Advances in Nanoparticles
Vol.3 No.3(2014), Article
ID:48842,7
pages
DOI:10.4236/anp.2014.33013
Extracting the Atomic Coordinates and Connectivity of Zirconia Nanotubes from PDB Files for Modelling in ANSYS
Ibrahim D. Muhammad*, Mokhtar Awang
Mechanical Engineering Department, Universiti Teknologi PETRONAS, Seri Iskandar, Malaysia
Email: *ibrahimuhd@gmail.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 1 June 2014; revised 3 July 2014; accepted 20 July 2014
ABSTRACT
Zirconia in the form of nanotubes has potential for application in various areas. However, information on structural and mechanical properties of zirconia nanotubes is not easily available and/ or limited in scope. This challenge requires multi-scale numerical modeling and simulation. As a way out, the structure of (10, 10) zirconia nanotube is modeled using available crystal and molecular software (Material Studio© and CrystalMaker©). The output in the form of PDB file is exported into ANSYS by using a script developed in Python. The output contains only the atomic coordinates and connectivity pattern, which make the conversion process faster and more efficient compared to manual option used when performing similar task.
Keywords:Zirconia Nanotubes, Molecular Modelling, Python, ANSYS

1. Introduction
Since 1991, carbon nanotubes have attracted widespread interest as the most important material for nanotechnology because of their semiconducting, electron-emitting, high-strength, and other unique properties [1] . In addition to carbon nanotubes, several inorganic nanotubes have been synthesized and/or modelled ranging from metal sulfides, nitrides, metal oxides, rare earth oxide nanotubes and metallic nanotubes [2] .
In recent years, Zirconia nanotubes (ZNT) are one of the most investigated inorganic nanotubes and can be synthesized using various techniques with anodization been the most widely used method due to its simplicity and ability to fabricate self-organized ZNT arrays [3] -[5] . Variations in morphology and geometrical parameters such as diameters, lengths and wall thickness have been observed in the ZNT synthesized [6] .
For better utilization of the potentials of ZNT, detailed information about its mechanical and other properties are required. However, the technical difficulties involved in the manipulation of these nanoscale structures make the direct determination of their mechanical properties difficult and/or impossible [6] . As an alternative, the numerical approach has been utilized; it is based on multi-scale modelling using molecular dynamics and continuum mechanics via finite element modelling [7] .
Modelling of inorganic nanotubes (INT) is based on the same concepts used for carbon nanotubes (CNT) where the tubes are assumed to be formed by rolling up of nanosheet (NNS) into a hollow cylinder and can be single or multi walled [8] . Thus, ZNT can be visualized as Zr-O hexagonal network, obtained by rolling up a CZNT sheet. The atoms are held together by bonds of definite length and the atoms of Zr and O2 can be conceived as material points which are connected by load bearing beam elements. Currently, theoretical studies on the structure and properties of ZNT are limited with emphasis on first principles calculations that are restricted to small dimensions [9] .
Structure of ZNT can be modelled using the Surface Builder of the Material Studio©, CrystalMaker© or any other software with capability for modelling inorganic nanotubes and exported in the form of Protein Data Bank (PDB) file. The PDB file(s) contain detailed information such as atomic coordinates, bibliographic citations, primary and secondary structure, information, and crystallographic structure factors [10] . PDB file format is a standard means of information interchange in macromolecular structure determination and is widely recognized by molecular modelling software. But PDB files cannot be exported directly into ANSYS© or other related software for finite element analysis (FEA) and not all the data are required for modelling the structure of the nanotube. The manual extraction of data required is time consuming and prone to error [10] . In this study, the model of ZNT is developed and exported as a PDB file. Thereafter, required data in the generated file for modelling of the nanotube in ANSYS are extracted using script from Phyton Wing IDE (version 101 v 5).
2. Computational Method
Using the geometrical parameters for zirconia nanosheet and nanotube [9] , a single walled ZNT having (10, 10) chirality was modelled using Material Studio©. The structure was then saved as PDB file. The generated PDB file was imported into the CrystalMaker© and another PDB file generated. The format of the PDB files generated are similar and the relevant sections related to atomic coordinates and connectivity identified.
Based on the files generated, specific data required were extracted using a script to carry out the following:
1) Open the PDB file and read each line.
2) Select out lines having the word “ATOM”.
3) Get the X, Y and Z coordinates of those atoms.
4) Store the coordinates as nodes.
5) Select out lines with the word “CONECT”.
6) Separate each line into pair connections only, with the first atom connected to others in twos serially.
7) Print out the results in a formatted table.
The output in txt form was thereafter exported into ANSYS for further simulation.
3. Results and Discussion
3.1. Geometrical Structure of Cubic Zirconia Nanotube
The symmetry and structure of a single-walled cubic zirconia nanotube (CZNT) is based on the concept of layer folding [9] of the nanosheet. The structural parameters for the armchair (10, 10) CZNT are given in Table1
The side and top views of the nanotube are shown in Figure 1 indicating the atoms and bonds.
3.2. Required Atomic Records from PDB File
The PDB files from Material Studio© consist of 80-character records, each with a left-justified keyword of up to six characters that indicates its record format type. The information available in the PDB output is divided into three sections:
1) REMARK section gives general information about the file.
2) Coordinate section contains the collection of atomic coordinates with fields divided into ten (10) columns.
 (a)
(a) (b)
(b) (c)
(c)
Figure 1. (a) Tilted; (b) front/side; and (c) top views of 10 × 10 cubic zirconia nanotube (O2 and Zr atoms are represented by red or darker and green or lighter balls respectively).
The fields are for “ATOM”, atom serial number and name, residue name, residue sequence number, X-coordinates, Y-coordinates, Z-coordinates, occupancy, temperature factor and charge on the atom.
3) CONECT section specifies connectivity between atoms for which coordinates are supplied. The connectivity is described using the atom serial number as found in the entry.
The Coordinate and CONECT sections are separated by the TER field/line, which indicates the end of a list of ATOM. TheCONECT section is terminated with END statement. Details of the PDB file from Material Studio© is shown in Figure 2.
Similar format for the PDB file was also obtained using CrystalMaker© with slight difference in the arrangement of the fields. In Material Studio© PDB files, atoms for each element are not arranged serially and the atom serial number and name are regarded as a single column. While in the PDB files from CrystalMaker©, arrangement is serial with all oxygen (O) atoms first and then the zirconium (Zr) atoms. Thus the PDB files for the same (10, 10) ZNT have different connectivity order format. Details of the PDB file from CrystalMaker© is shown in Figure 3.
3.3. Code for Extracting Required Information from PDB File
The important information needed for modelling nanotubes in ANSYS are the atomic coordinates and connectivity pattern. But arranging the required atomic data manually is time consuming and prone to error. Software such as MATLAB and others have been used, but Python have additional capabilities such as the availability of predefined modules for 3D structural format [10] [11] . The PDB format from CrystalMaker© was selected for the script as the Zr and O2 atoms were arranged separately. Based on the information needed, the following script was developed (Appendix).
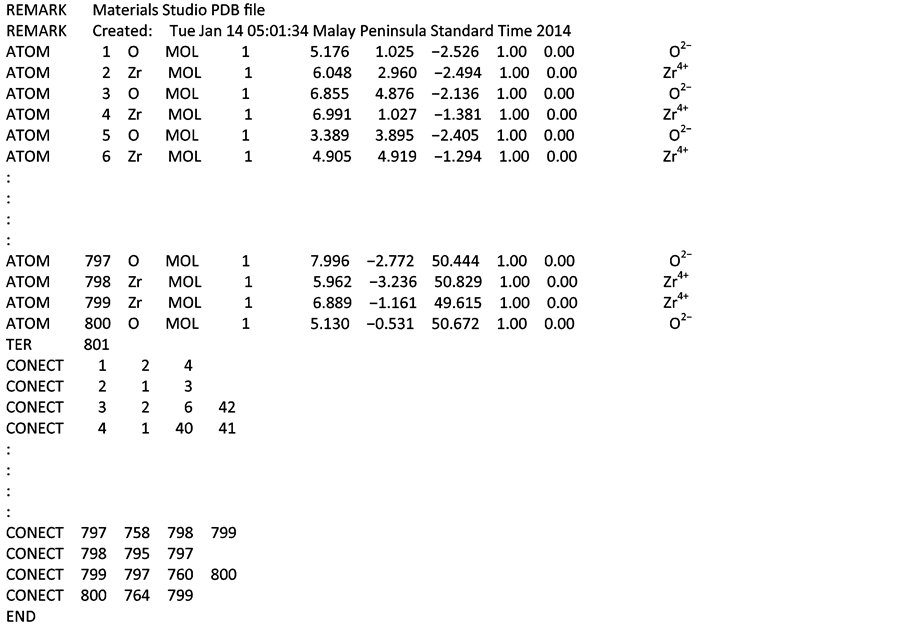
Figure 2. PDB File for 10 × 10 cubic zirconia nanotube from Material Studio.
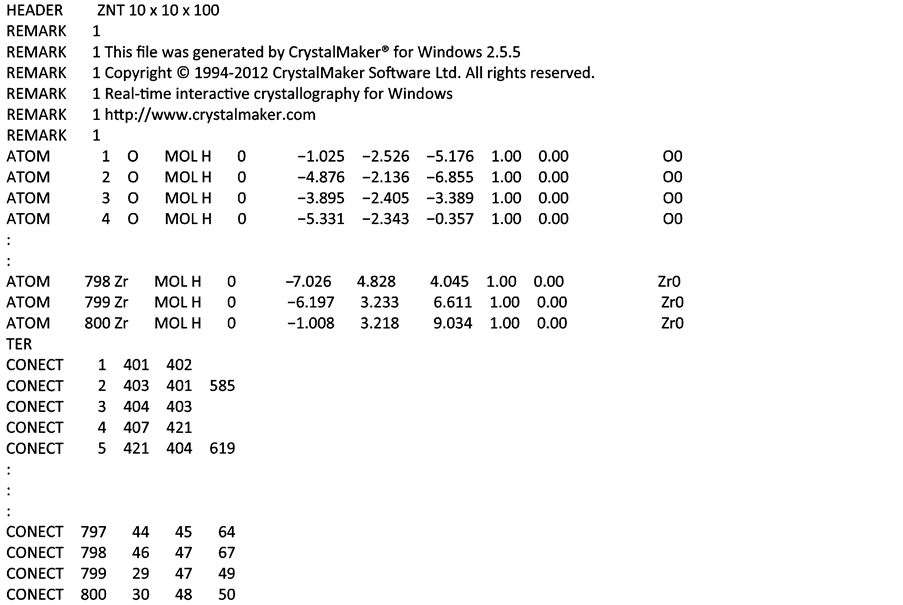
Figure 3. PDB File for 10 × 10 cubic zirconia nanotube from CrystalMaker.
The script opens the PDB file, read all lines to identify the atomic coordinates all starting with ATOM statement and print the output of each line in the following format: n, a, X, Y, Z where n represents nodes, a is the atomic serial number and XYZ are the values of the coordinates. The connectivity records are then split into the required pattern by listing the connection in pairs with the first atom on each CONECT line followed by the next atom; the process is repeated for each line until all atoms on the same line are listed in pairs with the first atom. The output for each line is in the following format: e, a, b where e represents the element, a and b are the serial numbers of the first atom and that of the attached atom respectively.
All the outputs for each line in relation to atomic coordinates and connectivity are recorded in a file and occur in less than 5 seconds, depending on the specifications of the computer and/or of the software used. The output file for a (10 × 10) cubic zirconia nanotube having a length of 52.50 Å is shown in Figure 4.
The /PREP7 is the ANSYS command used to create and setup the model, n defines the node and e defines the element by node connectivity. The data/file is imported in to ANSYS (APDL 14.0) by using the Read input from —option in the File Drop-down Menu. This can also be achieved by pasting the information as a macro in the command line. The model generated in ANSYS is illustrated in Figure 5.
With the model of the nanotube exported fully into ANSYS, further editing and simulation can be carried out as meshing has already been done, making the process faster and more efficient.
4. Conclusion
Due to challenges related to the size of nanotubes, creating their structural models with hexagonal layout directly in ANSYS is difficult and/or impossible. As an alternative, structure of zirconia nanotube was generated using available crystal and molecular software (Material Studio© and CrystalMaker©). The output was then

Figure 4. Output containing nodes and elements for (10 × 10) cubic zirconia nanotube.


Figure 5. Views of the structural model of (10 × 10) cubic zirconia nanotube in ANSYS.
converted to PDB format and the required data for modelling of the nanotube in ANSYS were extracted using a Python script. Only one file is processed at a time, thus generating required output within few seconds. The capability of the script can be improved by processing multiple files at a time and creation of interface with ANSYS.
Acknowledgements
The authors are grateful for the support from Universiti Teknologi PETRONAS and Ministry of Higher Education Malaysia (MOHE) through the Long-Term Research Grant Scheme (LRGS).
References
- Kijima, T. (2010) Introduction to Inorganic and Metallic Nanotubes. In: Kijima, T., Ed., Inorganic and Metallic Nanotubular Materials: Recent Technologies and Applications, Springer, Berlin, 5-16. http://dx.doi.org/10.1007/978-3-642-03622-4
- Tenne, R. and Seifert, G. (2009) Progress in the Study of Inorganic Nanotubes and Fullerene-Like Structures. Annual Review of Materials Research, 39, 387-413. http://dx.doi.org/10.1146/annurev-matsci-082908-145429
- Yan, C., Liu, J., Liu, F., Wu, J., Gao, K. and Xue, D. (2008) Tube Formation in Nanoscale Materials. Nanoscale Research Letters, 3, 473-480. http://dx.doi.org/10.1007/s11671-008-9193-6
- Lockman, Z., Ismail, S., Kawamura, G. and Matsuda, A. (2011) Formation of Zirconia and Titania Nanotubes in Fluorine Contained Glycerol Electrochemical Bath. Defect and Diffusion Forum, 76, 312-315.
- Li, L., Yan, D., Lei, J., He, J., Wu, S. and Pan, F. (2011) Fast Fabrication of Highly Regular and Ordered ZrO2 Nanotubes. Materials Letters, 65, 1434-1437. http://dx.doi.org/10.1016/j.matlet.2011.02.025
- Xia, R.O. and Catlow, R. (2009) Computational Modelling Study of Bulk and Surface of Yttria-Stabilized Cubic Zirconia. Chemistry of Materials, 21, 3576-358. http://dx.doi.org/10.1021/cm900417g
- Evarestov, R.A., Zhukovskii, Y.F., Bandura, A.V. and Piskunov, S. (2010) Symmetry and Models of Single-Wall BN and TiO2 Nanotubes with Hexagonal Morphology. The Journal of Physical Chemistry C, 114, 21061-21069. http://dx.doi.org/10.1021/jp106929f
- Guimaraes, L., Enyashin, A.N., Seifert, G. and Duarte, H.A. (2010) Structural, Electronic, and Mechanical Properties of Single-Walled Halloysite Nanotube Models. The Journal of Physical Chemistry C, 26, 11358-11363. http://dx.doi.org/10.1021/jp100902e
- Bandura, A.V. and Evarestov, R.A. (2012) Ab Initio Structural Modelling of ZrO2 Nanosheets and Single-Wall Nanotubes. Computational Materials Science, 65, 395-405. http://dx.doi.org/10.1016/j.commatsci.2012.08.001
- Nulaka, S., Allam, A.R. and Kopparthi, S. (2012) Python Program to Generate Atom Records from PDB Protein Files for Drug Design Studies. Journal of Bioinformatics & Research, 1, 36-40.
- Enyashin, A.N., Gemming, S. and Seifert, G. (2010) Simulation of Inorganic Nanotubes. In: Gemming, S., Schreiber, M. and Suck, J.-B. (Eds.), Materials for Tomorrow: Theory, Experiments and Modelling, Springer, Berlin, 33-57.
Appendix: Script/Code for Extracting Nodes and Elements to ANSYS
### get atoms coordinates from PDB file and write them as
###ANSYS nodes and elements with prep command.
line_value = 0 nodepoint_value = 0 file = open('C:\\xyz\\ZNT10x10CRYM.pdb', 'r')
igot = file.readlines()
output_file = open('C:\\xyz\\ZNT10x10CRYM.txt', "a") # needs input for distinct file output_file.write('/PREP7'+"\n") #open pre-processing menu for line in igot:
if line.find("ATOM") > -1:
nodepoint_value = nodepoint_value + 1
nv = str(nodepoint_value)
xyz = line.split()
#print nv,",", xyz[6], ",", xyz[7], ",", xyz[8]
line_i = "n" + ', '+ nv + ', ' + xyz[6] + ', ' + xyz[7] +', '+ xyz[8]+ "\n"
print(line_i)
output_file.write(line_i)
### reads connect records
elif line.find("CONECT") > -1:
con = line.split()
line_value = line_value + 1
#print line_value
#print con[2]
line_j = "e" + ', ' + str(line_value) + ', ' + con[2] + "\n"
output_file.write(line_j)
print(line_j)
line_i = "e" + ', ' + str(line_value) + ', ' + con[3] + "\n"
output_file.write(line_i)
print(line_i)
try:
line_k = "e"+ ', ' + str(line_value) + ', ' + con[4] + "\n"
print(line_k)
output_file.write(line_k)
except IndexError:
continue
NOTES

*Corresponding author.


