Agricultural Sciences
Vol.4 No.6A(2013), Article ID:33724,9 pages DOI:10.4236/as.2013.46A005
Are type IV pili involved in Vibrio anguillarum virulence towards sea bass (Dicentrarchus labrax L.) larvae?
![]()
1Laboratory for Process Microbial Ecology and Bioinspirational Management (PME&BIM), Thomas More Mechelen, Campus De Nayer, Department of Microbial and Molecular Systems (M2S), KU Leuven Association, Sint-Katelijne-Waver, Belgium; *Corresponding Author: hans.rediers@biw.kuleuven.be
2Scientia Terrae Research Institute, Sint-Katelijne-Waver, Belgium
3Centre for Food and Microbial Technology, M2S, KU Leuven, Leuven, Belgium
4Laboratory of Aquaculture & Artemia Reference Center, Department of Animal Production, Ghent University, Ghent, Belgium
Copyright © 2013 Ingeborg Frans et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 25 April 2013; revised 25 May 2013; accepted 9 June 2013
Keywords: Virulence; Vibrio anguillarum; Fish Pathogen; Type IV Pili
ABSTRACT
Vibrio anguillarum, an important bacterial fish pathogen, expresses a variety of virulence factors contributing to its ability to cause vibriosis in fish. Many virulence factors of this pathogen remain however unknown. For example, a type IV pilus system was previously reported to be potentially involved in the virulence of this bacterium but no experimental evidence was reported yet. In this study, complete genome sequencing of V. anguillarum strain VIB15, shown to be highly virulent towards sea bass (Dicentrarchus labrax L.) larvae, revealed the presence of a PilA pilin. A V. anguillarum VIB15 pilA mutant was constructed and the pathogenicity of this mutant was assessed in a gnotobiotic sea bass system developed for virulence screening. Our results suggest that the V. anguillarum pilA gene is not crucial for virulence towards sea bass larvae. Possibly, another type IV pilus system identified in V. anguillarum, showing homology to the mannose-sensitive hemagglutinin pilin of Vibrio cholerae, may complement the pilA mutation. Alternatively, the type IV pilus system has a role in infection of juvenile or adult fish, rather than in the larval phase. As such, further research is required to unravel the potential role of type IV pili in V. anguillarum virulence.
1. INTRODUCTION
Vibrio anguillarum is an important fish pathogen causing vibriosis in various cultured and wild fish as well as in bivalves and crustaceans. Because of its high morbidity and mortality rate, this hemorrhagic septicemic disease is responsible for severe economic losses in both larviculture and aquaculture worldwide [1-3]. Previous studies have identified several virulence-related genes in V. anguillarum, including genes involved in chemotaxis and motility, iron uptake, synthesis of lipopolysaccharides and extracellular products with proteolytic or haemolytic activity (reviewed in [4,5]). Much of the virulence mechanism of V. anguillarum remains however poorly understood. In order to get a better understanding of the virulence strategies employed by this pathogen, Rodkum et al. [6] identified 40 putative virulence-related genes by random genome sequencing. Focusing on the initial stages of infection, eighteen genes mediating adherence and colonization of bacteria to host cells were identified. Five of these genes were associated with type IV pili, flexible, hair-like filaments on the surface of a wide variety of Gram-negative and Gram-positive genera, making type IV pili the most widespread colonization factors [7]. In addition to the role as adherence factors to living and nonliving surfaces, these pili have an important function in other bacterial processes as well, including flagellum-independent twitching motility, DNA uptake and exchange, biofilm formation, and secretion of extracellular proteins [8,9]. Furthermore, type IV pili have been identified as important virulence factors in several human, plant and animal pathogens, including several fish pathogens [8,10-14]. Type IV pili possibly contribute to the pathogen’s virulence by their roles in adhesion, immune escape, surface motility, microcolony formation, and complement inhibition [8].
The type IV pilus is composed of hundreds of copies of the major pilin subunit and a low abundance of minor pilins required for function and/or assembly of the pilus. All type IV pilins are synthesized as prepilins comprising a leader peptide with an N-terminal consensus motif. Formation of the mature pilin involves cleavage of the leader peptide and methylation of the newly formed Nterminal residue by a specific prepilin peptidase/N-methyltransferase protein. After translocation across the inner membrane, the mature pilin is assembled into a multimeric pilus fiber on a base of minor pilins on the periplasmic side of the inner membrane (Figure 1). Minor pilins are pilin-like proteins involved in type II secretion system, which are required for secretion. Finally,
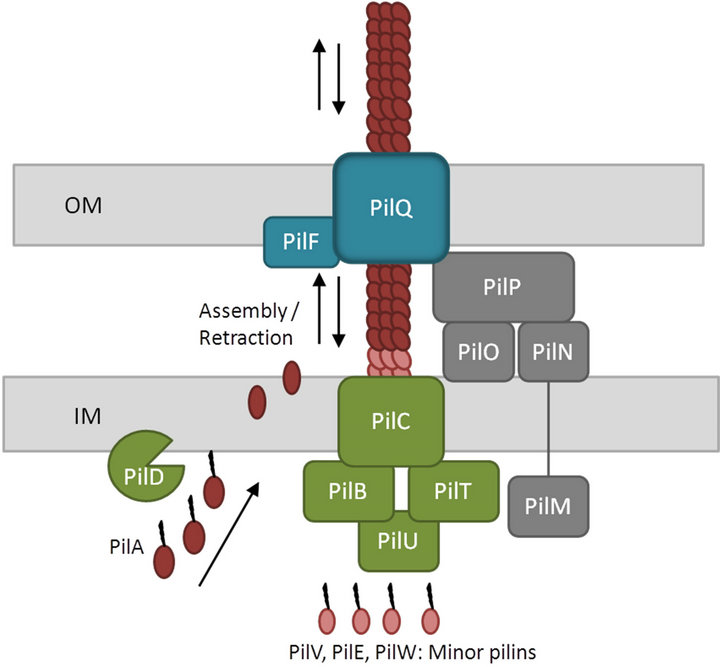
Figure 1. Schematic overview of the type IV PilA pilus system. The major type IV pilin, PilA, is synthesized as a prepilin consisting of a leader peptide with a consensus N-terminal motif. The leader peptide is cleaved and the resulting N-terminal residue is methylated by a specific prepilin peptidase/N-methyltransferase protein, PilD. After translocation across the inner membrane, the mature pilin is assembled into a multimeric pilus fiber on the periplasmic side of the inner membrane. Subsequently, the filament is secreted across the outer membrane through a secretin pore, PilQ. Two different ATPases, PilB and PilT, promote extension and retraction, respectively of the pilus. In addition, a transmembrane protein found in the inner membrane, PilC, and a number of pseudopilins, PilV, PilE and PilW, are involved in the pilin biogenesis. The four subcomplexes shown are the cytoplasmic motor subcomplex (green), the inner membrane alignment subcomplex (grey), the outer membrane secretin pore subcomplex (blue) and the pilus itself (brown) (adapted from [33-35]).
the major pilin is secreted across the outer membrane through a secretin pore.
In the current study, the potential role of the V. anguillarum type IV pili in virulence was investigated using a gnotobiotic sea bass (Dicentrarchus labrax L.) larvae system.
2. MATERIALS AND METHODS
2.1. Bacterial Strains
Bacteria and plasmids used in this study are listed in Table 1. The parental strain for the knockout mutant was V. anguillarum VIB15, an O1 serotype strain which was originally isolated from a diseased sea bass in Greece [15]. This strain was made rifampicin resistant (VIB15RifR) by natural selection, allowing survival of the bacterium in the gnotobiotic system used in this study. V. anguilllarum was grown in tryptic soy broth (TSB) (Oxoid, Erembodegem, Belgium) supplemented with 1% NaCl, or in marine broth (MB) (Difco Laboratories, Detroit, USA) for 24 hours at 28˚C. Escherichia coli strains were grown overnight in Luria Bertani (LB) broth or TSB at 37˚C. For solid media, 15 g/l of agar was added. Medium additives were used in the following concentrations when required: 40 mg/ml X-Gal (Invitrogen, Merelbeke, Belgium), 100 µg/ml ampicillin (Ap), 5 µg/ml chloramphenicol (Cm), 10 µg/ml, 100 µg/ml or 200 µg/ml rifampicin (Rif), and 50 µg/ml streptomycin (Sm) (Sigma Aldrich).
2.2. DNA Isolation and Manipulation
Plasmid DNA isolation and PCR purification was performed using QIAprep Spin Miniprep Kit and QIAquick PCR Purification Kit, respectively (Qiagen Inc., Valencia, CA, USA). Genomic DNA was extracted using the phenol/chloroform extraction method as described by Lievens et al. [16]. DNA quantity and quality was assessed using the Quant-iTTM PicoGreen® dsDNA reagent (Invitrogen).
Standard techniques for restriction endonuclease digestion, ligation, gel electrophoresis, and southern hybridization were performed as described by Sambrook and Russell [17]. Restriction and modification enzymes were obtained from Bioké (Leiden, The Netherlands) and Roche Diagnostics (Vilvoorde, Belgium). DNA quantity and quality was assessed using the Nanodrop spectrophotometer or the Quant-iTTM PicoGreen® dsDNA reagent (Invitrogen).
2.3. PCR
PCR amplification was performed in a total volume of 20 µl containing 0.5 µM of each primer, 0.15 mM of each deoxynucleoside triphosphate (Invitrogen), 2.0 U
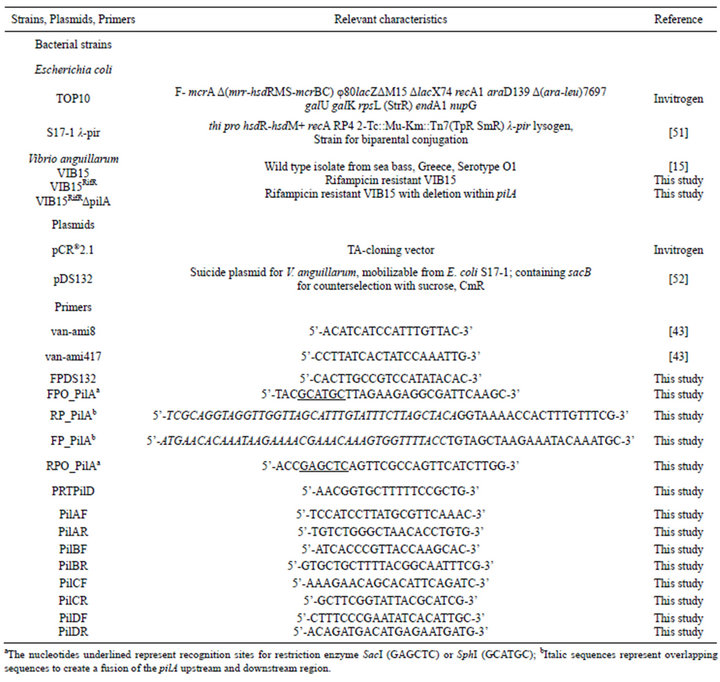
Table 1. Bacterial strains, plasmids and primers used in this study.
Taq DNA polymerase (Bioké), 10× ThermoPol Reaction Buffer (Bioké), and 1 µl of DNA solution (1 ng/µl, as measured by a Nanodrop spectrophotometer). All amplifications were performed using a Bio-Rad T100 thermal cycler with initial denaturation at 94˚C for 5 min, followed by 30 cycles of 30 sec at 94˚C, 30 sec at 59˚C, and 30 sec at 72˚C, with a final extension step at 72˚C for 10 min. In case Platinum® Pfx DNA Polymerase (Invitrogen) was used, elongation temperature was changed to 68˚C.
2.4. Genome Sequencing
V. anguillarum strain VIB15 was grown overnight on marine agar (MA) (Difco) at 28˚C. Following DNA isolation, a paired-end library (2 × 100 bp, 500 bp inserts) was prepared for sequencing on the Illumina HiSeq 2000 platform (Illumina, San Diego, CA, USA) at Genomics Core (UZ Leuven, Belgium) according to the manufacturer’s instructions. Sequencing reads were subjected to quality filtering using NGS-QC Toolkit [18]. Low quality reads (reads with 70% of the nucleotides having a Phred quality score below 30) were removed, and high quality reads were trimmed to remove the first and the last 13 nucleotides. Trimmed reads were de novo assembled using SOAPdenovo v1.05 (http://www.soap.genomics.org.cn/soapdenovo.html). Assembled contigs were subsequently scaffolded by SSPACE [19], and additional gaps were closed by GapFiller [20]. The assembled genome sequence was annotated by the RAST server [21]. Homology searches were performed by using the BLAST program [22].
2.5. Bacterial Mating
Plasmids were introduced into V. anguillarum VIB15RifR by conjugation procedures described previously [23]. Briefly, overnight cultures of V. anguillarum and E. coli S17-1 λ-pir were mixed at a ratio of 10:1 in 2.5 ml 10 mM MgSO4. The cell mixture was vacuum filtered onto a 0.22-µm-pore-size filter, which was placed on tryptic soy agar (TSA) and allowed to incubate overnight at 25˚C. Following incubation, the cells were harvested and plated on LB agar plates containing 200 µg/ml Rif and 5 µg/ml Cm and allowed to incubate at 25˚C until V. anguillarum colonies were observed.
2.6. Construction of Chromosomal Mutation
To generate a pilA in-frame deletion fragment of strain VIB15RifR, an asymmetric overlap extension PCR (AOEPCR) was performed (Figure 3). The upstream and downstream regions of the pilA gene were amplified asymmetrically using primers FPO_PilA/RP_PilA and FP_PilA/RPO_PilA, respectively (Table 1), at a 1:10 inner:outer primer ratio and Platinum® Pfx DNA Polymerase (Invitrogen) according to the manufacturer’s instructions. The inner primer for amplification of the region upstream of pilA, RP_PilA, includes 40 nucleotides at the 5’ end, which is complementary to the downstream region and vice versa for primer FP_PilA, generating an excess of single-stranded DNA with overlapping 3’ ends. Both fragments hybridized and functioned as a template for PCR amplification of the desired in-frame deletion fragment. For this PCR reaction 1µl of each reaction, Taq DNA polymerase and the outer primers FPO_PilA and RPO_PilA were used. The resulting PCR product was directly cloned into the pCR®2.1 cloning vector using the TA cloning kit (Invitrogen). Following SphI/SacI digestion of the resulting plasmid, gel purification and ethanol precipitation, the pilA knock-out fragment was ligated into the SphI/SacI site of the dephosphorylated pDS132 suicide vector. The ligation reaction was subsequently purified using the QIAquick PCR Purification Kit (Qiagen Inc.), electroporated into E. coli S17-1 λ-pir and introduced into VIB15RifR by bacterial mating. VIB15RifR single recombinants were selected on LB plates containing chloramphenicol and incorporation of the plasmid was confirmed with PCR (FPDS132/RPO_PilA). In the next step, double homologous recombinants were selected on LB5 (LB with 0.5% NaCl) containing 100 µg/ml Rif and 10% sucrose for sacB counter selection. Gene replacement in these strains was confirmed by PCR using pilA-specific primers PilAF and PilAR and southern hybridization.
2.7. RT-PCR
Total RNA was extracted from V. anguillarum VIB15RifR and the pilA knockout mutant after overnight growth on MA using the SpectrumTM Plant Total RNA kit (Sigma Aldrich). Subsequently, cDNA was synthesized using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s instructions, and the pil locus-specific reverse primer PRTPilD. The obtained cDNA was analyzed by PCR for the presence of transcripts from pilA, pilB, pilC and pilD. The primers used for amplification are listed in Table 1.
2.8. Virulence Testing
Both VIB15RifR strain and pilA knockout mutant were subjected to a virulence assay in a standardized gnotobiotic model system with axenic European sea bass (D. labrax L.) larvae as hosts, as described previously [24, 25]. Briefly, eggs of D. labrax were obtained from natural spawning at the hatchery of Ecloserie Marine de Gravelines (France) followed by a standard disinfection protocol generating germ-free eggs. After hatching, sea bass larvae were challenged with a suspension of 105 cfu/ml V. anguillarum. Prior to inoculation, these bacterial strains were grown in 10% MB with addition of NaCl and 10 µg/ml Rif, and incubated on a horizontal shaker at 120 rpm at 16˚C ± 0.5˚C for 2 days. For each challenge, 10 replicates were prepared and the vials were rotated at 4 rpm tangential to the axis of the vials. On DAH (days after hatching) 6, 8, 10 and 12 (i.e. 3 - 9 days post-exposure), survival of the sea bass larvae was monitored by microscopic analysis. Non-inoculated germ-free larvae that were kept under similar conditions served as a control. Throughout the experiment, eggs and larvae were kept at a salinity of 36 g/l in a temperature controlled room at 16˚C ± 0.5˚C in constant dim light (10 candela steradian/m2). The experiment was approved by the ethical committee of Ghent University (no. EC2012/ 071). Two independent repeats of this gnotobiotic sea bass challenge experiment were performed. Statistical analysis of the larval survival data was performed by means of R v2.12.1. Survival was reported as mean values (% survival) ± standard error of the mean (SEM). Data were tested for normality and subjected to non-parametric tests. Wilcoxon rank-sum test was used to compare the survival of the sea bass larvae. Significance was accepted at p < 0.05.
3. RESULTS
3.1. Identification of V. anguillarum Type IV Pilus Biogenesis Gene Cluster
To identify genes involved in the biogenesis of the V. anguillarum type IV pilus system, the complete genome sequence of V. anguillarum strain VIB15 was determined. In a previous study using a standardized gnotobiotic system, it was demonstrated that strain VIB15 is highly virulent towards sea bass (D. labrax L.) larvae [25].
The genome sequence revealed that the gene encoding the major pilin, pilA, comprises 427 bp and is arranged in a typical type IV pilus biogenesis cluster, pilABCD. pilB encodes a cytoplasmic protein required for ATP binding and hydrolysis, pilC an inner membrane protein, and pilD the prepilin peptidase/N-methyltransferase protein. Downstream of this cluster, a fifth open reading frame, yacE, has been identified, encoding a dephospho-CoA kinase. Although the same gene organization is found in many other bacteria, the role of yacE in the type IV pilus assembly is unknown [26,27]. Upstream of the V. anguillarum pilABCD cluster, the nadC gene, encoding a quinolinate phosphoribosyltransferase, was identified. The nadC-pilABCD-yacE gene organization was also observed in V. cholerae and V. vulnificus, indicating a conserved gene synteny [26,28]. The protein sequences of PilA to PilD are 99% - 100% identical to the corresponding proteins in V. anguillarum strain 775, and shows high amino acid identity to the V. vulnificus homologs (42%, 75%, 75%, and 71%, respectively) [28- 30].
Four additional clusters required for pilus biogenesis and assembly, pilMNOPQ, pilVWE, pilUT, and pilF, were identified on chromosome 1 (Figure 2). The pilVWE cluster encodes minor pilins which form the base for the major pilin, PilA. Recently, Giltner et al. [31] reported that these minor pilins are also incorporated into the growing pilus fiber, supporting an additional role of these minor pilins in optimal pilus assembly. The pilQ gene encodes a secretin through which the major pilin is secreted across the outer membrane. Assembly of this secretin requires its cognate pilotin, PilF, an outer membrane lipoprotein involved in the correct localization and multimerization of PilQ [32]. Furthermore, in contrast to PilB, which is involved in the assembly and extension of type IV pilin, PilT and PilU promote disassembly and retraction of the pilin. Finally, PilM, PilN, PilO and PilP form the bridge between the cytoplasmic and outer membrane components of the type IV pilus system [33, 34]. Figure 2 illustrates that the different genes and gene clusters involved in type IV pilus biogenesis, are scattered throughout chromosome I of V. anguillarum, which is also the case in other organisms [7,14,33].
Focusing on the major pilin, PilA, considerable homology to the type IV pilin precursor proteins of many other bacteria was observed [11,26,28,35,36]. Typically, the N-terminal region of the premature PilA of V. anguillarum VIB15 is dominated by a short, hydrophilic leader peptide and shows the presence of the conserved
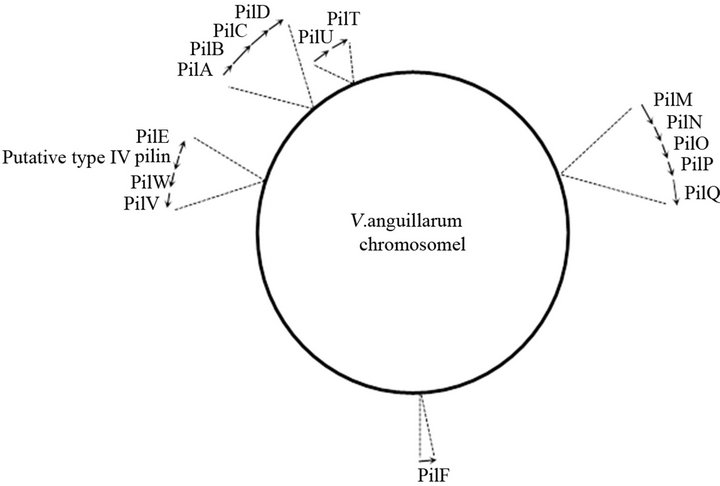
Figure 2. Schematic representation of chromosome I of V. anguillarum VIB15 mapping the genes involved in PilA type IV pilus system. The arrows indicate the approximate size and orientations of the genes.
consensus cleavage site with the GFTLIE amino acid motif, which is identical in PilA of several other Vibrio species such as V. cholerae [26]. After cleavage of the leader peptide, the mature PilA contains a very hydrophobic N-terminal segment of about 20 - 25 residues. The C-terminal domain of PilA is less conserved, although it contains a characteristic pair of cysteines, responsible for a disulfide-bonded loop containing a receptor-binding site. Involvement of this binding-site either in intersubunit contacts throughout the length of the pilus fiber or host-cell adherence have been proposed [37-40].
3.2. Construction of a Non-Polar pilA Mutant Strain
To examine the potential role of PilA in V. anguillarum infection, a non-polar knockout mutant of strain VIB15RifR was generated. Using AOE-PCR [41], an in-frame deletion of 378 bp of the pilA gene was constructed (Figure 3). First, it was verified that the growth in MB was not affected in the mutant strain (data not shown). Second, RT-PCR was used to verify that the pilus biogenesis genes downstream of pilA, i.e. pilB, pilC and pilD, are still expressed in the knockout strain. The cDNA generated by a reverse primer downstream of pilD (PRTPilD) was examined for the presence of pilA, pilB, pilC, and pilD transcripts using specific primers internal to each gene (Table 1). As can be seen in Figure 4, the presence of PCR fragments of the expected size for strain VIB15RifR, corresponding to transcription of pilA (364 bp), pilB (979 bp), pilC (882 bp), and pilD (567 bp), was confirmed. However, the pilA mutant strain was clearly deficient in pilA transcription, confirming that the deletion in the pilA gene resulted in the loss of transcription of the target gene but did not affect transcription of the downstream genes.
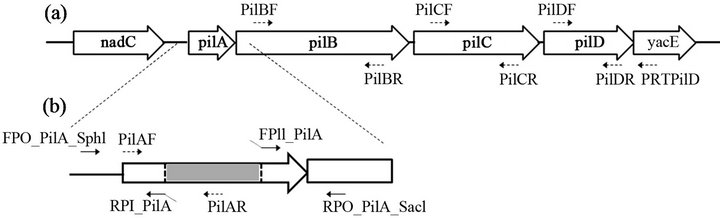
Figure 3. Gene organization of the type IV PilA pilus biogenesis gene cluster of V. anguillarum VIB15 (a); The primers used for fusing the pilA upstream and downstream regions are indicated solid arrows, while primers used for confirmation of the pilA mutation are indicated with dashed arrows (b).
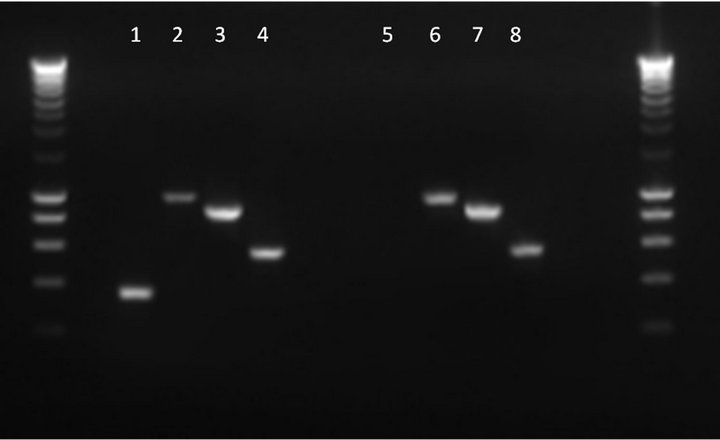
Figure 4. Detection of pilA, pilB, pilC and pilD transcripts by RT-PCR in parental V. anguillarum strain VIB15RifR (lanes 1 to 4, respectively) and pilA knockout strain (lanes 5 to 8) using specific internal primers for each gene. Smartladder (Eurogentec) was used as molecular weight marker.
3.3. Assessment of the Virulence of the pilA Mutant
To investigate the role of the PilA pilus in the virulence of V. anguillarum, both wild-type strain and the pilA knockout mutant were subjected to virulence assessment in the gnotobiotic sea bass larvae system. Previous studies have demonstrated the usefulness of this experimental system to assess the virulence of bacterial pathogens in vivo [24,25,42]. After challenging the sea bass larvae with V. anguillarum, larval survival was monitored at DAH 6, 8, 10 and 12. The experiment was terminated at DAH 12 as the sea bass larvae died of starvation in prolonged experiments. The averaged results of both challenging experiments are presented in Figure 5. Survival on DAH 12 of the sea bass larvae challenged with the parental strain or the PilA knockout mutant (16% ± 4% survival and 10% ± 3% survival, respectively) was significantly different from the control group (72% ± 5%). However, on DAH 12, no significant difference in survival could be observed between the wild-type VIB15 strain and the pilA knockout mutant. The same could be observed on DAH 6, DAH 8 and DAH 10. These results suggest that the PilA type IV pilus is not a crucial virulence factor in V. anguillarum VIB15. To confirm that mortality was caused by the tested V.
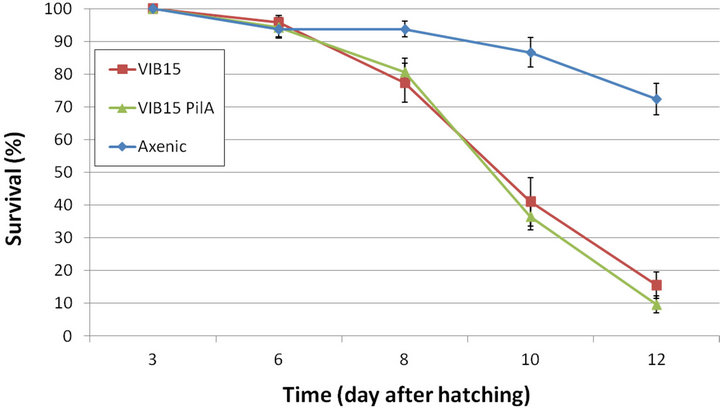
Figure 5. Survival (in percentage) of sea bass larvae during the gnotobiotic experiment after challenging the larvae with the wild-type V. anguillarum strain VIB15RifR and the pilA knockout strain. The averaged results of two independent repeats are presented. Error bars represent mean ± SEM (n = 20).
anguillarum strains and not by contamination, the identity of the inoculated strains was confirmed by isolating the bacteria again at the end of the experiment (DAH 12) followed by PCR using the V. anguillarum-specific primers van-ami8/van-ami417 and the pilA-specific primers PilAF/PilAR [43].
4. DISCUSSION
The PilA type IV pilus system has been previously identified as an important virulence factor in several fish pathogens. It has for example been reported that the Aeromonas salmonicida PilA homologue, TapA, was required for virulence in rainbow trout (Oncorhynchus mykiss (Walbaum)) and Atlantic salmon (Salmo salar L.) [11,14]. In addition, the pilA-encoded type IV pilus of V. vulnificus contributed significantly to the persistence in Crassostrea virginica oysters [13]. In several other pathogenic bacteria, the type IV pilus has been shown to be crucial for virulence [7,8,12,44]. Therefore, the putative role of this type IV pilus system in the virulence of V. anguillarum was investigated in the current study.
The complete genome sequence of V. anguillarum strain VIB15, previously reported to be highly virulent towards sea bass larvae, revealed the presence of genes and gene clusters involved in secretion and assembly of a type IV pilus system (Figure 2) [7,35]. PilA, the major pilin, is encoded by the pilABCD gene cluster, showing high similarity to the pilABCD gene cluster of V. vulnificus [28], and V. cholerae [26], and the tapABCD gene cluster of A. salmonicida [11]. To investigate the potential role of this type IV pilus system in the virulence of V. anguillarum, a non-polar knockout mutant of the major pilin, PilA, of strain VIB15 was successfully created. The virulence of the pilA mutant was assessed using a standardized gnotobiotic system with axenic European sea bass larvae as host organism [24]. This standardized gnotobiotic system has proven its strength as a model to evaluate the virulence of V. anguillarum in vivo [24,25, 42]. Unexpectedly, the virulence assay revealed no significant difference in larval survival between the wildtype strain and the pilA knockout mutant. A possible explanation for this observation is the identification of a second type IV pilus system in the V. anguillarum genome, the mannose-sensitive hemagglutinin pilus (MSHA) system, which may complement the PilA deficiency. This system is very similar to the V. cholerae MSHA pilus and has been previously identified in other Vibrio species such as V. vulnificus and V. parahaemolyticus [45]. However, in V. vulnificus, the presence of the MSHA pilus was not able to complement for the loss of the PilA pilus, as the pilA knock out mutant was severely attenuated in biofilm formation and virulence in mice [28]. Moreover, genes responsible for the biosynthesis of this MSHA pilus were repressed during V. cholerae infection in infant mice [46].
Alternatively, the PilA pilus might have a role in infection of juvenile or adult fish, rather than the fish larvae. In this regard, it has been reported that the expression of the pilA gene of V. cholerae and V. parahaemolyticus is induced by chitin, a polymer found in high abundance in fish scales and oyster shells [13,47,48]. This may indicate that in the gnotobiotic larvae system used in this study, the V. anguillarum pilA expression is lower, suggesting that the PilA pilus doesn’t play a crucial role in virulence towards sea bass larvae. The influence of type IV pilin on the virulence of A. salmonicida in rainbow trout and Atlantic salmon was examined using juveniles covered with scales instead of larvae without scales [11,14,49]. In addition, the presence of chitin in the shell of the oysters might explain the role of PilA of V. vulnificus in oysters [13]. Finally it is possible that the type IV pilin of V. anguillarum strain VIB15, PilA, is not involved in the virulence of the pathogen but in the persistence of the bacterium in the aquatic environment. Chiavelli et al. [50] for instance, reported that the type IV pilus of V. cholerae promotes attachment to zooplankton exoskeletons as a survival strategy in the aquatic environment.
In conclusion, although type IV pili have been identified as important virulence factors in several human, plant and animal pathogens, including several fish pathogens, the data presented here suggested that PilA had no crucial role in the virulence of V. anguillarum VIB15 towards sea bass (D. labrax L.) larvae. Some possible explanations have been proposed but further studies are necessary to elucidate the role of both type IV pilus systems in fish and the environment.
5. ACKNOWLEDGEMENTS
The authors would like to thank Brigitte Van Moffaert for her excellent technical support. This work was financially supported by the “Agency for Innovation by Science and Technology in Flanders (IWT)”, by the European FP7 project “Promicrobe—Microbes as positive actors for more sustainable aquaculture” (Project Reference: 227197), and by Ghent University project “Host microbial interactions in aquatic production” (BOF12/GOA/022).
REFERENCES
- Aguirre-Guzman, G., Ruiz, H.M. and Ascencio, F. (2004) A review of extracellular virulence product of Vibrio species important in diseases of cultivated shrimp. Aquaculture Research, 35, 1395-1404. doi:10.1111/j.1365-2109.2004.01165.x
- Paillard, C., Leroux, F. and Borrego, J.J. (2004) Bacterial disease in marine bivalves: Review of recent studies: Trends and evolution. Aquatic Living Resources, 17, 477- 498. doi:10.1051/alr:2004054
- Toranzo, A.E., Magarinos, B. and Romalde, J.L. (2005) A review of the main bacterial fish diseases in mariculture systems. Aquaculture, 246, 37-61. doi:10.1016/j.aquaculture.2005.01.002
- Frans, I., Michiels, C.W., Bossier, P., Willems, K.A., Lievens, B. and Rediers, H. (2011) Vibrio anguillarum as a fish pathogen: Virulence factors, diagnosis and prevention. Journal of Fish Diseases, 34, 643-661. doi:10.1111/j.1365-2761.2011.01279.x
- Naka, H. and Crosa, J.H. (2011) Genetic determinants of virulence in the marine fish pathogen Vibrio anguillarum. Fish Pathology, 46, 1-10. doi:10.3147/jsfp.46.1
- Rodkhum, C., Hirono, I., Stork, M., Lorenzo, M.D., Crosa, J.H. and Aoki, T. (2006) Putative virulence-related genes in Vibrio anguillarum identified by random genome sequencing. Journal of Fish Diseases, 29, 157-166. doi:10.1111/j.1365-2761.2006.00692.x
- Pelicic, V. (2008) Type IV pili: E pluribus unum? Molecular Microbiology, 68, 827-837. doi:10.1111/j.1365-2958.2008.06197.x
- Craig, L., Pique, M.E. and Tainer, J.A. (2004) Type IV pilus structure and bacterial pathogenicity. Nature Reviews Microbiology, 2, 363-378. doi:10.1038/nrmicro885
- Giltner, C.L., Nguyen, Y. and Burrows, L.L. (2012) Type IV pilin proteins: Versatile molecular modules. Microbiology and Molecular Biology Reviews, 76, 740-772. doi:10.1128/MMBR.00035-12
- Kang, Y., Liu, H., Genin, S., Schell, M.A. and Denny, T.P. (2002) Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Molecular Microbiology, 46, 427-437. doi:10.1046/j.1365-2958.2002.03187.x
- Masada, C.L., LaPatra, S.E., Morton, A.W. and Strom, M.S. (2002) An Aeromonas salmonicida type IV pilin is required for virulence in rainbow trout Oncorhynchus mykiss. Diseases of Aquatic Organisms, 51, 13-25. doi:10.3354/dao051013
- Essex-Lopresti, A.E., Boddey, J.A., Thomas, R., Smith, M.P., Hartley, M.G., Atkins, T., et al. (2005) A type IV pilin, PilA, contributes to adherence of Burkholderia pseudomallei and virulence in vivo. Infection and Immunity, 73, 1260-1264. doi:10.1128/IAI.73.2.1260-1264.2005
- Paranjpye, R.N., Johnson, A.B., Baxter, A.E. and Strom, M.S. (2007) Role of type IV pilins in persistence of Vibrio vulnificus in Crassostrea virginica oysters. Environmental Microbiology, 73, 5041-5044.
- Boyd, J.M., Dacanay, A., Knickle, L.C., Touhami, A., Brown, L.L., Jericho, M.H., Johnson, S.C. and Reith, M. (2008) Contribution of type IV pili to the virulence of Aeromonas salmonicida subsp. salmonicida in Atlantic salmon (Salmo salar L.). Infection and Immunity, 76, 1445-1455. doi:10.1128/IAI.01019-07
- Austin, B., Alsina, M., Austin, D.A., Blanch, A.R., Grimont, F., Grimont, P.A.D., et al. (1995) Identification and typing of Vibrio anguillarum: A comparison of different methods. Systematic and Applied Microbiology, 18, 285- 302. doi:10.1016/S0723-2020(11)80400-5
- Lievens, B., Brouwer, M., Vanachter, A.C.R.C., Levesque, C.A., Cammue, B.P.A. and Thomma, B.P.H.J. (2003) Design and development of a DNA array for rapid detection and identification of multiple tomato vascular wilt pathogens. FEMS Microbiology Letters, 223, 113-122. doi:10.1016/S0378-1097(03)00352-5
- Sambrook, J. and Russell, D.W. (2001) Molecular cloning: A laboratory manual. 3rd Edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor.
- Patel, R.K. and Jain, M. (2012) NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PloS One, 7, e30619. doi:10.1371/journal.pone.0030619
- Boetzer, M., Henkel, C.V., Jansen, H.J., Butler, D. and Pirovano, W. (2011) Scaffolding pre-assembled contigs using SSPACE. Bioinformatics, 27, 578-579. doi:10.1093/bioinformatics/btq683
- Boetzer, M. and Pirovano, W. (2012) Toward almost closed genomes with GapFiller. Genome Biology, 13, R56. doi:10.1186/gb-2012-13-6-r56
- Aziz, R.K., Bartels, D., Best, A.A., Dejongh, M., Disz, T., Edwards, et al. (2008) The RAST server: Rapid annotations using subsystems technology. BMC Genomics, 9, 75. doi:10.1186/1471-2164-9-75
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389-3402. doi:10.1093/nar/25.17.3389
- Milton, D.L., O’Toole, R., Horstedt, P. and Wolf-Watz, H. (1996) Flagellin A is essential for the virulence of Vibrio anguillarum. Journal of Bacteriology, 178, 1310-1319.
- Dierckens, K., Rekecki, A., Laureau, S., Sorgeloos, P., Boon, N., Van den Broeck, W. and Bossier, P. (2009) Development of a Listonella (Vibrio) anguillarum challenge test for gnotobiotic sea bass (Dicentrarchus labrax) larvae. Environmental Microbiology, 11, 526-533. doi:10.1111/j.1462-2920.2008.01794.x
- Frans, I., Dierckens, K., Crauwels, S., Van Assche, A., Leisner, J., Larsen, M.H., Michiels, C.W., Bossier, P., Willems, K.A., Lievens, B. and Rediers, H. (2013) Does virulence assessment of Vibrio anguillarum using sea bass (Dicentrarchus labrax) larvae correspond with genotypic and phenotypic characterization? Plos One, submitted.
- Fullner, K.J. and Mekalanos, J.J. (1999) Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infection and Immunity, 67, 1393-1404.
- Browne-Silva, J. and Nishiguchi, M.K. (2008) Gene sequences of the pil operon reveal relationships between symbiotic strains of Vibrio fischeri. International Journal of Systematic and Evolutionary Microbiology, 58, 1292- 1299. doi:10.1099/ijs.0.65370-0
- Paranjpye, R.N. and Strom, M.S. (2005) A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infection and Immunity, 73, 1411-1422. doi:10.1128/IAI.73.3.1411-1422.2005
- Naka, H., Dias, G.M., Thompson, C.C., Dubay, C., Thompson, F.L. and Crosa, J.H. (2011) Complete genome sequence of the marine fish pathogen Vibrio anguillarum harboring the pJM1 virulence plasmid and genomic comparison with other virulent strains of V. anguillarum and V. ordalii. Infection and Immunity, 79, 2889-2900. doi:10.1128/IAI.05138-11
- Paranjpye, R.N., Lara, J.C., Pepe, J.C., Pepe, C.M. and Strom, M.S. (1998) The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice. Infection and Immunity, 66, 5659- 5668.
- Giltner, C.L., Habash, M. and Burrows, L.L. (2010) Pseudomonas aeruginosa minor pilins are incorporated into Type IV pili. Journal of Molecular Biology, 398, 444- 461. doi:10.1016/j.jmb.2010.03.028
- Koo, J., Tammam, S., Ku, S.Y., Sampaleanu, L.M., Burrows, L.L. and Howell, P.L. (2008) PilF is an outer membrane lipoprotein required for multimerization and localization of the Pseudomonas aeruginosa Type IV pilus secretin. Journal of Bacteriology, 190, 6961-6969. doi:10.1128/JB.00996-08
- Burrows, L.L. (2012) Pseudomonas aeruginosa twitching motility: Type IV pili in action. Annual Review of Microbiology, 66, 493-520. doi:10.1146/annurev-micro-092611-150055
- Tammam, S., Sampaleanu, L.M., Koo, J., Manoharan, K., Daubaras, M., Burrows, L.L. and Howell, P.L. (2013) PilMNOPQ from the Pseudomonas aeruginosa Type IV Pilus System form a transenvelope protein interaction network that interacts with PilA. Journal of Bacteriology, 195.
- Mattick, J.S. (2002) Type IV pili and twitching motility. Annual Review of Microbiology, 56, 289-314. doi:10.1146/annurev.micro.56.012302.160938
- Chattopadhyay, S., Paranjpye, R.N., Dykhuizen, D.E., Sokurenko, E.V. and Strom, M.S. (2009) Comparative evolutionary analysis of the major structural subunit of Vibrio vulnificus type IV pili. Molecular Biology and Evolution, 26, 2185-2196. doi:10.1093/molbev/msp124
- Hazes, B., Sastry, P.A., Hayakawa, K., Read, R.J. and Irvin, R.T. (2000) Crystal structure of Pseudomonas aeruginosa PAK pilin suggests a main-chain-dominated mode of receptor binding. Journal of Molecular Biology, 299, 1005-1017. doi:10.1006/jmbi.2000.3801
- Audette, G.F., Irvin, R.T., and Hazes, B. (2004) Crystallographic analysis of the Pseudomonas aeruginosa strain K122-4 monomeric pilin reveals a conserved receptorbinding architecture. Biochemistry, 43, 11427-11435. doi:10.1021/bi048957s
- Craig, L., Volkmann, N., Arvai, A.S., Pique, M.E., Yeager, M., Egelman, E.H. and Tainer, J.A. (2006) Type IV pilus structure by cryo-electron microscopy and crystallography: Implications for pilus assembly and functions. Molecular Cell, 23, 651-662. doi:10.1016/j.molcel.2006.07.004
- Harvey, H., Habash, M., Aidoo, F. and Burrows, L.L. (2009) Single residue changes in the C terminal disulfidebonded loop of the Pseudomonas aeruginosa type IV pilin influence pilus assembly and twitching motility. Journal of Bacteriology, 191, 6513-6524. doi:10.1128/JB.00943-09
- Xiao, Y.H., Yin, M.H., Hou, L., Luo, M. and Pei, Y. (2007) Asymmetric overlap extension PCR method bypassing intermediate purification and the amplification of wildtype template in site-directed mutagenesis. Biotechnology Letters, 29, 925-930. doi:10.1007/s10529-007-9327-4
- Rekecki, A., Gunasekara, R.A.Y.S.A., Dierckens, K., Laureau, S., Boon, N., Favoreel, H., et al. (2012) Bacterial host interaction of GFP-labelled Vibrio anguillarum HI- 610 with gnotobiotic sea bass, Dicentrarchus labrax L., larvae. Journal of Fish Diseases, 35, 265-273. doi:10.1111/j.1365-2761.2011.01342.x
- Hong, G.E., Kim, D.G., Bae, J.Y., Ahn, S.H., Bai, S.C. and Kong, I.S. (2007) Species specific PCR detection of the fish pathogen Vibrio anguillarum, using the amiB gene, which encodes N-acetylmuramoyl-L-alanine amidase. FEMS Microbiology Letters, 269, 201-206. doi:10.1111/j.1574-6968.2006.00618.x
- Forslund, A.L., Salomonsson, E.N., Golovliov, I., Kuoppa, K, Michell, S., Titball, R., et al. (2010) The type IV pilin, PilA, is required for full virulence of Francisella tularensis subspecies tularensis. BMC Microbiology, 10, 227. doi:10.1186/1471-2180-10-227
- Aagesen, A.M. and Häse, C.C. (2012) Sequence analyses of type IV pili from Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus. Microbial Ecology, 64, 509- 524. doi:10.1007/s00248-012-0021-2
- Hsiao, A., Liu, Z., Joelsson, A., Zhu, J. (2006) Vibrio cholerae virulence regulator-coordinated evasion of host immunity. Proceedings of the National Academy of Sciences USA, 103, 14542-14547. doi:10.1073/pnas.0604650103
- Meibom, K.L., Li, X.B., Nielsen, A.T., Wu, C.Y., Roseman, S. and Schoolnik, G.K. (2004) The Vibrio cholerae chitin utilization program. Proceedings of the National Academy of Sciences USA, 101, 2524-2529. doi:10.1073/pnas.0308707101
- Frischkorn, K., Stojanovski, A. and Paranjpye, R. (2013) Vibrio parahaemolyticus type IV pili mediate interactions with diatom-derived chitin and point to an unexplored mechanism of environmental persistence. Environmental Microbiology. doi:10.1111/1462-2920.12093
- Zaku, S.G., Emmanuel, S.A., Aguzue, O.C. and Thomas, S.A. (2011) Extraction and characterization of chitin; a functional biopolymer obtained from scales of common carp fish (Cyprinus carpio L.): A lesser known source. African Journal of Food Science, 5, 478-483.
- Chiavelli, D.A., Marsh, J.W. and Taylor, R.K. (2001) The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Applied and Environmental Microbiology, 67, 3220-3225. doi:10.1128/AEM.67.7.3220-3225.2001
- Simon, R., Priefer, U. and Pühler, A. (1983) A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram-negative bacteria. Biotechnology, 1, 784-791. doi:10.1038/nbt1183-784
- Philippe, N., Alcaraz, J.P., Coursange, E., Geiselmann, J. and Schneider, D. (2004) Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid, 51, 246-255. doi:10.1016/j.plasmid.2004.02.003

