Journal of Agricultural Chemistry and Environment
Vol.2 No.3(2013), Article ID:35682,9 pages DOI:10.4236/jacen.2013.23010
Biphenyland carvone-induced protein expression patterns in Rhodococcus sp. ACS
![]()
Received 7 May 2013; revised 21 June 2013; accepted 1 July 2013
Copyright © 2013 Jong-Shik Kim. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ABSTRACT
Protein expression patterns in the polychlorinated biphenyl (PCB)-degrading Rhodococcus sp. strain ACS were examined following growth on two substrates capable of inducing the enantioselective biotransformation of PCBs via different degradation pathways. Eleven inducible proteins were identified by SDS-PAGE and characterized by LC-MS/MS. Four of the peptides, a spore coat protein, an extracellular serine protease, a spoVP, and a molecular chaperonin from Bacillus subtilis, were identified as being unique to biphenyl-induced cells, whereas an extracellular serine protease from B. subtilis was identified as being unique to carvone-induced cells. None of the peptides identified had sequences that corresponded to known dioxygenases or other PCB-degrading enzymes of this Grampositive bacterium, suggesting that the identified induced proteins may be involved in either PCB degradation or adaptive responses that protect cells from toxicity.
Keywords: Polychlorinated Biphenyl (PCB); Carvone; Rhodococcus; Protein Expression
1. INTRODUCTION
Polychlorinated biphenyls (PCBs) were once widely used as coolants and lubricants in transformers, capacitors, and dielectric insulators; however, the production and use of PCBs have since been outlawed due to severe toxicity and environmental contamination. It is estimated that approximately 1.5 million tons of PCBs were produced worldwide before 1988 [1]. Although the production of these compounds was halted due to their longterm persistence, PCBs continue to threaten human health in contaminated areas, as well as through metabolic products that persist in the environment [2,3].
PCBs are commonly degraded by co-metabolic processes involving dioxygenases, which are induced by growth on medium containing biphenyl or other inducing substrates as the sole carbon source [4,5]. Carvone, a plant-derived monoterpene and the principal component of spearmint oil, has several properties that promote PCB biodegradation [6]. Recent evidence suggests that Grampositive and -negative bacteria respond differently to various inducing substrates, and that Gram-positive bacteria are strongly induced to degrade PCBs following exposure to plant-derived monoterpenes [4,6-10]. A Gram-positive bacterium, Rhodococcus sp. ACS, was isolated by Andrew Singer from PCB-contaminated soil obtained from a site in Staten Island, New York, in 1997. The bacterium is similar in morphology to Arthrobacter sp. strain B1B [11] and has the ability to co-metabolize PCBs when grown in the presence of carvone, as well as other terpenes, including citral and cineole [12]. An analysis of the enantioselectivity of Rhodococcus sp. ACS further suggested that multiple degradation pathways are induced depending on the inducing substrate [12]. The key enzymes involved in PCB degradation are dioxygenases, which are polymeric enzymes. To date, most dioxygenase enzymes have been studied in Gramnegative bacteria [13-15]; relatively few dioxygenases have been characterized from Gram-positive bacteria [16-19]. Previously characterized dioxygenase enzymes contain a large subunit that ranges in size from 51,000 to 65,000 Da [13-15] and multiple small subunits that range in size from 22,000 to 27,300 Da [15]. The large and small subunits assemble into holoenzymes that are approximately 200,000 to 250,000 Da in size [13,15].
2. MATERIALS AND METHODS
As a first step toward identifying PCB-degrading enzymes, we compared the protein expression patterns of Rhodococcus sp. strain ACS grown on two different inducing substrates. The bacteria were cultured on either 1000 mg/L biphenyl or 250 mg/L carvone in a mineral salt medium containing 1% fructose. The concentration of carvone used was the maximum concentration that could be tolerated without negatively affecting growth. Soluble proteins were separated by SDS-PAGE and the protein expression patterns analyzed to identify inducible proteins as compared to control cells. The proteins were separated by 15% Laemmli SDS-PAGE, and then stained with Coomassie blue. Proteins that were differentially expressed were excised from the gel. The gel slices were dried in a vacuum and then hydrated in 40 mL of 20 mg/mL trypsin (Promega, Madison, WI) [20]. In-gel digestion was performed for 16 h at 37˚C. Peptides were eluted from the gel slices with 50% (v/v) acetonitrile/5% (v/v) trifluoroacetic acid, and the eluent was dried in a vacuum [20]. A mass spectrometric analysis was performed by LC-MS/MS (Waters, Milford, MA) at the Analytical Chemistry Instrumentation Facility of the University of California, Riverside (Riverside, CA) to determine similarities to other previously characterized proteins in the NCBI protein database. The nucleotide sequence data for Rhodococcus sp. ACS will appear in the GenBank/EMBL/DDBJ nucleotide sequence databases under accession number DQ286393.
3. RESULTS AND DISCUSSION
One-dimensional separation will allow the detection of differentially expressed proteins only if they are not at the same molecular weight as a highly expressed constitutive protein and as long as the bands that are excised are likely to contain multiple proteins, not just one. Coomassie staining will only detect those proteins that are highly expressed in at least one of the treatments. Proteins with lower expression levels may be functionally important but undetectable by these techniques. Thus, the lack of dioxygenases among the identified peptides may reflect these limitations.
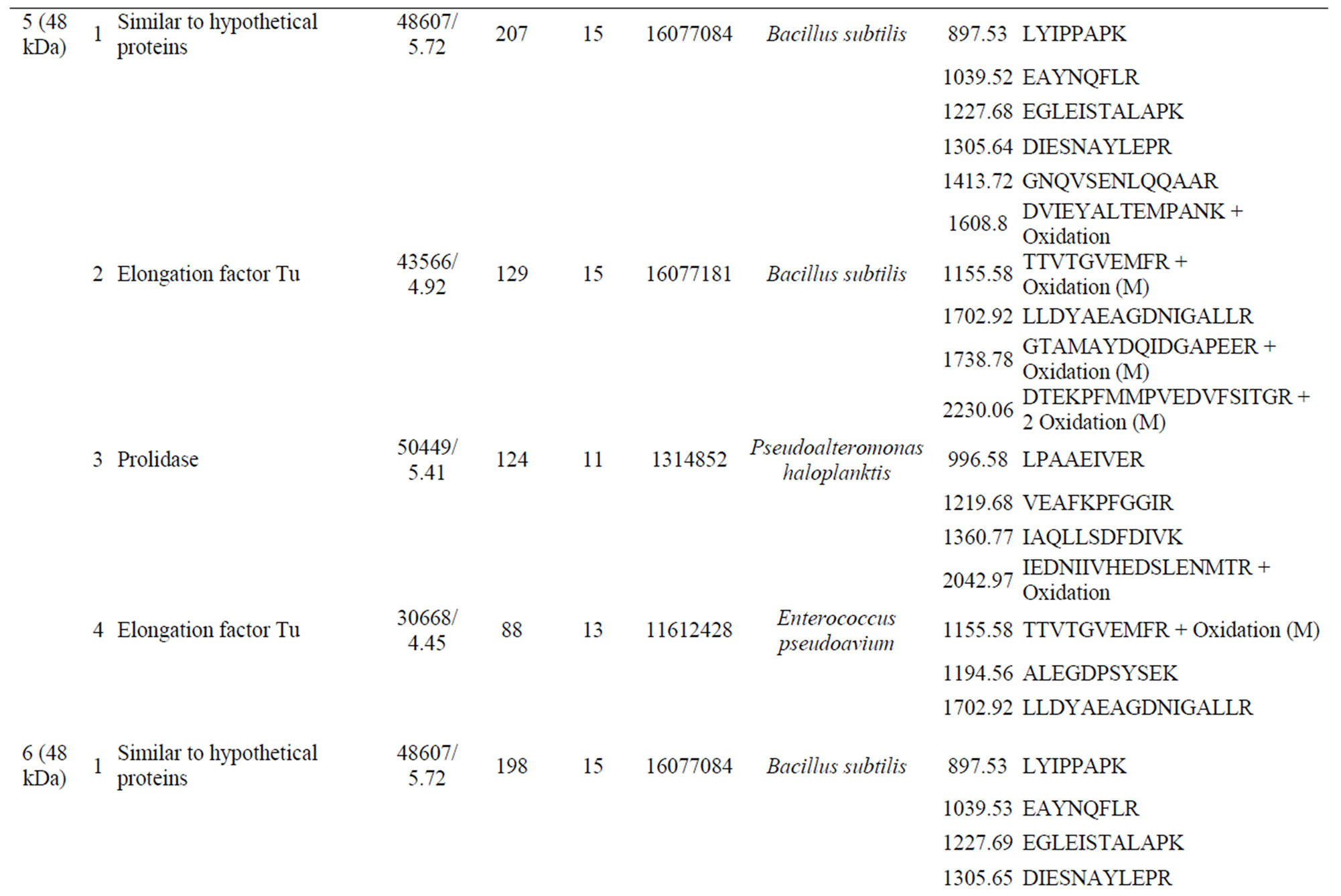
SDS-PAGE revealed differences in the expression levels of at least eleven peptides; these peptides were in the approximate size range of previously described large and small subunits of known dioxygenases [13-15]. The eleven peptides selected for analysis included one that was down-regulated in cells grown on biphenyl and carvone, four peptides that were unique to biphenyl-grown cells, and one peptide that was unique to carvone-grown cells; the remaining four peptides were induced by both biphenyl and carvone (Figure 1).
The eleven proteins were analyzed by LC-MS/MS and compared against the NCBI database using the Mascot algorithm [21] for identification. Most of the peptide sequences exhibited significant homology with previously identified proteins in both Gram-positive and -negative bacteria; however, most of these proteins were either hypothetical or poorly characterized [22]. None of the eleven major inducible proteins corresponded to

Figure 1. SDS-PAGE analysis of extracellular proteins from Rhodococcus sp. strain ACS induced by biphenyl and carvone.
amino acid sequences of dioxygenases previously associated with PCB degradation. The two peptides (Figure 1, peptide bands 3 and 8) expressed by cells during growth on carvone were a strong match to a known extracellular protease. The protein down-regulated by growth on biphenyl and carvone had matches that corresponded to a variety of very different enzymes, including an alcohol dehydrogenase, a serine protease from Bacillus subtilis, and Omp C from Escherichia coli (band 8, Supplementary Table S1). Proteins induced by both biphenyl and carvone (Figure 1, peptide bands 1, 2, 5, and 6) included a small acid-soluble protein from B. subtilis (bands 1 and 2) and an uncharacterized hypothetical protein from B. subtilis (bands 5 and 6).
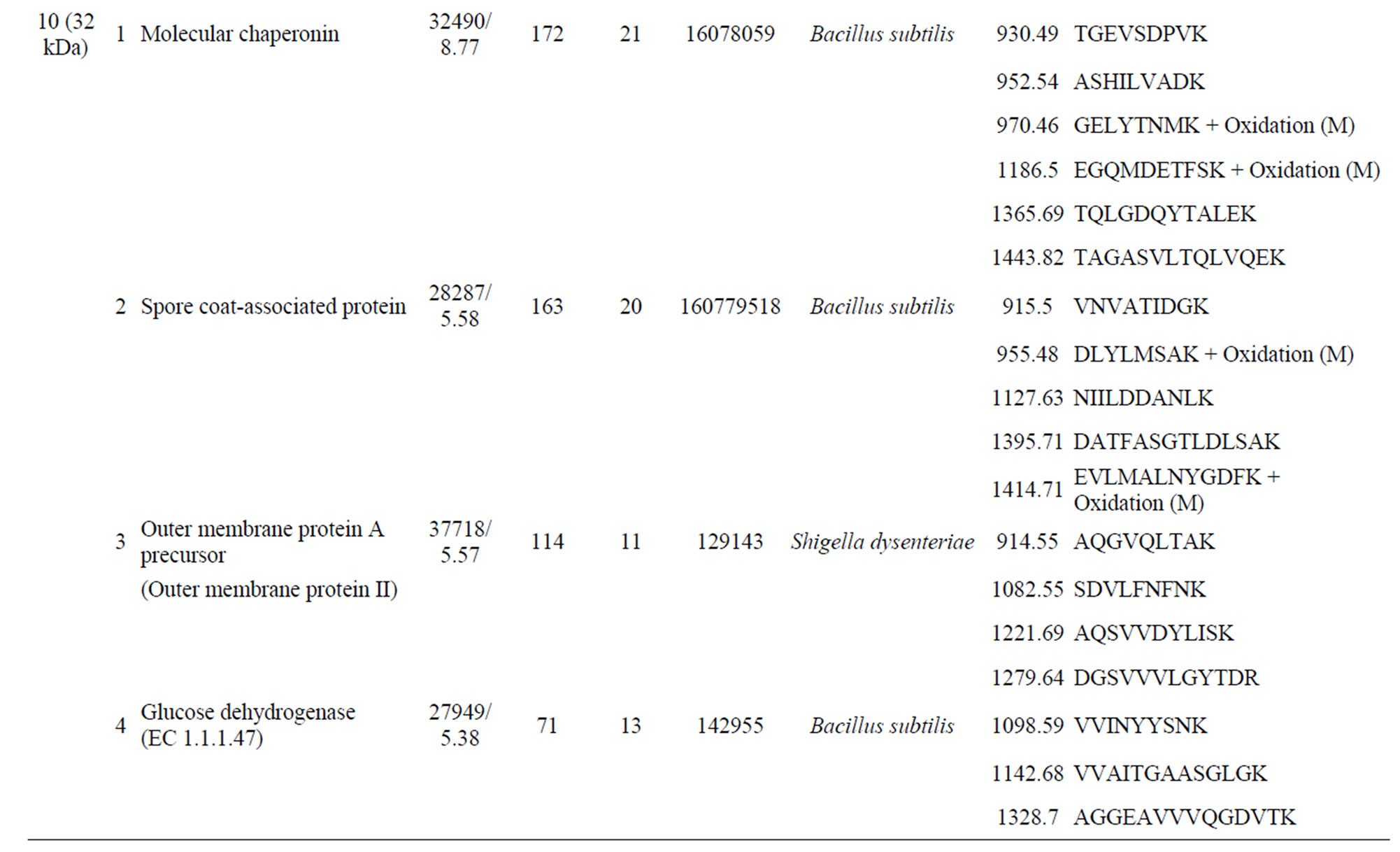
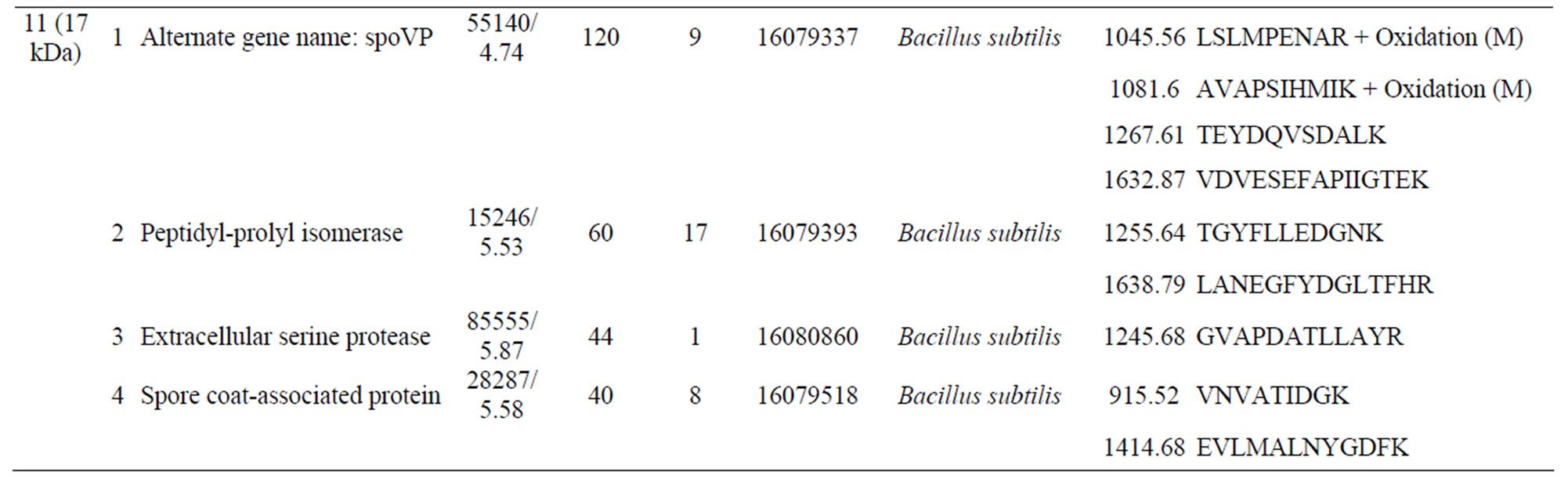
Four peptides were identified as being unique to the biphenyl-grown cells. The first peptide (band 11, 17 kDa) had a match of 120 (Mascot probability score) with spoVP and a match of 60 with peptidyl-prolyl isomerase from B. subtilis; there was also a weak match for a serine protease from B. subtilis (Supplementary Table S1). The second peptide (band 10, 32 kDa) had a probability score of 172 for a molecular chaperonin from B. subtilis, and lower scores for several other peptides, including a spore coat-associated protein from B. subtilis, outer membrane protein II from Shigella dysenteriae, and glucose dehydrogenase (EC 1.1.1.47) from B. subtilis (Sup-
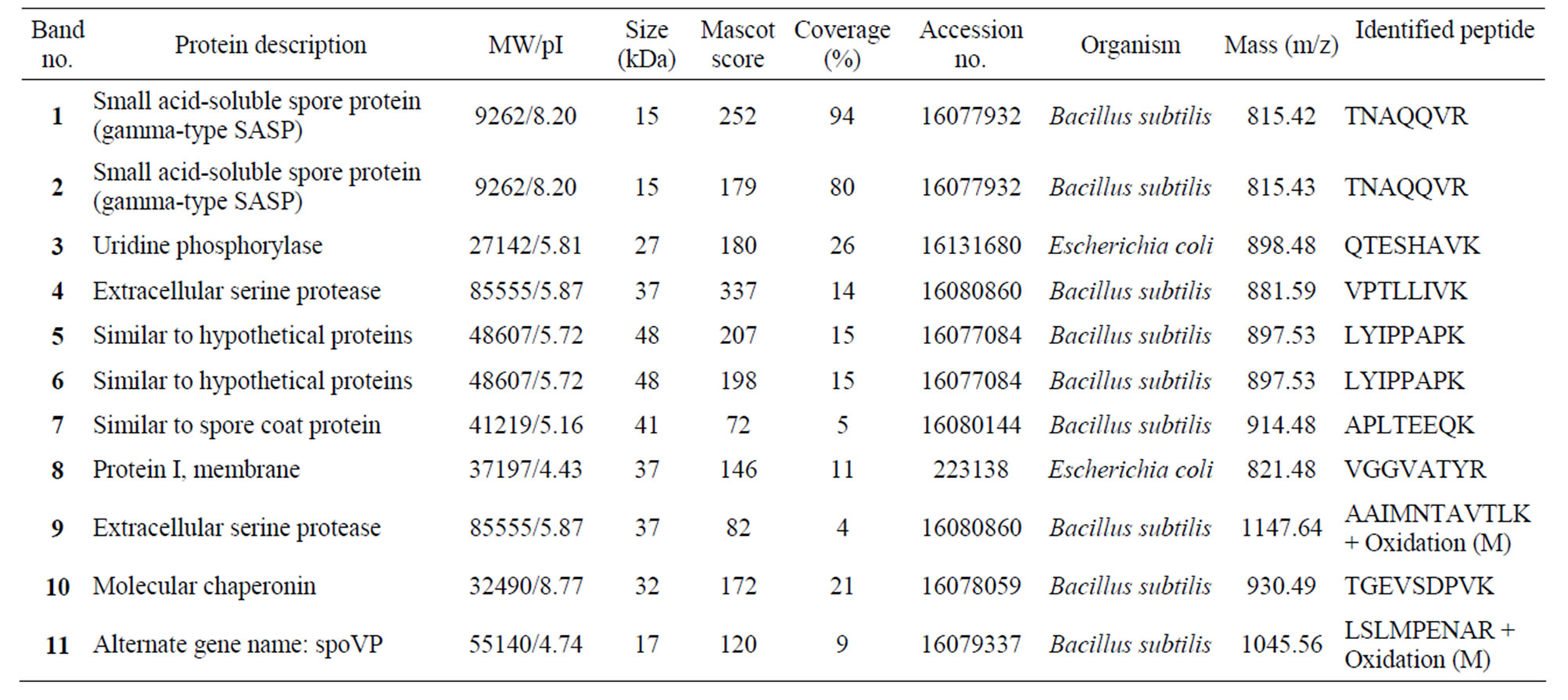
Table 1. List of identified peptide of bands.
plementary Table S1). The third peptide (band 9, 37 kDa) had a match of 82 with extracellular serine protease from B. subtilis, and outer-membrane protein A (outer membrane protein) from E. coli (Supplementary Table S1). The forth peptide (band 7, 41 kDa) had a probability score of 72 for spore coat protein from B. subtilis (Supplementary Table S1). The peptide unique to the carvone-grown cells (band 4) had strong match to an extracellular serine protease from B. subtilis (337) (Table 1).
Generally, a Mascot probability score of >30 indicates similarity to a known enzyme [9]. The results of this study, however, suggest that the proteins from Rhodococcus sp. ACS induced by growth on either carvone or biphenyl have functions unrelated to those from other microorganisms exhibiting high probability scores. The lack of dioxygenases in the protein matches may be due to methodological limitations; moreover, many other proteins may also be important for the response of the bacterium to biphenyl and carvone. Functional analyses, through methods such as transposon mutagenesis, will be necessary to determine the activity of these proteins and their role in PCB degradation. In addition to dioxygenases, PCB degradation pathways contain numerous enzymes involved in stepwise transformations of PCBs and their intermediates. Monoterpenes, including carvone, are toxic at high concentrations and cause cell lysis above 500 ppm [6]. There are minimal estimates of several hundred million genes present in environmental samples, the majority of which code for unknown proteins. It is therefore not surprising that protein sequence alignments are currently unable to provide insight into the degradative enzymes of poorly characterized microorganisms. The identification of major inducible proteins is nonetheless a first step in identifying important degradative enzymes, which can now be targeted for further study.
REFERENCES
- Holoubek, I. (2001) Polychlorinated biphenyl (PCB) contaminated sites worldwide. In: Robertson, L.W. and Hansen, L.G.. Ed., PCBs: Recent Advances in Environmental Toxicology and Health Effects, University of Kentucky Press, Lexington, 17-26.
- Park, J.S., Linderholm, L., Charles, M.J., Athanasiadou, M., Petrik, J., Kocan, A., Drobna, B., Trnovec, T., Bergman, A. and Hertz-Picciotto, I. (2007) Polychlorinated biphenyls and their hydroxylated metabolites (OH-PCBs) in pregnant women from eastern Slovakia. Environmental Health Perspectives, 115, 20-27. doi:10.1289/ehp.8913
- Rezek, J., Macek, T., Mackova, M., Triska, J. and Ruzickova, K. (2008) Hydroxy-PCBs, methoxy-PCBs and hydroxyl-methoxy-PCBs: Metabolites of polychlorinated biphenyls formed in vitro by tobacco cells. Environmental Science & Technology, 42, 5746-5751. doi:10.1021/es800445h
- Zorádová-Murínová, S., Dudášová, H., Lukáčová, L., Certík, M., Silharová, K., Vrana, B. and Dercová, K. (2012) Adaptation mechanisms of bacteria during the degradation of polychlorinated biphenyls in the presence of natural and synthetic terpenes as potential degradation inducers. Applied Microbiology and Biotechnology, 94, 1375-1385. doi:10.1007/s00253-011-3763-8
- Focht, D.D. (1995) Strategies for the improvement of aerobic metabolism of polychlorinated biphenyls. Current Opinion in Biotechnology, 6, 341-407. doi:10.1016/0958-1669(95)80057-3
- Gilbert, E.S. and Crowley, D.E. (1997) Plant compounds that induce polychlorinated biphenyl biodegradation by Arthrobacter sp. strain B1B. Applied and Environmental Microbiology, 63, 1933-1938.
- Gilbert, E.S. and Crowley, D.E. (1998) Repeated application of carvone-induced bacteria to enhance biodegradation of polychlorinated biphenyls in soil. Applied Microbiology and Biotechnology, 50, 489-494. doi:10.1007/s002530051325
- Singer, A.C., Gilbert, E.S., Luepromchai, E. and Crowley, D.E. (2000) Bioremediation of polychlorinated biphenylcontaminated soil using carvone and surfactant-grown bacteria. Applied Microbiology and Biotechnology, 54, 638-843. doi:10.1007/s002530000472
- Singer, A.C., Wong, C.S. and Crowley, D.E. (2002) Differential enantioselective transformation of atropisomeric polychlorinated biphenyls by multiple bacterial strains with different inducing compounds. Applied and Environmental Microbiology, 68, 5756-5759. doi:10.1128/AEM.68.11.5756-5759.2002
- Tandlich, R., Brezna, B. and Dercova, K. (2001) The effect of terpenes on the biodegradation of polychlorinated biphenyls by Pseudomonas stutzeri. Chemosphere, 44, 1547-1555. doi:10.1016/S0045-6535(00)00523-3
- Kohler, H.-P.E., Kohler-Staub, D. and Focht, D.D. (1988) Cometabolism of polychlorinated biphenyls: Enhanced transformation of Aroclor 1254 by growing bacterial cells. Applied and Environmental Microbiology, 54, 1940-1945.
- Singer, A.C., Jury, W., Luepromchai, E., Yahng, C.S. and Crowley, D.E. (2001) Contribution of earthworms to PCB bioremediation. Soil Biology & Biochemistry, 33, 765-776. doi:10.1016/S0038-0717(00)00224-8
- Furukawa, K. and Arimura, N. (1987) Nucleotide sequence of the 2,3-dihydroxybiphenyl dioxygenase gene of Pseudomonas pseudoalcaligenes. Journal of Bacteriology, 169, 924-927.
- Haddock, J.D. and Gibson, D.T. (1995) Purification and characterization of the oxygenase component of biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. Journal of Bacteriology, 177, 5834-5839.
- Hurtubise, Y., Barriault, D., Powlowski, J. and Sylvestre, M. (1995) Purification and characterization of the Comamonas testosteroni B-356 biphenyl dioxygenase components. Journal of Bacteriology, 177, 6610-6618.
- Leigh, M.B., Prouzova, P., Mackova, M., Macek, T., Nagle, D.P. and Fletcher, J.S. (2006) Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Applied and Environmental Microbiology, 72, 2331-2342. doi:10.1128/AEM.72.4.2331-2342.2006
- Benvenuti, M., Briganti, F., Scozzafava, A., Golovleva, L., Travkin, V.M. and Mangani, S. (1999) Crystallization and preliminary crystallographic analysis of the hydroxyquinol 1,2-dioxygenase from Nocardioides simplex 3E: A novel dioxygenase involved in the biodegradation of polychlorinated aromatic compounds. Acta Crystallographica Section D: Biological Crystallography, D55, 901-903. doi:10.1107/S0907444998017715
- Ohmori, T., Morita, H., Tanaka, M., Miyauchi, K., Kasai, D., Furukawa, K., Miyashita, K., Ogawa, N., Masai, E. and Fukuda, M. (2011) Development of a strain for efficient degradation of polychlorinated biphenyls by patchwork assembly of degradation pathways. Journal of Bioscience and Bioengineering, 111, 437-442. doi:10.1016/j.jbiosc.2010.12.002
- Patrauchan, M.A., Miyazawa, D., LeBlanc, J.C., Aiga, C., Florizone, C., Dosanjh, M., Davies, J., Eltis, L.D. and Mohn, W.W. (2012) Proteomic analysis of survival of Rhodococcus jostii RHA1 during carbon starvation. Applied and Environmental Microbiology, 78, 6714-6725. doi:10.1128/AEM.01293-12
- Chang, I.F., Szick-Miranda, K., Pan, S. and Bailey-Serres, J. (2005) Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiology, 137, 848-862. doi:10.1104/pp.104.053637
- Perkins, D. N., Pappin, D.J.C., Creasy, D.M. and Cottrell, J.S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis, 20, 3551-3567. doi:10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2
- Lai, E.M., Phadke, N.D., Kachman, M.T., Giorno, R., Vazquez, S., Vazquez, J.A., Maddock, J.R. and Driks, A. (2003) Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. Journal of Bacteriology, 185, 1443-1454. doi:10.1128/JB.185.4.1443-1454.2003
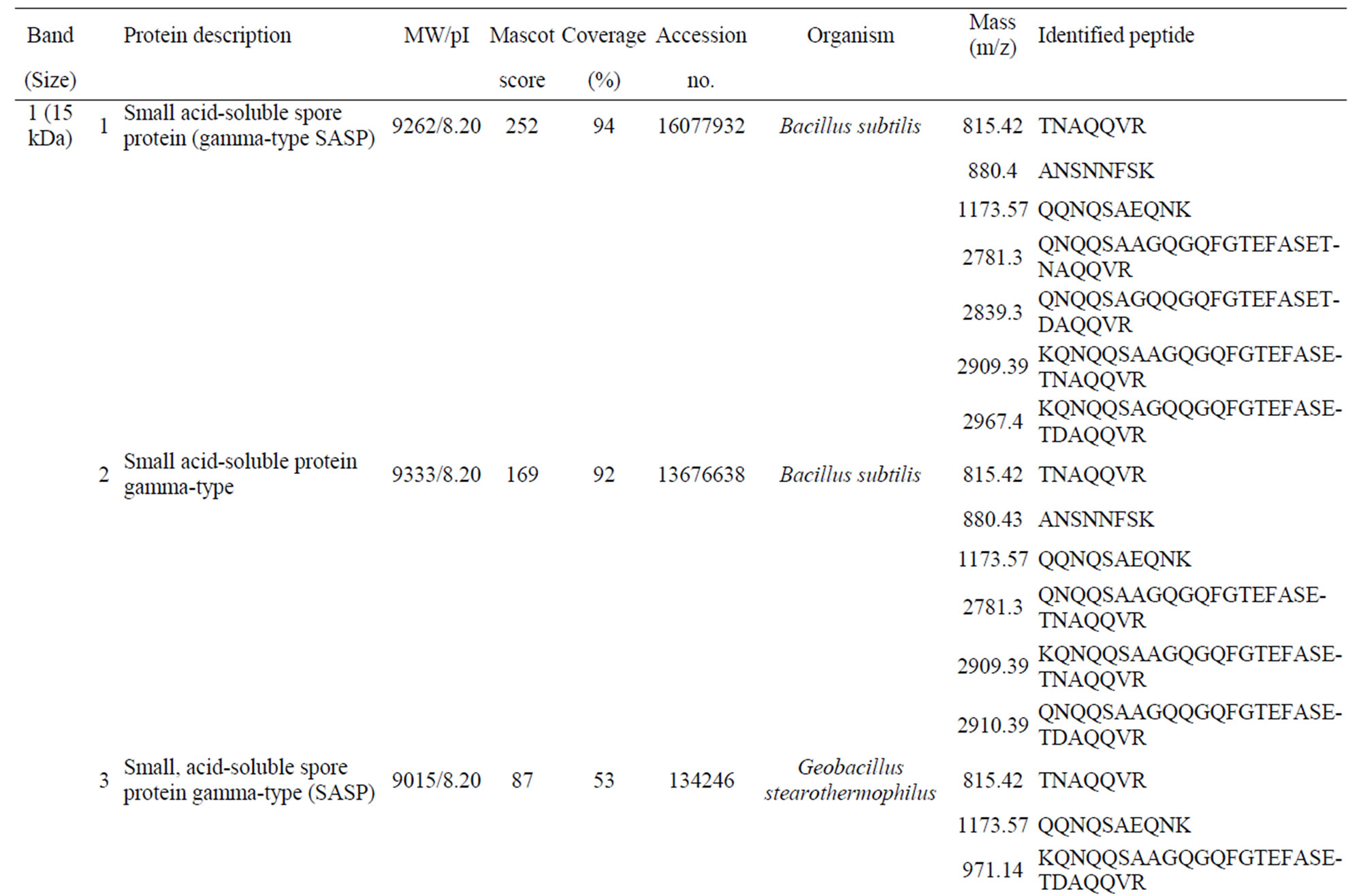
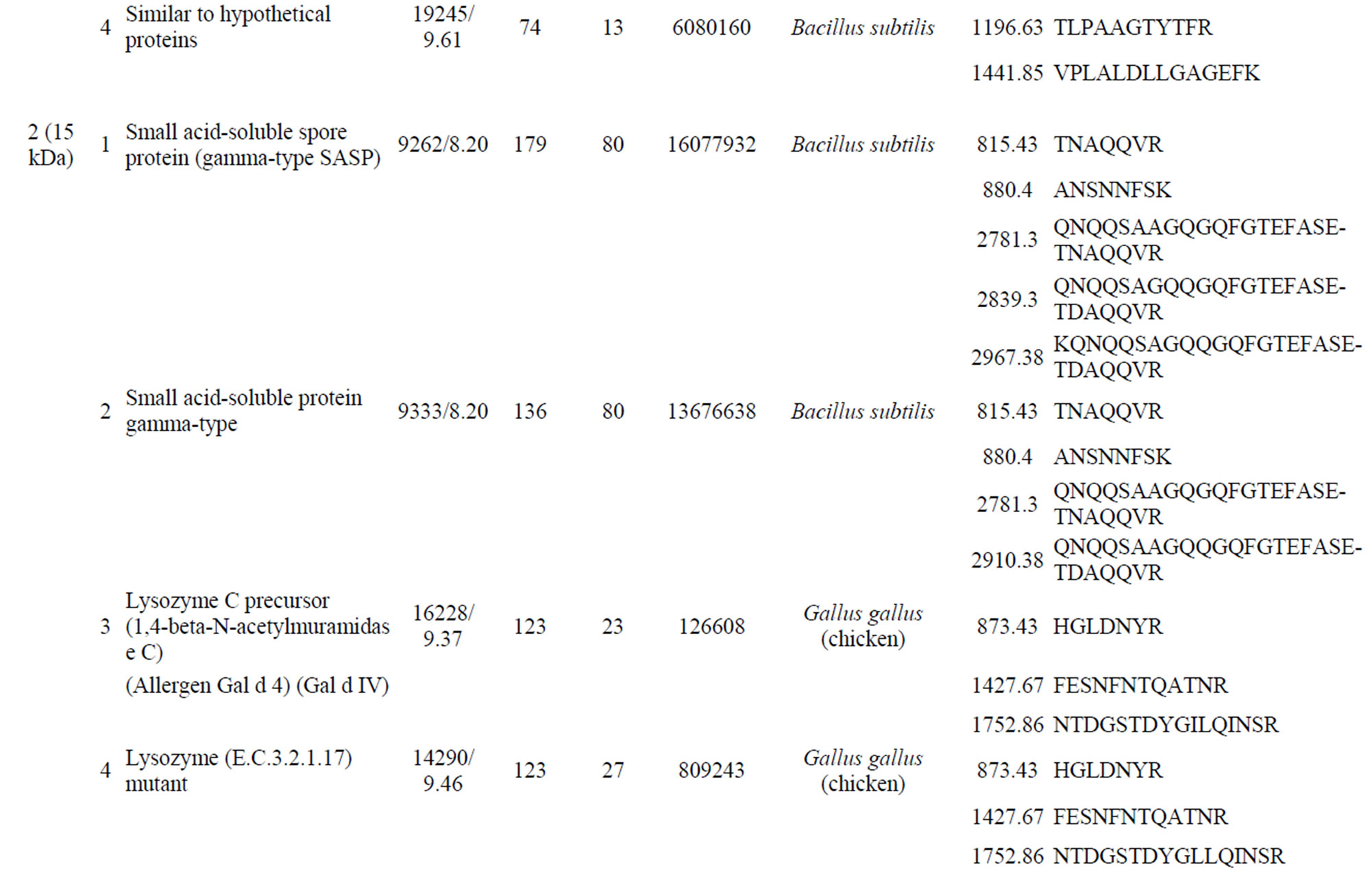
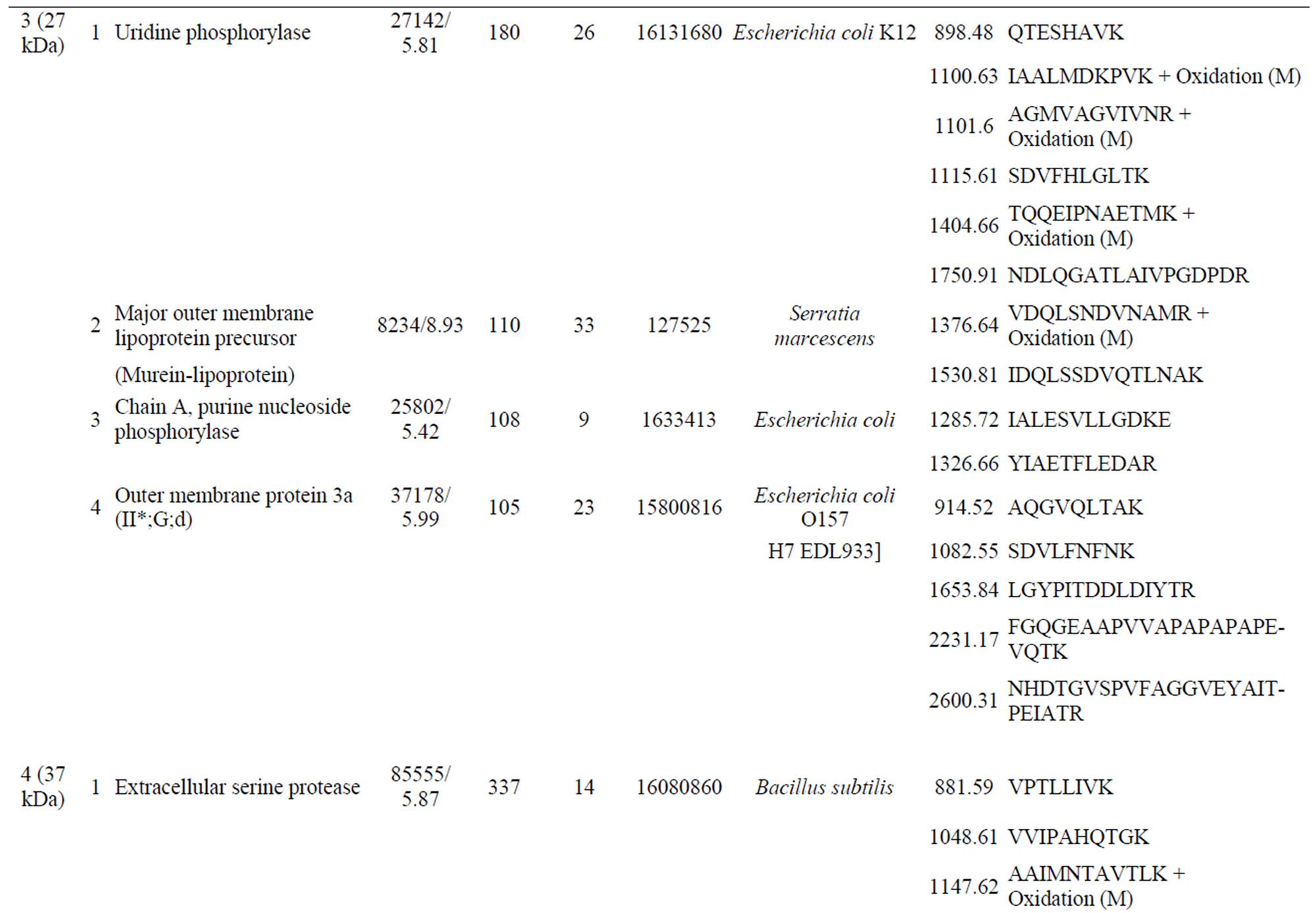



Supplementary Table S1. List of the identified peptide bands.

