Agricultural Sciences
Vol. 4 No. 2 (2013) , Article ID: 28198 , 6 pages DOI:10.4236/as.2013.42013
Expression and localization of the spore wall protein SWP26 of Nosema bombycis in the silkworm BmN cell line
![]()
1Jiangsu University of Science and Technology, Zhenjiang, China
2Sericultural Research Institute, Chinese Academy of Agricultural Sciences, Zhenjiang, China; *Corresponding Author: guoxijie@126.com
Received 26 December 2012; revised 25 January 2013; accepted 31 January 2013
Keywords: Bacmid; Expression; Microsporidia; Nosema bombycis; Spore Wall Protein
ABSTRACT
The microsporidian spore wall proteins, as the main components of the spore wall, play a key role in spore adherence to host cells and in recognition of the parasite by the host during the invasion process. In this study, we used the Bac-to-Bac baculovirus expression system to express the spore wall protein SWP26, fused to enhanced green fluorescent protein (EGFP), in the silkworm BmN cell line. The SWP26 and EGFP genes were inserted into the baculovirus transfer vector pFastBac1. The transfer vector pFastBac1-swp26-egfp was transformed into the bacterium Escherichia coli DHl0Bac/Bombyx mori nucleopolyhedrovirus (BmNPV) to construct the recombinant vBmswp26-egfp bacmid. The vBmswp26-egfp bacmid DNA was then used to transfect BmN cells to obtain the recombinant baculovirus. Western blotting analysis of total protein lysates in BmN cells infected by the recombinant virus showed a protein band of approximately 51 kDa, which corresponded to the deduced molecular weight of the swp26-egfp fusion protein. In addition, a fluorescence signal was observed in the cytoplasm and nucleoplasm of transfected cells, indicating that SWP26 had been successfully expressed in BmN cells. The SWP26 expression system established in this study lays the foundation for additional molecular and cellular studies, especially those focused on the interaction between the SWP26 protein of Nosema bombycis and the proteins of the silkworm, Bombyx mori.
1. INTRODUCTION
Microsporidia are a large group of highly adapted obligate intracellular parasites, which can infect a wide range of vertebrate and invertebrate hosts, including insects, fish, crustaceans, and mammal groups [1]. Nearly all insects are susceptible to this pathogen, but over half of the susceptible insect hosts, such as the silkworm Bombyx mori and the mosquito Anopheles stephensi, belong to one of the following two orders, Lepidoptera and Diptera [2,3]. Most of the entomopathogenic microsporidia belong to the genus Nosema, with more than 150 described species from 12 insect orders [2,3]. Nosema bombycis, the microsporidia first described in the literature, is a silkworm parasite that causes severe economic losses in the sericulture industry.
Microsporidia can survive outside the host for several years as dormant microscopic spores. Key to the protection of these spores is a thick spore wall composed of a double layer of a chitin-rich endospore and a proteinbased exospore [4]. The spore wall provides structural rigidity and protects the mature spore from the environment. Recent research suggested that the spore wall proteins (SWPs) are involved in the complex infection mechanism, and are responsible for adherence of the microsporidian spores to host cells, which is an integral part of activation and host cell infection [5]. To date, three exosporal proteins of Encephalitozoon species have been reported: ExP1 in E. cuniculi [6], SWP1 in both E. cuniculi and E. intestinalis, and SWP2 in E. intestinalis alone [7,8]. Three endosporal proteins (EnP1/SWP3, EnP2, and EcCDA) have been identified only in E. cuniculi [9-11]. In addition to these, five spore wall proteins have been studied in N. bombycis: three exosporal proteins (SWP5, SWP26, and SWP32) and two endosporal protein (SWP25 and SWP30) [12-16].
Previous studies showed that SWP26 of N. bombycis contains a C-terminal heparin-binding motif (HBM) that is known to interact with extracellular glycosaminoglycans. Transmission immuno-electron microscopy revealed that SWP26 protein was expressed at high levels in the endospore and the plasma membrane during endospore development, and that it was distributed in the exospore of mature spores [13]. It was suggested that SWP26 is involved in endospore formation and host cell adherence [13]. In this study, we used the Bac-to-Bac baculovirus expression system to express SWP26 fused to enhanced green fluorescent protein (EGFP) in BmN cells, to establish a system for screening for proteins in the silkworm that interact with SWP26.
2. MATERIALS AND METHODS
2.1. Materials
The spores of N. bombycis (Zhenjiang isolate) were preserved at the Sericultural Research Institute, Chinese Academy of Agricultural Sciences (SRI-CAAS). The BmN cells were cultured at 27˚C in TC-100 insect medium (AppliChem Inc., Germany) supplemented with 10% fetal calf serum (Gibco BRL, USA).The bacterial strain Escherichia coli DH10Bac/Bombyx mori nucleopolyhedrovirus (BmNPV) was kindly provided by Professor Zhang ChuanXi (Zhejiang University, China). The pFastBac1 donor vector and the Cellfectin®ⅡReagent were purchased from Invitrogen Corporation, USA. The LA-Taq enzyme, restriction enzymes, pMD18-T vectors, and T4 DNA ligase were purchased from Takara Company (Takara Bio Inc., Japan).
2.2. DNA Preparation and Gene Cloning
Genomic DNA of N. bombycis was extracted as described by Dong et al. [17]. The full length SWP26 gene, including the signal peptide region, was amplified using the primers SWP26-F (5’-CGGAATTCATGAATATTATCATCTTCAGCT-3’, the EcoR I site is underlined) and SWP26-R (5’-CGCTCGAGATCATCAATTCCAGAAATAC-3’, the Xho I site is underlined). The EGFP gene was amplified from the pLEGFP-C1 plasmid with primers EGFP-F (5’-AACTCGAGATGGTGAGCAAGG-3’, the Xho I site is underlined) and EGFP-R (5’-CGAAGCTTTTACTTGTACAGCTCG-3’, the Hind Ш site is underlined). Polymerase chain reactions were performed according to standard conditions with an annealing temperature of 56˚C. The amplified products were subjected to 1.0% agarose gel electrophoresis, excised and purified, cloned into pMD18-T vectors, and the cloned fragments were then sequenced.
2.3. Construction of the Recombinant Bacmid
The SWP26 gene PCR product was digested with EcoR I and Xho I, and inserted into the multiple cloning site (MCS) of the donor plasmid, pFastBac1, within the same restriction enzyme sites to obtain the pFastBac1- swp26 plasmid. Then, the Xho I/Hind Ш-digested EGFP gene was inserted into pFastBac1-swp26 to generate the transfer plasmid, pFastBac1-swp26-egfp, containing the SWP26 gene fused to the EGFP reporter gene, so that the resulting protein product would have SWP26 fused to EGFP at the C-terminus. The transfer plasmid was sequenced and was transformed into E. coli DH10Bac/ BmNPV to construct the recombinant bacmid baculovirus. According to the instructions in the manual for the Bac-to-Bac® baculovirus expression systems (Invitrogen), recombinant bacmid DNA was isolated from the DH10- Bac/BmNPV cells using the QIAGEN large-construct kit (Qiagen, Valencia, CA) and quantified by spectrometry. The recombinant bacmids were identified by blue/white selection and PCR analysis using the primers M13+ and M13–. The verified recombinant bacmid was designated as vBmswp26-egfp.
2.4. Transfection and Fluorescence Detection
Recombinant bacmid DNA (vBmswp26-egfp) was transfected into BmN cells (2 × 106 cells/flask) using the Cellfectin®II Reagent. After 3 days, EGFP fluorescence in BmN cells was observed and photographed with an inverted fluorescence microscope (Olympus IX7, Japan). After 5 days, the cells were harvested by centrifuging at 500 × g for 5 min and the supernatant was saved for the next cycle of infection.
2.5. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Western Blotting Analysis
The collected cells were lysed using RIPA lysis buffer (50 mM Tris, 150 Mm NaCl, 1% NP-40, and 0.25% sodium deoxycholate, at pH 7.4) and centrifuged. The lysate was centrifuged and the proteins in the supernatant were separated by SDS-PAGE on 12% polyacrylamide gels, transferred onto polyvinylidene difluoride (PVDF) membranes and blocked under standard conditions. The primary antibody (GFP Epitope Tag; Invitrogen) and secondary antibody (anti-rabbit horse radish peroxidase [HRP]; Sigma) were used at 1:2000 and 1:25,000, respectively. The protein-antibody complexes were visualized using diaminobenzidine tetrahydrochloride (DAB) as the chromogenic HRP substrate.
2.6. Confocal Laser-Scanning Microscopy
BmN cells were infected with vBmswp26-egfp and collected at 12, 24, and 48 h post infection. The cells were rinsed three times with phosphate-buffered saline (PBS) and fixed in a 1:1 mixture of methanol and acetone on ice for 15 min, followed by three washes with PBS. After washing, cells were incubated with the nuclear DNAspecific stain DAPI (Sigma, USA) for 1 h. Cells were directly observed using a Leica TCS SP5 confocal laserscanning microscope.
3. RESULTS
3.1. Cloning of the SWP26 Gene and EGFP Gene
A 685-bp DNA fragment (including the restriction sites) encompassing the SWP26 gene of N. bombycis and a 736-bp DNA fragment (including the restriction sites) encompassing the EGFP gene were synthesized by PCR using primer sets specific to the corresponding genes (Figure 1(a)).
3.2. Construction of the Recombinant Bacmid
Sequencing and restriction endonuclease digestion (Figure 1(b)) were performed to verify the insertion of the SWP26 and EGFP gene sequences in the recombinant donor plasmid.
The recombinant bacmids were selected by blue/white selection, and the white clones were further identified by PCR with the M13+ and M13– primers. A 3.7 kb specific band was detected in the positive clones, while a 300-bp band was detected in the negative control (blue clones) (Figure 1(c)). These results indicated that all the white
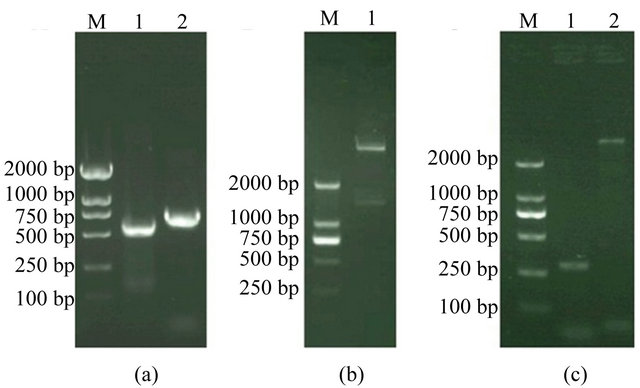
Figure 1. (a) Electrophoresis of the products of PCR. Lane M, marker DL2000; Lane 1, PCR product for SWP26 gene. Lane 2, PCR product for EGFP gene; (b) Electrophoretic identification of the recombinant DNA. Lane M, marker DL2000; Lane 1, recombinant DNA (pFastBac1-swp26-egfp) cleaved by EcoR I and Hind Ш; (c) The recombinant bacmid baculoviruses vBmegfp-swp26 was analyzed by PCR with M13+ and M13– primers. Lane M, marker DL2000; Lanes 1, PCR product of the recombinant bacmid vBmegfp-swp26; Lane 2, PCR product of blank bacmid.
clones were positive clones. Therefore, we extracted bacmid DNA from the positive clones for the following cell transfection.
3.3. Expression of the swp26-egfp Fusion Protein in BmN Cells
The recombinant bacmid baculovirus, vBmswp26-egfp, was transfected into BmN cells and the EGFP fluorescence was monitored (Figure 2). The cells transfected with the recombinant bacmid exhibited cytopathic effects characteristic of baculovirus infections.The nucleus of the transfected cell enlarged, and some of the cells started floating at later stages. At 96 h after infection with the recombinant baculoviruses at a multiplicity of infection (MOI) of 10, the cells were collected and lysed and the clarified lysates were subjected to Western blotting analysis. A GFP-specific antibody was used to identify the swp26-egfp fusion protein, with an approximate molecular weight of 51 kDa (Figure 3). Fluorescence detection and Western blotting analysis indicated that the swp26-egfp fusion protein was successfully expressed in the transfected BmN cells.
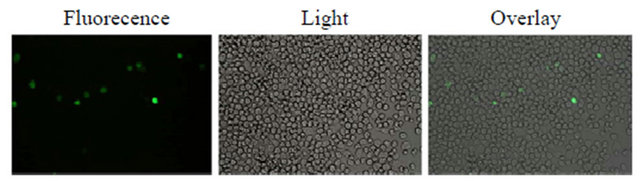
Figure 2. Images of BmN cells transfected with vBmegfp-swp26 obtained using inverted fluorescence and light micro-scopes.
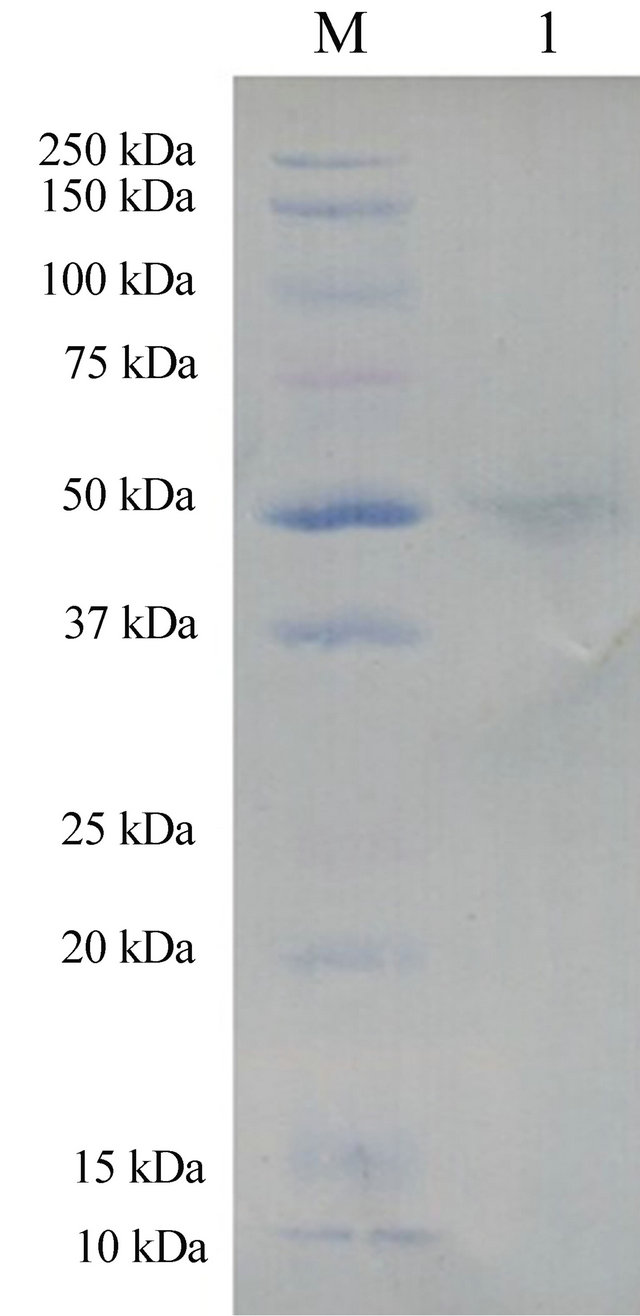
Figure 3. Recombinant swp26- egfp protein expression analysis by western blotting.
3.4. Localization of SWP26 in vBmswp26-egfp-Infected Cells
To detect the subcellular localization of swp26-egfp protein, BmN cells were infected with vBmswp26-egfp at an MOI of 5. Cells were then collected for fluorescence observation at 12, 24, and 48 h post infection (p.i.). From 12 to 48 h p.i., fluorescence was observed within the cytoplasm and the nucleoplasm. Moreover, fluorescence was mainly distributed in the cytoplasm at 12 h p.i., and high levels of fluorescence could also be observed within the nucleoplasm with the passage of time (at 48 h p.i.) (Figure 4). These data indicate that the swp26-egfp fusion protein was localized mainly in the cytoplasm at 12 h, and was subsequently localized in both the cytoplasm and nucleoplasm.
4. DISCUSSION
The interaction between the host and parasite is an important factor that determines the course of the infection, by influencing how effectively the parasite invades the host and develops in the host cells. Recent studies indicated that some microsporidian spores adhere to the surface of the host cell through special mechanisms during the infection process of microsporidia and that adherence precedes spore activation [5,18]. This sequence of events can induce a more efficient rate of infection. During the infection process, the spore wall of microsporidia directly contacts the host cell. Furthermore, the spore wall proteins, as the main components of the spore wall, play a key role in spore adherence to host cells and in recognition of the pathogen by the host during the invasion process. Therefore, the presence or absence of spore wall proteins will directly influence spore germination and virulence of microsporidia [19-22]. Immunoelectron microscopy analysis showed that N. bombycis SWP26 is present in both immature and mature spores. The in vitro binding assays showed that the SWP26 protein has an ability to bind directly to the host cell surface. Li et al. (2009) suggested that SWP26 might be involved in the interaction between spores and host cells, via the C-terminal HBM.
The most direct research methods available for functional gene analysis, such as transgenic models, gene knockouts, and RNA interference (RNAi), cannot be used for microsporidia because of the peculiarity of microsporidian species. The role of microsporidian spore wall proteins and other important functional proteins in the infection process are not clear yet. In this study, we have established a silkworm cultured BmN cell linebased system for expressing GFP-tagged SWP26, to address the function of SWP26 of N. bombycis and screen for SWP26-interacting host proteins. We successfully used the Bac-to-Bac baculovirus expression system [23] to express swp26-egfp fusion protein in BmN cells. The use of natural host cells of microsporidia to express mi-
Figure 4. Intracellular localization of swp-egfp protein in vBmswp26-egfp-infected BmN cells. Cells were collected at 12, 24 and 48 h p.i. and observed using a confocal laser-scanning microscope.
crosporidia-specific proteins not only allows the expressed proteins to retain their natural activity, but also enables us to screen for interacting proteins in host cells by co-immunoprecipitation.
To determine the subcellular localization of SWP26 in BmN cells, we cloned the full-length SWP26 gene (GenBank accession number: EU677842) along with the signal peptide region and expressed it as a swp26-egfp fusion protein in BmN cells. The results showed that SWP26 was distributed in the cytoplasm and the nucleoplasm, and not limited to the cytoplasm or the nucleoplasm alone. These data also suggest that cytoplasmic and nuclear proteins involved in the host-parasite interaction and in the immunological responses of the host cell to N. bombycis may interact with SWP26. The successful application of this SWP26 expression system lays the foundation for further studies probing the interactions between SWP26 of N. bombycis with proteins of the silkworm.
5. ACKNOWLEDGEMENTS
This work was supported by the National Basic Research Program of China (Grant No. 2012CB114600) and the earmarked fund for Modern Agro-industry Technology Research System. We are grateful to everyone who generously provided us with access to the software used in this work. We also thank all our partners and laboratory members for their kind help and criticism.
REFERENCES
- Wittner, M. and Weiss, L.M. (1999) The microsporidia and microsporidiosis. ASM Press, Washington DC.
- Becnel, J.J. and Andreadis, T.G. (1999) Microsporidia in insects. In: Wittner, M. and Weiss, L.M. Eds., The Microsporidia and Microsporidiosis, ASM Press, Washington DC, 447-501.
- Tanada, Y. and Kaya, H.K. (1993) Insect pathology. Academic Press, New York.
- Vávra, J. and Larsson, J.I.R. (1999) Structure of the microsporidia. In: Wittner, M. and Weiss, L.M., Eds., The Microsporidia and Microsporidiosis, ASM Press, Washington, DC, 7-84.
- Southern, T.R., Jolly, C.E., Lester, M.E. and Hayman, J.R. (2007) EnP1, a microsporidian spore wall protein that enables spores to adhere to and infect host cells in vitro. Eukaryotic Cell, 6, 1354-1362. doi:10.1128/EC.00113-07
- Brosson, D., Kuhn, L., Delbac, F., Garin, J., Vivares, C.P. and Texier, C. (2006) Proteomic analysis of the eukaryotic parasite Encephalitozoon cuniculi (microsporidia): A reference map for proteins expressed in late sporogonial stages. Proteomics, 6, 3625-3635. doi:10.1002/pmic.200500796
- Bohne, W., Ferguson, D.J., Kohler, K. and Gross, U. (2000) Developmental expression of a tandemly repeated, glycineand serine-rich spore wall protein in the microsporidian pathogen Encephalitozoon cuniculi. Infection and Immunity, 68, 2268-2275. doi:10.1128/IAI.68.4.2268-2275.2000
- Hayman, J.R., Hayes, S.F., Amon, J. and Nash, T.E. (2001) Developmental expression of two spore wall proteins during maturation of the microsporidian Encephalitozoon intestinalis. Infection and Immunity, 69, 7057-7066. doi:10.1128/IAI.69.11.7057-7066.2001
- Brosson, D., Kuhn, L., Prensier, G., Vivares, C.P. and Texier, C. (2005) The putative chitin deacetylase of Encephalitozoon cuniculi: A surface protein implicated in microsporidian spore-wall formation. FEMS Microbiology Letters, 247, 81-90. doi:10.1016/j.femsle.2005.04.031
- Peuvel-Fanget, I., Polonais, V., Brosson, D., Texier, C., Kuhn, L., Peyret, P., Vivares, C. and Delbac, F. (2006) EnP1 and EnP2, two proteins associated with the Encephalitozoon cuniculi endospore, the chitin-rich inner layer of the microsporidian spore wall. International Journal for Parasitology, 36, 309-318. doi:10.1016/j.ijpara.2005.10.005
- Xu, Y., Takvorian, P., Cali, A., Wang, F., Zhang, H., Orr, G. and Weiss, L.M. (2006) Identification of a new spore wall protein from Encephalitozoon cuniculi. Infection and Immunity, 74, 239-247. doi:10.1128/IAI.74.1.239-247.2006
- Cai, S., Lu, X., Qiu, H., Li, M. and Feng, Z. (2011) Identification of a Nosema bombycis (Microsporidia) spore wall protein corresponding to spore phagocytosis. Parasitology, 138, 1102-1109. doi:10.1017/S0031182011000801
- Li, Y., Wu, Z., Pan, G., He, W., Zhang, R., Hu, J. and Zhou, Z. (2009) Identification of a novel spore wall protein (SWP26) from microsporidia Nosema bombycis. International Journal for Parasitology, 39, 391-398.
- Wu, Z., Li, Y., Pan, G., Tan, X., Hu, J., Zhou, Z. and Xiang, Z. (2008) Proteomic analysis of spore wall proteins and identification of two spore wall proteins from Nosema bombycis (Microsporidia). Proteomics, 8, 2447- 2461. doi:10.1002/pmic.200700584
- Wu, Z., Li, Y., Pan, G., Zhou, Z. and Xiang, Z. (2009) SWP25, a novel protein associated with the Nosema bombycis endospore. Journal of Eukaryotic Microbiology, 56, 113-118. doi:10.1111/j.1550-7408.2008.00375.x
- Li, Z., Pan, G., Li, T., Huang, W., Chen, J., Geng, L., Yang, D., Wang, L. and Zhou, Z. (2012) SWP5, a spore wall protein, interacts with polar tube proteins in the parasitic microsporidian Nosema bombycis. Eukaryotic Cell, 11, 229-237. doi:10.1128/EC.05127-11
- Dong, S., Shen, Z., Xu, L. and Zhu, F. (2009) Sequence and phylogenetic analysis of SSU rRNA gene of five microsporidia. Current Microbiology, 60, 30-37. doi:10.1007/s00284-009-9495-7
- Hayman, J.R., Southern, T.R. and Nash, T.E. (2005) Role of sulfated glycans in adherence of the microsporidian Encephalitozoon intestinalis to host cells in vitro. Infection and Immunity, 73, 841-848. doi:10.1128/IAI.73.2.841-848.2005
- Cali, A. and Takvorian, P.M. (1999) Developmental morphology and life cycles of the microsporidia. In: Wittner, M. and Weiss, L. Eds., The Microsporidia and Microsporidiosis, American Society of Microbiology, Washington DC, 85-128.
- Enriquez, F.J., Wagner, G., Fragoso, M. and Ditrich, O. (1998) Effects of an anti-exospore monoclonal antibody on microsporidial development in vitro. Parasitology, 117, 515-520. doi:10.1017/S0031182098003345
- Sak, B., Sakova, K. and Ditrich, O. (2004) Effects of a novel anti-exospore monoclonal antibody on microsporidial development in vitro. Parasitology Research, 92, 74- 80. doi:10.1007/s00436-003-0988-1
- Zhang, F., Lu, X., Kumar, V.S., Zhu, H., Chen, H., Chen, Z. and Hong, J. (2007) Effects of a novel anti-exospore monoclonal antibody on microsporidial Nosema bombycis germination and reproduction in vitro. Parasitology, 134, 1551-1558. doi:10.1017/S0031182007002934
- Motohashi, T., Shimojima, T., Fukagawa, T., Maenaka, K. and Park, E.Y. (2005) Efficient large-scale protein production of larvae and pupae of silkworm by Bombyx mori nuclear polyhedrosis virus bacmid system. Biochemical and Biophysical Research Communications, 326, 564-569. doi:10.1016/j.bbrc.2004.11.060

