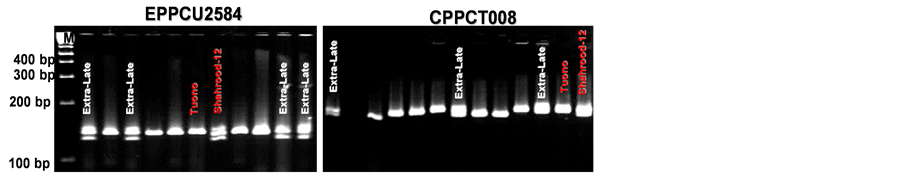
Figure 4. Metaphor® agarose gels showing de polymorphism observed in the application of SSR markers UDP96003, EPPCU2584, BPPCT035 and CPPCT008 to the modified bulk segregant analayis of the F1 almond population “Tuono” × “Shahrood-12”.
Results also indicated the suitability of SSR markers for the analysis of QTL linked to flowering time and chill and heat requirements as has been described before in almond [18] [39] and other related species such as apricot [40] [41] , rose [42] and peach [43] and the future application in marker assisted selection as has been described in the case of bitterness in almond [20] .
Other opportunities of this study derive from the recent sequencing of the complete genome of the peach. The International Peach Genome Initiative (IPGA) has recently released the complete peach genome sequence which is available on http://www.rosaceae.org/peach/genome [44] . This complete peach genome sequence will be of great interest in the future molecular studies in Prunus species such as apricot. In this sense, due to the easy location of SSR markers in this genome sequence we can found the position of our SSR locus linked to flowering time focusing our future studies in the nearest DNA regions in linkage groups 5 and 6.
The construction of linkage maps and QTL analysis takes a considerable amount of time and effort and may be very expensive. Therefore, alternative methods that can save time and money would be very useful, especially if resources are limited. One of these methods is the BSA where only bulks of selected genotypes are analyzed [13] . The disadvantages of this method is that they are not efficient in determining the effects of QTL and that only one trait can be tested at a time since the individuals selected for extreme phenotypic values will usually nor represent extreme phenotypic values for other traits.
These results can be considered as an initial point to search inside the genome for the flowering time expression and hold promise for speeding up the fine mapping and identification of region responsible for the variation of the trait. Fine mapping consists of saturating the identified genome region near the SSR identified as linked to the tait assayed. SNP markers could be the most suitable markers for increasing the resolution of the initial maps developed with SSRs or for increasing the resolution of determined regions of the map [11] using the same populations. SNPs are the most abundant molecular markers (estimated at more than 1 per 1000 bp) and are widely distributed throughout the genome (although their occurrence and distribution varies among species). In addition, extremely degraded DNA samples can be used, and multiplexing hundreds of markers in a chip is possible in several Prunus species, including peach [45] and cherry [46] . In addition, the markers identified in are seldom suitable for marker assisted selection without further testing, validation and additional development. Generally, the steps required for validation include testing the markers in correctly phenotyped germplasm.
3.3. RAMP Application in Almond and Apricot
Regarding the optimization of the application of RAMP markers for the molecular characterization of almond and apricot (Figure 5) genotypes, annealing temperature is the critical step as has been previously described in peach [9] and pomegranate [10] . Annealing temperature of the anchored primers were usually 10˚C - 15˚C higher than those of the RAPD primers; thus, at higher annealing temperature, only the anchored primers would anneal efficiently, whereas in PCR cycles at low annealing temperatures, both anchored SSR and RAPD primers would anneal. The optimization of the annealing conditions represents the main limitation in the application of this type of DN markers [11] .
The PCR program could be modified such that there is switching between high and low annealing temperatures during reaction. Most fragments obtained with RAMP primers alone disappear when RAPD primer are included, and different patterns are obtained with the same RAMP primer and different RAPDs, indicating that RAPD primer compete with RAMP primer during the low annealing temperature cycle.
Regarding the application of RAMP markers in almond and apricot (Figure 5) progenies, results showed the dominant nature of these markers although with a reduced polymorphism. In addition, the great number of combinations of markers makes these markers very abundant.
The easy application and cheap analysis in regular agarose represent the most prominent advantage when compared with others DNA markers as RFLPs or SSRs.
Finally, these results also confirm the transferability of these RAMP markers across the genus Prunus and
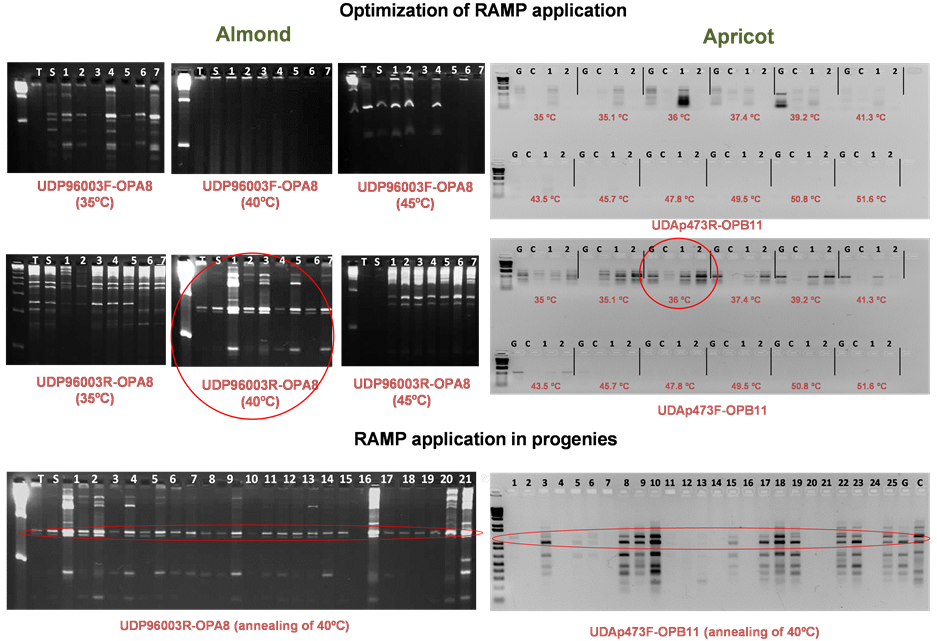
Figure 5. RAMP optimization and application: A) in the F1 almond progeny “Tuono” (T) × “Shahrood-12” (S) using SSR primers [forward (F) and reverse (R)] from UDP96003 marker and RAPD primer OPA8, and assaying different annealing temperatures (from 35˚C to 45˚C), and B) in the F1 apricot progeny “Goldrich” (G) × “Currot” (C) using SSR primers [forward (F) and reverse (R)] from UDAp473 marker and RAPD primer OPB11, and assaying different annealing temperatures (from 35˚C to 51.6˚C).
their suitability for molecular characterization of mapping of population later QTL or BSA analysis.
Acknowledgements
This study has been partially supported by the following projects from the Spanish Ministry of Economy and Competiveness: “Almond breeding” (AGL2010-22197-C02-02), “Apricot breeding” (AGL2010-21903) and “Gene expression analysis of the resistance to Plum pox virus, PPV (Sharka) in apricot by transcriptome deep-sequencing (RNA-Seq)” (AGL2010-16335).
References
- Dönmez, A.A. and Yildirimli, S. (2000) Taxonomy of Genus Prunus L. (Rosaceae) in Turkey. Turkish Journal of Botany, 14, 187-202.
- Potter, D. (2012) Basic Information on the Stone Fruit Crops. In: Kole, C. and Abbott, A.G., Eds., Genetics, Genomics and Breeding of Stone Fruits, CRC Press, New York, 1-21. http://dx.doi.org/10.1201/b13104-2
- Martínez-Gómez, P., Sozzi, G.O., Sánchez-Pérez, R., Rubio, M. and Gradziel, T.M. (2003) New Approach to Prunus Tree Crop Breeding. Journal of Food Agriculture and Environment, 1, 52-63.
- Martínez-Gómez, P., Sánchez-Pérez, R. and Rubio, M. (2012) Clarifying Omics Concepts, Challenges and Opportunities for Prunus Breeding in the Post-Genomic Era. OMICS: Journal of Integrative Biology, 16, 268-283. http://dx.doi.org/10.1089/omi.2011.0133
- Martínez-Gómez, P., Arulsekar, S., Potter, D. and Gradziel, T.M. (2003) An Extended Inter-Specific Gene Pool Available to Peach and Almond Breeding as Characterized Using Simple Sequence Repeat (SSR) Markers. Euphytica, 131, 313-322. http://dx.doi.org/10.1023/A:1024028518263
- Campoy, J.A., Martínez-Gómez, P., Ruiz, D., Rees, J. and Celton, J.M. (2010) Developing Microsatellite Multiplex and Megaplex PCR Systems for High Throughput Characterization of Breeding Progenies and Linkage Maps Spanning the Apricot Genome. Plant Molecular Biology Reporter, 28, 560-568. http://dx.doi.org/10.1007/s11105-010-0186-0
- Wu, K.S., Danneberger, L. and Scolnik, P. (1994) Detection of Microsatellite Polymorphisms without Cloning. Nucleic Acid Research, 22, 3257-3258. http://dx.doi.org/10.1093/nar/22.15.3257
- Provan, J., Thomas, W.T.B., Forster, B.P. and Powel, W. (1999) Copia-SSR: A Simple Marker Technique Which Can Be Used on Total Genomic DNA. Genome, 42, 363-366. http://dx.doi.org/10.1139/g98-121
- Cheng, H.Y., Yang, W.C. and Hsiao, Y. (2001) Genetic Diversity and Relationship among Peach Cultivars Based on Random Amplified Microsatellite Polymorphism (RAMP). Botanical Bulletin of the Academia Sinica, 42, 201-206.
- Lihua, Z., Mingyang, L., Guangze, C., Tianchun, P. and Chenghai, S. (2013) Assessment of the Gentic Diversity and Genetic Relationships of Pomegranate (Punica granatum L.) in China Using RAMP Markers. Scientia Horticulturae, 151, 63-67. http://dx.doi.org/10.1016/j.scienta.2012.12.015
- Huang, H.L., Huang, I.Y., Lin, C.Y. and Huang, M.C. (2013) Effective Strategies for Identifying Novel Genetic Markers Based on DNA Polymorphisms. Journal of Molecular Biomarkers & Diagnosis, 5, 1. http://dx.doi.org/10.4172/2155-9929.1000156
- Boopathi, N.M. (2013) Genetic Mapping and Marker Assisted Selection: Basics, Practice and Benefits. Springer, New York, 293 p. http://dx.doi.org/10.1007/978-81-322-0958-4
- Michelmore, R.W., Paran, I. and Kesseli, R.V. (1991) Identification of Markers Linked to Disease-Resistance Genes by Bulked Segregant Analysis: A Rapid Method to Detect Markers in Specific Genomic Regions by Using Segregating Populations. Proceedings of the National Academy of Science USA, 88, 9828-9832. http://dx.doi.org/10.1073/pnas.88.21.9828
- Warburton, M.L., Becerra-Velasquez, V.L., Goffreda, J.C. and Bliss, F.A. (1996) Utility of RAPD Markers in Identifying Genetic Linkages to Genes of Economic Interest in Peach. Theoretical and Applied Genetics, 93, 920-925. http://dx.doi.org/10.1007/BF00224094
- Ballester, J., Socias i Company, R., Arús, P. and de Vicente, M.C. (2001) Genetic Mapping of a Major Gene Delaying Blooming Time in Almond. Plant Breeding, 120, 268-270. http://dx.doi.org/10.1046/j.1439-0523.2001.00604.x
- Doyle, J.J. and Doyle, J.L. (1987) A Rapid Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochemistry Bulletin, 19, 11-15.
- Sánchez-Pérez, R., Ballester, J., Dicenta, F., Arús, P. and Martínez-Gómez, P. (2006) Comparison of SSR Polymorphisms Using Automated Capillary Sequencers, and Polyacrylamide and Agarose Gel Electrophoresis: Implications for the Assessment of Genetic Diversity and Relatedness in Almond. Scientia Horticulturae, 108, 310-316. http://dx.doi.org/10.1016/j.scienta.2006.02.004
- Sánchez-Pérez, R., Howad, W., Dicenta, F., Arús, P. and Martínez-Gómez, P. (2007) Mapping Major Genes and Quantitative Trait Loci Controlling Agronomic Traits in Almond. Plant Breeding, 126, 310-318. http://dx.doi.org/10.1111/j.1439-0523.2007.01329.x
- Dondini, L., Lain, O., Geuna, F., Banfi, R., Gaiotti, F., Tartarini, S., Bassi, D. and Tesolin, R. (2007) Development of a New SSR-Based Linkage Map in Apricot and Analysis of Synteny with Existing Prunus Map. Tree Genetics & Genomes, 3, 239-249. http://dx.doi.org/10.1007/s11295-006-0059-8
- Sánchez-Pérez, R., Howad, W., García-Mas, J., Arús, P., Martínez-Gómez, P. and Dicenta, F. (2010) Molecular Markers for Kernel Bitterness in Almond. Tree Genetics & Genomes, 6, 237-247. http://dx.doi.org/10.1007/s11295-009-0244-7
- Dirlewanger, E., Crosson, A., Tavaud, P., Aranzana, M.J., Poizat, C., Zanetto, A., Arús, P. and Laigret, L. (2002) Development of Microstellite Markers in Peach and Their Use in Genetic Diversity Analysis in Peach and Sweet Cherry. Theoretical and Applied Genetics, 105, 127-138. http://dx.doi.org/10.1007/s00122-002-0867-7
- Mnejja, M., Garcia-Mas, J., Howad, W. and Arús, P. (2005) Development and Transportability across Prunus Species of 42 Polymorphic Almond Microsatellites. Molecular Ecology Notes, 5, 531-535. http://dx.doi.org/10.1111/j.1471-8286.2005.00977.x
- Aranzana, M.J., García-Mas, J., Carbó, J. and Arús, P. (2002) Development and Variability Analysis of Microsatellite Markers in Peach. Plant Breeding, 121, 87-92. http://dx.doi.org/10.1046/j.1439-0523.2002.00656.x
- Howad, W., Yamamoto, T., Dirlewanger, E., Testolin, R., Cosson, P., Cipriani, G., Monforte, A.J., Georgi, L., Abbott, A.G. and Arús, P. (2005) Mapping with a Few Plants: Using Selective Mapping for Microsatellite Saturation of the Prunus Reference Map. Genetics, 171, 1305-1309. http://dx.doi.org/10.1534/genetics.105.043661
- Yamamoto, T., Mochida, K., Imai, T., Shi, I.Z., Ogiwara, I. and Hayashi, T. (2002) Microsatellite Markers in Peach [Prunus persica (L.) Batsch] Derived from an Enriched Genomic and cDNA Libraries. Molecular Ecology Notes, 2, 298-302. http://dx.doi.org/10.1046/j.1471-8286.2002.00242.x
- Downey, L.D. and Iezzoni, A.F. (2000) Polymorphic DNA Markers in Cherry Are Identified Using Sequences from Sweet Cherry, Peach, and Sour Cherry. Journal of the American Society of Horticultural Science, 125, 76-80.
- Sosinski, B., Gannavarapu, M., Hager, L.E., Beck, L.E., King, G.J., Ryder, C.D., Rajapakse, S., Baird, W.V., Ballard, R.E. and Abbott, A.G. (2000) Characterization of Microsatellite Markers in Peach (Prunus persica (L) Basch). Theoretical and Applied Genetics, 101, 421-428. http://dx.doi.org/10.1007/s001220051499
- Cantini, C., Iezzoni, A.F., Lamboy, W.L., Boritzki, M. and Struss, D. (2001) DNA Fingerprinting of Tetraploid Cherry Germplasm Using SSR. Journal of the American Society of Horticultural Science, 126, 205-209.
- Testolin, R., Messina, R., Lain, O., Marrazo, T., Huang, G. and Cipriani, G. (2004) Microsatellites Isolated in Almond from an AC-Repeat Enriched Library. Molecular Ecology Notes, 4, 459-461. http://dx.doi.org/10.1111/j.1471-8286.2004.00700.x
- Messina, R., Lain, O., Marrazo, T., Cipriano, G. and Testolin, R. (2004) New Set of Microsatellite Loci Isolated in Apricot. Molecular Ecology Notes, 4, 432-434. http://dx.doi.org/10.1111/j.1471-8286.2004.00674.x
- Cipriani, G., Lot, G., Huang, H.G., Marrazzo, M.T., Peterlunger, E. and Testolin, R. (1999) AC/GT and AG/CT Microsatellite Repeats in Peach (Prunus persica (L) Basch): Isolation, Characterization and Cross-Species Amplification in Prunus. Theoretical and Applied Genetics, 99, 65-72. http://dx.doi.org/10.1007/s001220051209
- Sánchez-Pérez, R., Ortega, E., Duval, H., Martínez-Gómez, P. and Dicenta, F. (2007) Inheritance and Correlation of Important Agronomic Traits in Almond. Euphytica, 155, 381-391. http://dx.doi.org/10.1007/s10681-006-9339-5
- Socias i Company, R., Felipe, A.J. and Gómez-Aparisi, J. (1999) A Major Gene for Flowering Time in Almond. Plant Breeding, 118, 443-448. http://dx.doi.org/10.1046/j.1439-0523.1999.00400.x
- Dicenta, F., García, J.E. and Carbonell, E.A. (1993) Heritability of Flowering, Productivity and Maturity in Almond. Journal of Horticultural Science, 68, 113-120.
- Vargas, F.J. and Romero, M.A. (2001) Blooming Time in Almond Progenies. Options Méditerranéennes, 56, 29-34.
- Colic, S., Rakonjac, V., Zec, G., Nikolic, D. and Fotiric-Aksic, M. (2012) Morphological and Biochemical Evaluation of Selected Almond [Prunus dulcis (Mill.) D.A. Webb] Genotypes in Northern Serbia. Turkish Journal of Agriculture and Forestry, 36, 429-438.
- Kester, D.E., Raddi, P. and Asay, R. (1977) Correlation of Chilling Requirements for Germination, Blooming and Leafing within and among Seedling Populations of Almond. Journal of the American Society of Horticultural Science, 102, 145-148.
- Dicenta, F., García-Gusano, M., Ortega, E. and Martínez-Gómez, P. (2005) The Possibilities of Early Selection of Late-Flowering Almonds as a Function of Seed Germination or Leafing Time of Seedlings. Plant Breeding, 124, 305- 309. http://dx.doi.org/10.1111/j.1439-0523.2005.01090.x
- Sánchez-Pérez, R., Dicenta, F. and Martínez-Gómez, P. (2012) Inheritance of Chilling and Heat Requirements for Flowering in Almond and QTL Analysis. Tree Genetics & Genomes, 8, 379-389. http://dx.doi.org/10.1007/s11295-011-0448-5
- Olukolu, B., Trainin, T., Fan, S., Kole, C., Bielenberg, D., Reighard, G., Abbott, A. and Holland, D. (2009) Genetic Linkage Mapping for Molecular Dissection of Chilling Requirement and Budbreak in Apricot (Prunus armeniaca L.). Genome, 52, 819-828. http://dx.doi.org/10.1139/G09-050
- Campoy, J.A., Ruiz, D., Egea, J., Rees, J., Celton, J.M. and Martínez-Gómez, P. (2011) Inheritance of Flowering Time in Apricot (Prunus armeniaca L.) and Analysis of Linked Quantitative Trait Loci (QTLs) Using Simple Sequence Repeat Markers. Plant Molecular Biology Reporter, 29, 404-410. http://dx.doi.org/10.1007/s11105-010-0242-9
- Hibrand-Saint Oyant, L., Crespel, L., Rajapakse, S., Zhang, L. and Foucher, F. (2008) Genetic Linkage Maps of Rose Constructed with New Microsatellite Markers and Locating QTL Controlling Flowering. Tree Genetics & Genomes, 4, 11-23. http://dx.doi.org/10.1007/s11295-007-0084-2
- Fan, S., Bielenberg, D.G., Zhebentyayeva, T.N., Reighard, G.L., Okie, W.R., Holland, D. and Abbott, A.G. (2010) Mapping Quantitative Trait Loci Associated with Chilling Requirement, Heat Requirement and Bloom Date in Peach (Prunus persica). New Phytology, 185, 917-930. http://dx.doi.org/10.1111/j.1469-8137.2009.03119.x
- Verde, I., Abbott, A.G., Scalabrin, S., Jung, S., Shu, S., Marroni, F., Zhebentyayeva, T., Dettori, M.T., Grimwood, J., Cattanoro, F., Zuccolo, A., Rossini, L., Jenkins, J., Vendramin, E., Meisel, L.A., Decroocq, V., Sosininski, B., Prochnik, S., Mitros, T., Policriti, A., Cipriani, G., Dondini, L., Ficklin, S., Goodstein, D.M., Xuan, P., Del Fabbro, C., Aramini, V., Copeti, D., Barakat, A., Testolin, R., Stella, A., Tartarin, S., Arús, P., Orellana, A., Wells, C., Main, D., Vizzotto, G., Silva, H., Salamani, F., Schmutz, J., Morgante, M. and Rokhsar, D. (2013) The High-Quality Draft of Peach (Prunus persica) Identifies Unique Patterns of Genetic Diversity, Domestication and Genome Evolution. Nature Genetics, 45, 487-494. http://dx.doi.org/10.1038/ng.2586
- Verde, I., Bassil, N., Scalabrin, S., Gilmore, B., Lawley, C.T., Gasic, K., Micheleti, D., Rosyara, U.R., Cattonaro, F., Vendramin, E., Main, D., Aramini, V., Blas, A.L., Mockler, T.C., Bryant, D.W., Whilelm, L., Troggio, M., Sosinski, B., Aranzana, M.J., Arús, P., Iezzoni, A., Morgante, N. and Peace, C. (2012) Development and Evaluation of a 9K SNP Array for Peach by Internationally Coordinated SNP Detection and Validation in Breeding Germplasm. PLoS ONE, 7, e35668. http://dx.doi.org/10.1371/journal.pone.0035668
NOTES

*Equal contribution.
#Corresponding author.


