Agricultural Sciences
Vol.3 No.7(2012), Article ID:24489,5 pages DOI:10.4236/as.2012.37111
The genetic variation of the backcross modified lines developed from the maize line 08-641 selected by different directions
![]()
1Maize Research Institute, Sichuan Agricultural University, Key Laboratory of Crop Genetic Resources and Improvement, Ya’an, China; *Corresponding Author: fman4027@163.com
2Xinjin Rural Development Burean, Chengdu, China
3College of Agronomy, Sichuan Agricultural University, Chengdu, China
Received 1 September 2012; revised 11 October 2012; accepted 17 October 2012
Keywords: Backcross Breeding; Select Direction; Genetic Variation; SSR
ABSTRACT
In this study, two donaors CN962 and 8065 were used to improve the resistance to northern leaf blight of the recurrent parent inbred line 08-641 (R08). A total of 79 lines (BC2F4) were developed by a bidirectional selection based on the similarity and dissimilarity in the shape and color of seeds to R08.The genetic variation of these lines were analyzed by 44 pairs of SSR molecular markers, the result showed that a total of 272 alleles were detected in the improved lines and R08, 123 out of them were detected in the modified lines but discarded in R08. The modified line selected based on dissimialry in the shape and corlor of seeds to R08 have lower genetic similary between R08 than that between the lines selected based on similary in the shape and color of seeds and R08, and the genetic variation of these lines were wider. It concluded that when the backcross breeding were used to improve the maize inbred lines, multidirectional selection based on phenotypic value were contribute to create and keep genetic variation.
1. INTRODUCTION
Backcross breeding is one of the main methods for crop breeding, which can quickly and effectively improve the undesirable traits of elite cultivar, and accelerate the breeding process [1,2]. Backcross breeding in maize (Zea mays L.) has been extensively used to transfer favorable alleles for monogenic traits from donor genotypes to elite inbred lines but high heritability polygenic traits have also been transferred through this method [3-5]. Using backcross breeding to improve the elite maize inbred lines of minor imperfection was an effective method to increase efficiency. Many of the parents of the commercial hybrids were developed from backcross of early elite maize inbred lines [6].
The maize line 08-641(R08) was an extensively used inbred lines in southwest China, it have high general combining abilities, high resistance to adversities, wide adaptabilities and the superior comprehensive agronomic properties, but the resistance to northern leaf blight was inferior relatively [7]. In this study, two donaors namely CN962 and 8065, which have high resistance to northern leaf blight, high general combining abilities and extensively used in north China , were used to improve the resistance to northern leaf blight of the recurrent parent inbred line R08, and 2 typical ears coded as A and B was selected from their BC2F2 families, respectively. The A was similar to R08, and B was dissimilar to R08 in the shape and color of the seed. These two ears were selfcrossed and a bidirectional selection based on the similarity and dissimilarity in the shape and color of seeds to R08 was carried out, respectively. A total of 79 lines (BC2F4) were developed and grouped as AR, ANR, BR and BNR. The AR and ANR present the similar and dissimilar groups developed from A, BR and BNR present the similar and dissimilar groups developed from B, respectively. The genetic variation of these lines was analyzed by SSR molecular markers with the objective to offer some theoretical reference to raise efficiency by using b across breeding in maize.
2. MATERIALS AND METHODS
2.1. The Procedure of the Backcross
Two donaors CN962 and 8065 were used to improve the germination rate, resistance to northern leaf blight of the recurrent parent inbred line R08, and 2 typical ears coded as A and B was selected from their BC2F2 families, respectively. The A was similar to R08, and B was dissimilar to R08 in the shape and color of the seed. These two ears were selfcrossed twice and a bidirectional selection based on the similarity and dissimilarity in the shape and color of seeds to R08 was carried out, respectively. A total of 79 lines (BC2F4) were developed and grouped as AR, ANR, BR and BNR. AR and ANR present the similar and dissimilar groups developed from A, BR and BNR present the similar and dissimilar groups developed from B, respectively (Table 1).
2.2. SSR Analyses
Genomic DNA was isolated following CTAB procedure described by Scott O. R. [8] with minor modification. The PCR amplification was carried out in a PCR amplifier (PTC-100, Bio-Rad, USA). The PCR protocol began with a denaturing setup at 95˚C for 5 minutes; followed by 35 cycles of 1 min at 95˚C (denature), 2 min at 55˚C (anneal), and 2 min at 72˚C (extend), and end with a 10 min extension at 72˚C. The PCR amplification products were separated on a 6% (w/v) denatured polyacrylamide gel and visualized using silver staining.
2.3. Statistical Analyses
The SSR bands were scored as present (1) or absent (0), each of which was treated as an independent character. The data were translated to the format that the software could read according to the user’s manual of software package employed to analyze the data. The indexes were conducted based on the scores. The number and percentage of polymorphic loci, Polymorphism information content (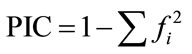 , where fi is the frequency of ith allel), mark index coefficient (MI = A × PIC, where A is the munber of allels in each loci), genetic similarity (
, where fi is the frequency of ith allel), mark index coefficient (MI = A × PIC, where A is the munber of allels in each loci), genetic similarity (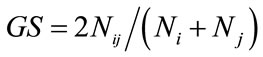 , where
, where  is the number of SSR alleles common to individual i and j, whereas
is the number of SSR alleles common to individual i and j, whereas  and
and 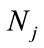 are the total number of SSR alleles observed for individual i and j, respectively) ,were calculated by NTSYS software version pc2.1 [9,10].
are the total number of SSR alleles observed for individual i and j, respectively) ,were calculated by NTSYS software version pc2.1 [9,10].
3. RESULTS AND ANALYSIS
3.1. The Results of SSR Amplification
The results of SSR amplification (Figure 1) showed that the mean PIC and the MI of 44 pairs of markers (Table 2) was 0.780 and 4.91, respectively. On the 44 SSR loci, a total of 272 alleles had been detected in these backcross modified lines. At each locus, the number of alleles varied from 3 to 11, with an average of 6.18. In the 272 alles, out of 123 were detected in the backcross
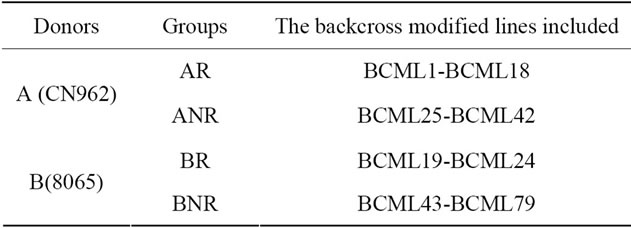
Table 1. The group of the tested backcross modified lines.
modified line while absent in the recurrent parent R08. This result showed that there were genetic difference between R08 and the backcross modified lines, indicated that the gentic variations were produced when the backcrossing were carried out to improve the recurrent parent.
3.2. The Genetic Similarity of the Backcross Modified Lines
The genetic similarity (Table 3) between the 79 backcross modified lines and recurrent parent R08 range from 0.689 (BCML46) to 0.857 (BCML64), with a mean of 0.784, the range was narrow and the mean was large, this results showed that after backcross for twice, the genetic similarity between the 79 backcross modified lines and recurrent parent R08 was close, these lines have obtain most components of genome contents of the recurrent parent.
The average genetic similarity between the AR group and recurrent parent R08 was larger than that between ANR group and R08, and the range was narrow which indicated that AR group was more similar to R08 than ANR group and the genetic variation of ANR group was richer. The average genetic similarity between the BR group and recurrent parent R08 was larger than that between BNR group and R08, and the range was narrow which indicated that BR group was more similar to R08 than BNR group and the genetic variation of BNR group was richer. The results show that the lines develop from the strains have similar shape and color in seeds to R08 were more close to R08 and the lines developed based on dissimilarity in the shape and color of seeds to R08 have generate new genetic variation.
3.3. Cluster Analysis
From UPGMA clusters analysis, the 79 backcross modified lines and R08 were distinctly divided into 5 groups at the similarity level of 0.765 (Figure 2). The first group contains the recurrent parent R08 and 65 backcross modified lines, all the lines from AR and BR group were clustered in this group, and there were 5 and 9 lines from ANR and BNR group did not cluster to this group. The results showed that the lines have dissimilary color and shape in seed have more genetic difference between R08, and more genetic various were detected in
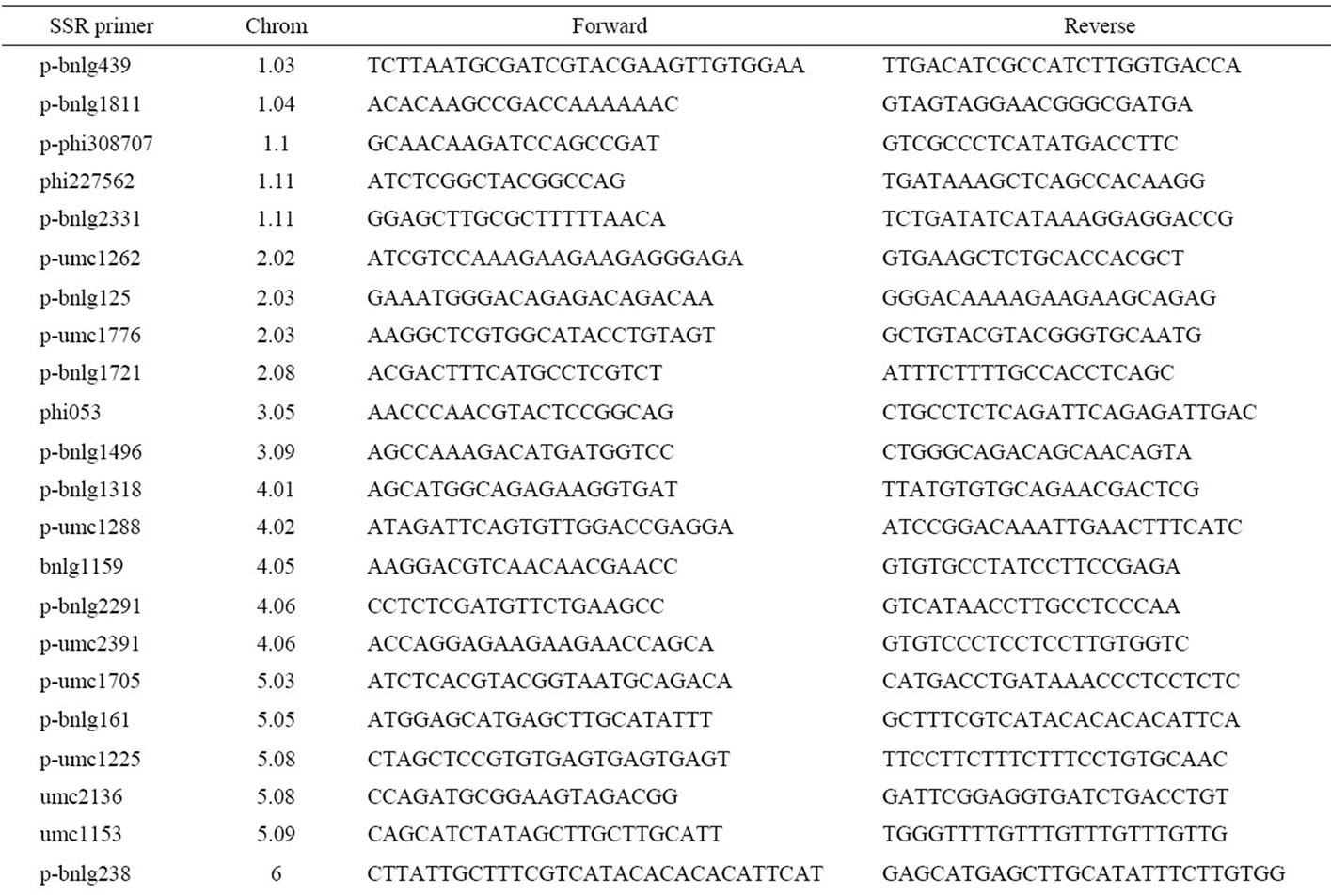

Table 2. The information of the 44 pairs of SSR markers.
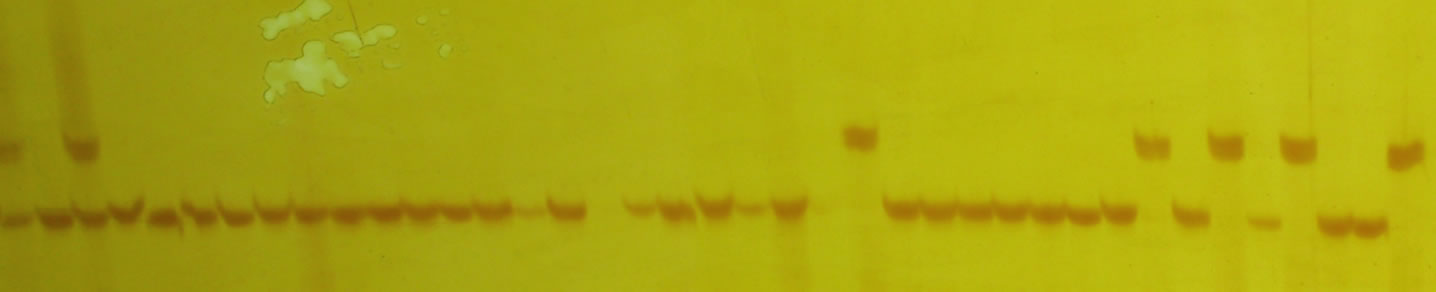
Figure 1. Electrophoresis of part PCR products by primer umc1277.
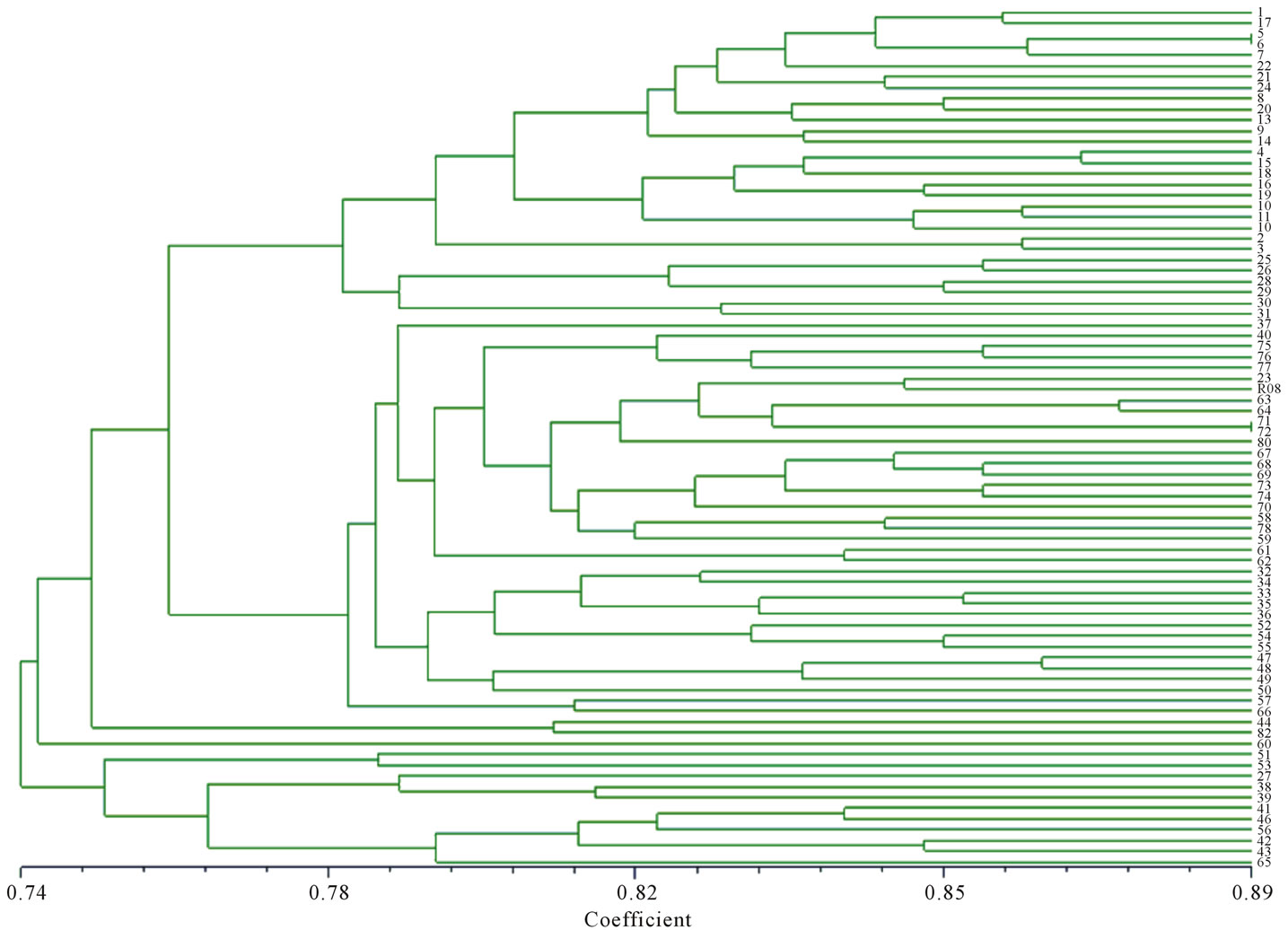
Figure 2. Dendrogram by of the genetic similarity identified with SSR markers.
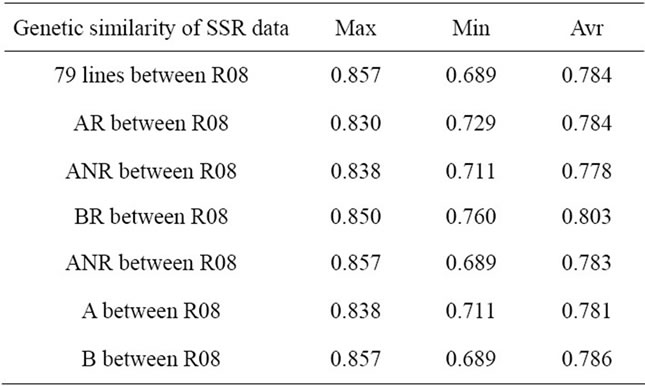
Table 3. Genetic similarity of SSR data.
the dissimilar groups.
4. DISCUSSION
Back crossing is carried out to eliminate the undesirable characteristics and develop useful ones in maize breeding programme, the feasibility of using the method of backcross to breed new disease resistant varieties during the process of pedigree breeding was investigated by different researchers [3-7,10-12]. In this study, two donaors CN962 and 8065 were used to improve the, resistance to northern leaf blight of the recurrent parent inbred line 08-641 (R08). A total of 79 lines (BC2F4) were obtained by a bidirectional selection based on the similarity and dissimilarity in the shape and color of seeds to R08. The result of SSR analyze showed that a total of 272 alleles were detected in the improved lines and R08, and 123 of them were detected in the modified lines but discarded in R08. The modified line selected based on dissimilary in the shape and color of seeds to R08 have lower genetic similary between R08 than that between the lines selected based on similary in the shape and color of seeds and R08, and the genetic varioation of these lines were wider. There were 2 possible reasons, the first reason was that the lines have dissimilary color and shape of seed to R08 accumulate more genetic variation differ from R08, so that more differ loci were detected by SSR markers, and the second reason was that after backcross twice and inbred four generation, rearrangement have happen between different allelic genes, so that new genotype differ from both the donaor and the recurrent parent generated. In that case, when a back cross was carried out to improve the trait controlled by multigene, the type that differ from the recurrent parent in phenotype may be the favorable ones. It concluded that when the backcross breeding were used to improve the maize inbred lines, multidirectional selection based on phenotypic value were contribute to create and keep new genetic variation.
5. ACKNOWLEDGEMENTS
This study was supported by the China Agriculture Research System (CARS-02-07) and the National Key R&D Technologies Program of China (2011AA10A103-2).
![]()
![]()
REFERENCES
- Ho, J.C., Mccouch, S.R. and Smith, M.E. (2002) Improvement of hybrid yield by advance backcross QTL analysis in elite maize. Theoretical and Applied Genetics, 105, 440-480. doi:10.1007/s00122-002-0945-x
- Chen, H.M., Zhang, Y.D., Chen, W., Kang, M.S., Tan, J., Wang, Y.F., Yang, J.Y. and Fan, X.M. (2010) Improving grain yield and yield components via backcross procedure. Maydica, 55, 145-153.
- Melching, A.E. (1990) Use of molecular markers in breeding for oligogenic disease resistence. Plant Breed, 104, 1-9. doi:10.1111/j.1439-0523.1990.tb00396.x
- Hospital, F. and Charcosset, A. (1997) Marker-assisted iotrogression of quantitative trait loci. Genetics, 147, 1469-1485.
- Stuber, C.W. and Sisco, P.H. (1991) Marker facilitated transfer of QTL alleles between elite inbred lines and responses in hybrid. Process 46th Annual Corn and Sorghum Industry Research Conference, American Seed Trade Association, 41, 70-83.
- Duvick, D.N. (1974) Continuous backcrossing to transfer prolificacy to a single-eared inbred line of maize. Crop Science, 14, 69-71. doi:10.2135/cropsci1974.0011183X001400010021x
- Qiao, S.B., Wang, Y.H., Yang, K.C., Rong, T.Z., Pan, G.T. and Gao, S.B. (2009) Effects contributed by different donor parents and backcross times on R08 improvement. Acta Agronomica Sinica, 35, 2187-2196. doi:10.3724/SP.J.1006.2009.02187
- Smith, J.S.C., Chin, E.C.L. and Shu, H. (1997) An evalution of the utility of SSR loci as molecular markers in maize (Zea mays L.): Comparison with data from RFLPs and pedigree. Theoretical and Applied Genetics, 95, 163- 173. doi:10.1007/s001220050544
- Nei, M. and Li, W.H. (1979) Mathemacal model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences of the United States of America, 76, 5256-5273. doi:10.1073/pnas.76.10.5269
- Rolf, J.F. (1998) NTSYS—pc2 Numerical taxonomy and multivariate analysis system (CP). Version 2.0 Exeter Software, Setauket, New York.
- Shang, A.L., Wang, Y.M. and Xi, Z.Y. (2010) Analysis on donor genome contents of backcross progenies detected by ssr markers on maize. Journal of Maize Science, 18, 24-28.
- Chen, H.M., Wang, Y.F., Yao, W.H., Luo, L.M., Li, J.L., Xu, C.X., Fan, X.M. and Guo, H.C. (2011) Utilization potential of the temperate maize inbreds integrated with tropical germplasm. Acta Agronomica Sinica, 37, 1785- 1793. doi:10.3724/SP.J.1006.2011.01785

