American Journal of Plant Sciences
Vol.4 No.8A(2013), Article ID:35946,4 pages DOI:10.4236/ajps.2013.48A005
Evaluation of Transgenic Nicotiana tabacum with dehE Gene Using Transposon Based IRAP Markers
![]()
1Faculty of Biosciences and Medical Engineering, Universiti Teknologi Malaysia, Johor Bahru, Malaysia; 2Molecular Biology and Genetics Department, Istanbul University, Vezneciler-Istanbul, Turkey.
Email: yilmazkaya55@hotmail.com, sibelyilmaz22@hotmail.com, *nermin@istanbul.edu.tr, fzhutm@gmail.com
Copyright © 2013 Yilmaz Kaya et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received May 20th, 2013; revised June 21st, 2013; accepted July 15th, 2013
Keywords: Nicotiana tabacum; Dehalogenase E; Tto1 Retrotransposon; IRAP; Transgenics
ABSTRACT
In the present study, five genetically modified herbicide tolerant Nicotiana tabacum cv. TAPM24 plants with a constructed vector pCAMBIA1301a carrying dehalogenase E (dehE) gene were compared with three non-transgenic controls using Tto1 retrotransposon specific IRAP markers. dehE gene was originally characterized in Rhizobium sp. and it produced an enzyme which degraded the Dalapon herbicide. IRAP protocol was applied on transgenic and non-transgenic plants to investigate the retrotransposon based genetic variation which may appear during transformation. Polymorphism rates were calculated as 0% - 20% from IRAP-PCR products among all plant samples. These results show that transformation of tobacco plant with the dehE gene may cause Tto1 retrotransposon alterations appearing as different band profiles. The findings are expected to contribute to genetic engineering studies to obtain better results and also to understand how transposons contribute to features such as transgenesis. In our knowledge, this is one of the first experimental data of transgenic N. tabacum engineered with dehE gene originated Rhizobium sp. in terms of retrotranposon based variation.
1. Introduction
Tto1 is one of the few active retrotransposons in tobacco. The sequence analysis suggests that Tto1 carries all functions required for autonomous transposition through reverse transcription. The copy number of Tto1 increases 10-fold during protoplast culture, regenerated plants from tissue cultures [1]. There are reports for the increase of retrotransposition during tissue culture and regenerated plants with other retrotransposons in barley [2,3]. Tto1 transposition is regulated mainly at the transcriptional level. It is shown that expression of Tto1 can be induced in leaves of tobacco by wounding stress [4]. Tto1 has been implicated in the expression of phenylpropanoid synthetic genes in response to defense-related stresses [5]. Tto1 is also re-activated by the implantation of the lowenergy ion beam ion [6].
In crop production, the use of bacterial genes has created new approaches for herbicide tolerance [7]. Bacterial dehalogenases cleave carbon-halogen bonds with different stereo-configurations. Three dehalogenases are produced by Rhizobium sp. collectively known as dehD, dehL and dehE. The dehE from Rhizobium sp. RC1 is non-stereo specific dehalogenase that can act on D-2- chloropropionate (D-2-CP), L-2-chloropropionate (L-2- CP), 2,2-dichloropropionate (2,2-DCP)-Dalapon, trichloroacetate (TCA), dichloroacetate (DCA) and monochloroacetate (MCA). The dehE is further characterised [8,9]. Dalapon is a chemical with herbicide properties only when active ingredient of 2,2-DCP is present. It is used selectively to control weed grasses [10,11]. The high concentration of Dalapon may kill desired plant that lacks of herbicide tolerance. To improve the Dalapon resistance level, production of transgenic plants is needed. Our group developed transgenic tobacco plant via A. tumefaciens introduction and expression of the herbicidetolerance gene, dehE [12].
Several molecular marker systems based on retrotransposons have been developed. All rely on the principle that a joint is formed, during retrotransposon integration, between genomic DNA and the retrotransposon. These joints may be detected by amplification between a primer corresponding to the retrotransposon and a primer matching a nearby motif in the genome [13]. The interretrotransposon amplified polymorphism (IRAP) method [14] detects retrotransposon insertional polymorphisms by amplifying the portion of DNA between two retroelements. It uses one or two primers pointing outwards from an LTR and amplifies the tract of DNA between two nearby retroelements. IRAP can be carried out with a single primer matching either the 5’ or 3’ end of the LTR but oriented away from the LTR itself, or with two primers.
There are very limited reports on transgenic plants in terms of genomic variations [15]. In the present research, we aimed to analyse transgenic tobacco plants carrying herbicide-tolerance gene, dehE gene in terms of Tto1 retrotransposition using IRAP technique.
2. Materials and Methods
2.1. Plant Material, Gene Construct and Transformation
N. tabacum cv. TAPM24 plants transformed with pCAMBIA1301a vector were tested whether they carry dehE gene or not. In addition, expression of dehE gene was determined by qPCR with dehE specific primers [12]. To analyses Tto1 retrotransposon caused genetic variations, 5 transgenic plants were randomly selected and used for DNA isolation. DNA isolations were performed from leaves by TRI Reagent® (Sigma, T9424) according to the manufacturer’s instructions. We also isolated DNA from 3 non-transformed tobacco plants and used them as control. Spectrophotometric assays were conducted to determine the quantity and quality of the isolated DNA.
2.2. Inter-Retrotransposon Amplified Polymorphism (IRAP) PCR
IRAP was performed with one primer (5’-TCCGCTGTGCAGTAGTGTTTAGTG-3’) designed from LTRsequences of Tto1 retrotransposon [16]. Amplification were carried out in 20 μl reaction volume containing 10 μl 2× Sapphire Enzyme Mix (Takara, RR350), 2 μl primer (10 pmol), 2 μl template genomic DNA (1 ng/μl) and 6 μl dH2O. The amplification conditions were one initial denaturation step at 94˚C for 3 min followed by 35 cycles at 94˚C (20 s), 55 (20 s) and 72˚C (1 min); the reactions were completed by a final extension step at 72˚C for 10 min.
2.3. Evaluation of PCR Products
Ten-μl IRAP-PCR products were loaded to 6% nondenaturing polyacrylamide (29:1 Acrylamide:Bis) gel and run at 150 V for 5 h in 1× TBE buffer. Gel were stained in 1× TBE buffer containing ethidium bromide for 15 minutes and scored visually. Both homomorphic and polymorphic bands were scored and similarity index was calculated using Jaccard’s coefficient [17].
3. Results and Discussion
In this study, IRAP marker technique was applied to 5 transgenic tobacco plants, transformed with dehE gene. In addition, we used 3 non-transformed control plants to find out whether there is a naturally occurred polymorphism or not between individuals. As result of IRAPPCR, we did not detect any polymorphic band among control plants (Figure 1, line C1-3). Hence, we conclude that Tto1 retrotransposon do not cause natural polymorphism.
The IRAP results of 5 dehE transformed tobacco plants showed that all samples have different band patterns (Figure 1, line T1-5). Although there is no polymorphism between control plants, transgenic lines showed 0% - 20% (Table 1). These polymorphism rates proved that dehE gene transformation could be caused retrotransposition events.
T1-5 indicate transgenic plants; C1-3 indicate control plants; NC indicates negative control; first and tenth lines are marker (arrows indicate some polymorphic and missing bands).
As shown Figure 1, all the transgenic lines have different band patterns. First transgenic plant has two novel bands (arrows a) while T5 missed a band (arrow b).
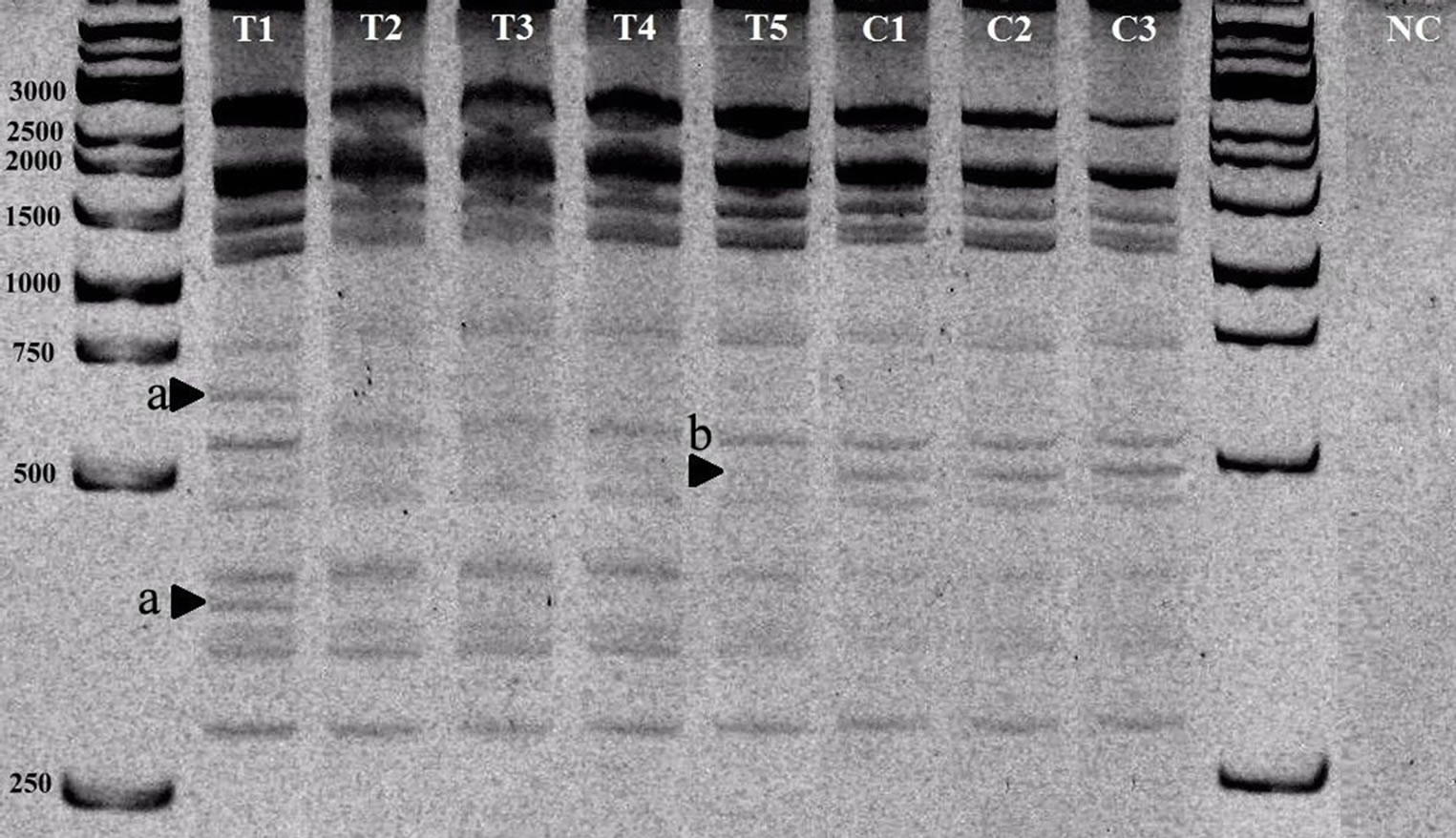
Figure 1. Tto1 IRAP-PCR result.
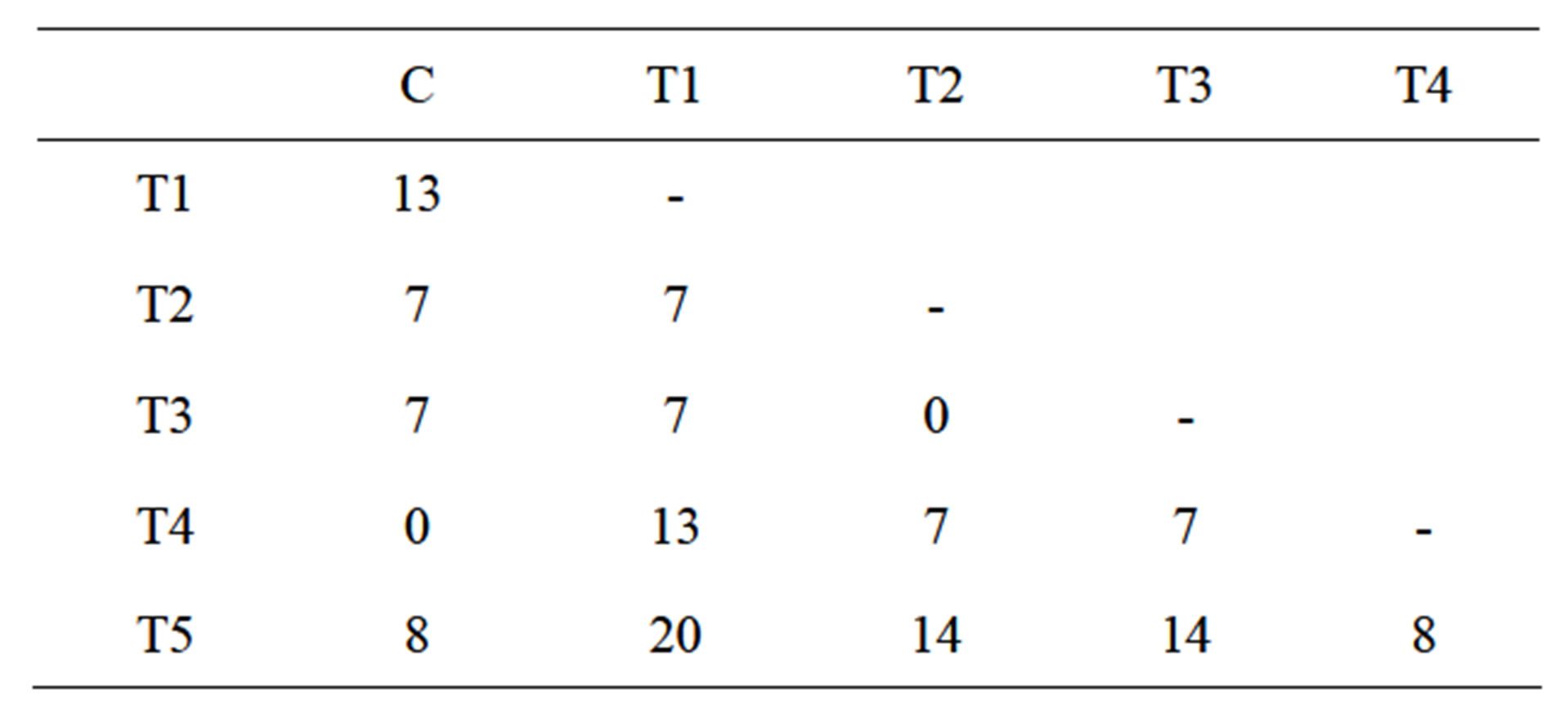
Table 1. IRAP polymorphism percentage between transgenic and control plants. T1-5 indicate transgenic plants; C indicates control plant.
Both T2 and T3 showed polymorphic patterns compared with control plants although they were homomorphic with each other. On the other hand, T4 line has identical band patterns with control plants. This results show that transformation of tobacco plants might cause retrotransposon movement and this effect is different for every individual.
Campbell et al. [18] used IRAP technique to determine somaclonal variation in regenerated plantlets from barley calli cultures. They obtained novel non-parental bands during analysis of DNA polymorphisms in parental controls and regenerants. They stated that retrotransposon-based marker systems, such as IRAP, based on retrotransposons such as BARE1, are valuable tools for the characterization of mutations that arise during tissue culture. Muhammad and Othman [19] studied banana somaclones with Random Amplified Polymorphic DNA (RAPD) and IRAP markers. They observed that RAPD markers were more polymorphic than IRAP markers [15]. IRAP was also recently used to study genetic diversity of cultivated flax (Linum usitatissimum L.) germplasm [20]. Retrotransposons show unique patterns of environmental and/or developmental regulation, e.g. tobacco Tnt1 and barley BARE1 copies were detected in roots and leaves, respectively [21,22]. Many retrotransposons were showed to be activated by biotic and abiotic stresses [23]. Copy numbers of tobacco Tto1 [1] and rice Tos17 [24] elements increased during tissue culture procedures. In our previous studies on barley tissue cultures we also observed retrotransposon movement increase in relation to the callus age and regenerated plantlets [2,3]. Although 10-fold increase in copy number of Tto1 during protoplast culture and regenerated plants from tobacco tissue cultures [1], this is the first report on transgenic lines in terms of retrotransposon movements.
4. Conclusion
In this study, retrotransposon integration events in transgenic tobacco plants were investigated. 0% - 20% polymorphisms were observed between transgenic individuals and controls. It was clear that some retrotransposition events occured during transformation. However, sequence characterization of the regions, which exhibit polymorphism, is crucial for the exploration of transformation development and retrotransposition.
5. Acknowledgements
Authors would like to thank The Malaysian Government for partly sponsor the work under research grant schemes FRGS 4F008 and GUP (QJ130000.7135.00H34 and Istanbul University, Molecular Biology and Genetics Department).
REFERENCES
- H. Hirochika, “Activation of Tobacco Retrotransposons during Tissue Culture,” The EMBO Journal, Vol. 12, No. 6, 1993, pp. 2521-2528.
- C. Evrensel, S. Yılmaz, A. Temel and N. Gozukirmizi, “Variations in BARE-1 Insertion Patterns in Barley Callus Cultures,” Genetics and Molecular Research, Vol. 10, No. 2, 2011, pp. 980-987. doi:10.4238/vol10-2gmr965
- E. Bayram, S. Yilmaz, H. Hamad-Mecbur, G. Kartal and N. Gozukirmizi, “Nikita Retrotransposon Movements in Barley (Hordeum vulgare L.) Callus Culture,” Plant Omics Journal, Vol. 5, No. 3, 2012, pp. 211-215.
- S. Takeda, K. Sugimoto, H. Otsuki and H. Hirochika, “Transcriptional Activation of the Tobacco Retrotransposon Tto1 by Wounding and Methyl Jasmonate,” Plant Molecular Biology, Vol. 36, No. 3, 1998, pp. 365-376. doi:10.1023/A:1005911413528
- K. Sugimoto, S. Takeda and H. Hirochika, “MYB-Related Transcription Factor Ntmyb2 Induced by Wounding and Elicitors Is a Regulator of the Tobacco Retrotransposon Tto1 and Defense-Related Genes,” Plant Cell, Vol. 12, No. 12, pp. 2511-2527.
- Y. Hui-yuan, W. Wei-dong, C. Wan-guang, Q. Guangyong and H. Yu-ping, “Retrotransposon Tto1 in Tobacco Was Activated by the Implantation of LowEnergy N+ Ion Beam,” Life Science Journal, Vol. 7, No. 3, 2010, pp. 141-147.
- Y. Kaya, S. Marakli, N. Gozukirmizi, E. Mohamed, M. A. Javed and F. Huyop, “Herbicide Tolerance Genes Derived from Bacteria,” The Journal of Animal and Plant Sciences, Vol. 23, No. 1, 2013, pp. 85-91.
- F. Huyop, T. Y. Yusn, M. Ismail, R. A. Wahab, R. A. Cooper, “Overexpression and Characterisation of NonStereospecific Haloacid Dehalogenase E (DehE) of Rhizobium sp.,” Asia Pacific Journal of Molecular Biology and Biotechnology, Vol. 12, No. 1-2, 2004, pp. 15-20.
- F. Huyop, N. A. A. Rashid, R. A. Wahab and R. A. Cooper, “Purification and Properties of Rhizobial DehL Expressed in Escherichia coli,” African Journal of Biotechnology, Vol. 7, No. 12, 2008, pp. 1944-1949.
- W. D. Burge, “Populations of Dalapon-Decomposing Bacteria in Soil as Influenced by Additions of Dalapon or Other Carbon Source,” Applied and Environmental Microbiology, Vol. 17, No. 4, 1969, pp. 545-550.
- S. H. Tseng, Y. J. Lin, P. C. Chang, S. S. Chou and H. M. Chang, “Determination of Residual Dalapon in Sugarcane by Gas Chromatograph Equipped with Electron Capture Detector (GC-ECD),” Journal of Food and Drug Analysis, Vol. 12, 2004, pp. 238-243.
- Y. Kaya, S. Yilmaz, S. Marakli, N. Gozukirmizi and F. Huyop, “Transformation of Nicotiana tabacum with DehE Gene,” Journal of Food Agriculture and Environment, Vol. 11, No. 3-4, 2013, in Press.
- A. H. Schulman, A. J. Flavell and T. H. N. Ellis, “The Application of LTR Retrotransposons as Molecular Markers in Plants,” Methods in Molecular Biology, Vol. 260, 2004, pp. 145-173.
- R. Kalendar, T. Grob, M. Regina, A. Suoniemi and A. H. Schulman, “IRAP and REMAP: Two New Retrotransposon-Based DNA Fingerprinting Techniques,” Theoretical and Applied Genetics, Vol. 98, No. 5, 1999, pp. 704- 711. doi:10.1007/s001220051124
- R. M. Ismail, “Evaluation of Genetically Modified Sugarcane Lines Carrying Cry1AC Gene Using Molecular Marker Techniques,” GM Crops and Food: Biotechnology in Agriculture and the Food Chain, Vol. 4, No. 1, 2013, pp. 58-66. doi:10.4161/gmcr.24299
- B. C. Yang, B.G. Xiao, X. J. Chen and C. H. Shi, “Assessing the Genetic Diversity of Tobacco Germplasm Using Intersimple Sequence Repeat and Inter-Retrotransposon Amplification Polymorphism Markers,” Annals of Applied Biology, Vol. 150, No. 3, 2007, pp. 393-401. doi:10.1111/j.1744-7348.2007.00139.x
- P. Jaccard, “Nouvelles Recherches sur la Distribution Florale,” Bulletin de la Societe Vaudoise des Sciences Naturelles, Vol. 44, 1908, pp. 223-270.
- B. C. Campbell, S. LeMare, G. Piperidis and I. D. Godwin, “IRAP, a Retrotransposon-Based Marker System for the Detection of Somaclonal Variation in Barley,” Molecular Breeding, Vol. 27, No. 2, 2010, pp. 193-206. doi:10.1007/s11032-010-9422-4
- A. J. Muhammad and F. Y. Othman, “Othman Characterization of Fusarium Wilt-Resistant and Fusarium WiltSusceptible Somaclones of Banana Cultivar Rastali (Musa AAB) by Random Amplified Polymorphic DNA and Retrotransposon Markers,” Plant Molecular Biology Reporter, Vol. 23, No. 3, 2005, pp. 241-249. doi:10.1007/BF02772754
- P. Smykal, N. Bacova-Kerteszova, R. Kalendar, J. Corander, A. H. Schulman and M. Pavelek, “Genetic Diversity of Cultivated Flax (Linum Usitatissimum L.) Germplasm Assessed by Retrotransposon-Based Markers,” Theoretical and Applied Genetics, Vol. 122, No. 7, 2011, pp. 1385-1397. doi:10.1007/s00122-011-1539-2
- S. Pouteau, E. Huttner and M. A. Grandbastien, “Specific Expression of the Tobacco Tnt1 Retrotransposon in Protoplasts,” The EMBO Journal, Vol. 10, No. 7, 1991, pp. 1911-1918.
- A. Suoniemi, K. Anamthawat-Jonsson, T. Arna and A. H. Schulman, “Retrotransposon BARE-1 Is a Major, Dispersed Component of the Barley (Hordeum vulgare L.) Genome,” Plant Molecular Biology, Vol. 30, No. 6, 1996, pp. 1321-1329. doi:10.1007/BF00019563
- M. L. Grandbastien, “Activation of Plant Retrotransposons under Stress Conditions,” Trends in Plant Science, Vol. 3, No. 5, 1998, pp. 181-187. doi:10.1016/S1360-1385(98)01232-1
- H. Hirochika, K. Sugimoto, Y. Otsuki, H. Tsugawa and M. Kanda, “Retrotransposons of Rice Involved in Mutations Induced by Tissue Culture,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 93, No. 15, 1996, pp. 7783-7788. doi:10.1073/pnas.93.15.7783
NOTES
*Corresponding author.

