American Journal of Plant Sciences
Vol.3 No.4(2012), Article ID:18696,12 pages DOI:10.4236/ajps.2012.34055
A Comparative Testing of Cucumber mosaic virus (CMV)-Based Constructs to Generate Virus Resistant Plants
![]()
1Hunan Plant Protection Institute, Changsha, China; 2Biocentre Klein-Flottbek and Botanical Garden, University of Hamburg, Hamburg, Germany.
Email: *cornelia.heinze@uni-hamburg.de
Received October 28th, 2011; revised November 29th, 2011; accepted February 20th, 2012
Keywords: Cucumber mosaic virus; Transgenic Plant; Resistance; Comparison
ABSTRACT
Among the viruses Cucumber mosaic virus (CMV) has been rated worldwide as one of the five most important viruses infecting vegetable species. CMV is a tripartite virus with high sequence variability, classified into three subgroups with 80% to 97% identical nucleotides in their coat protein. Due to the absence of natural resistance CMV is the plant virus with longest history in genetic engineering using pathogen induced approaches. However, the transformation and regeneration for some very important crops like chili is difficult. Therefore it will be an advantage to screen in model plants for gene constructs which might be independent of the target of final transformation and other parameters having an influence on the efficiency of a biotechnological approach. In our study we compared the resistance for all combinations of five different antiviral constructs, two different transformation vectors and two model host plants. From these approaches we identified the most effective construct which might also be applicable to transform eventually chili plants.
1. Introduction
The chili production has an economical impact in local as well as export markets in Asia and other parts of the world. More than one billion people consume chili in one or another form on a daily basis. The major diseases contributing to low yield and a reduced quality of fruits include bacterial wilt (Ralstonia solanacearum), phytophthora blight (Phytophthora capsici Leon.) powdery mildew (Leceillula rurica) and anthracnose (Colletotrichum sp.) [1]. In addition, several viruses are an important threat to chili production [2,3]. Due to its worldwide distribution and polyphagous vectors Cucumber mosaic virus (CMV) is one of the five most important viruses infecting vegetable species worldwide [4-6].
CMV is the type species of the genus Cucumovirus, family Bromoviridae. It has a tripartite ssRNA genome coding for one structural and four functional proteins. The RNA dependent RNA-polymerase (RdRP) is encoded on RNA 1 (ORF 1a) and RNA 2 (ORF 2a). The gene silencing suppressor (ORF 2b) overlaps with ORF 2a. RNA 3 encodes the cell to cell movement protein (ORF 3a) and the CP, which forms together with the RNA icosahedral particles. CMV is classified into subgroups Ia, Ib and II with 97% and 80% identical amino acids (aa) in the coat protein (CP), respectively [5,7,8].
Almost all known resistance genes found in several natural sources of Capsicum sp. are partial or polygenic [9-11] only few of them confer sustainable resistance [12,13]. Kang et al. [14] described a single dominant gene controlling CMV resistance in peppers which was effective against two virus isolates, however, a third one caused infection due to the extreme high variability of CMV. Lee et al. [15] identified the CMV strain CMVP1 that infected commercially available CMVP0 resistant pepper plants in the mid 1990th. Another promising resistant chili variety, breeding line VC246 from the World Vegetable Research and Development Center (WVRDC, Taiwan), revealed upon screening with several isolates from different serogroups five out of 28 isolates that overcame this resistance [16].
Several transgenic CMV resistant or tolerant plants were reported using the coat protein and the RNA replicase gene [17] and references therein [18]. In addition, extensive studies to induce resistance against CMV with truncated CP or 2a protein expressed in transgenic plants have been reported [19].
For chili, reports of tolerant plants using the coat protein gene are available [20-23]. However, the biotechnological production of CMV resistant chili plants is difficult, because the efficient transformation on chili pepper is inhibited since the shot regeneration rate is genotype specific [24] and the gene transfer via Agrobacterium infection into cotyledon and hypocotyls tissue is partly blocked for unknown reasons [25]. Therefore testing the efficiency of constructs directly in the chili lines is useless since the transformation efficiency is very low and/or not reproducible [26-28].
To circumvent the problem of the low transformation and regeneration efficiency we used as model plants N. benthamiana and N. tabacum cv. Samsun for transformation with five constructs each in two different vectors and screened all combinations with up to five CMV isolates from all subgroups Ia, Ib and II to identify constructs that conferred the immunity type of resistance independent of model plant, virus isolate and transformation vector.
2. Materials and Methods
2.1. CMV Isolates, Maintenance and Purification
In this study five CMV isolates representing all subgroups were included. CMVAN (subgroup Ib) was used for generating the constructs for plant transformation. Lines were challenged with the homologous isolate CMVAN as well as with the heterologous isolates CMVP3613 and CMVKS44 (subgroup Ib), CMVRT52 (subgroup Ia) and the subgroup II isolate CMVPV0420.
Virus isolates were propagated on N. glutinosa. For inoculum preparation virus particles were purified as described [29]. Particles were checked for specific infectivity on the local lesion hosts Vigna unguiculata and Chenopodium quinoa by mechanical inoculation [16]. All plants were incubated in the greenhouse at 25˚C +/– 1˚C with a photoperiod of 16 hours light and 8 hours dark.
2.2. Plant Inoculation and Resistance Screening
Two leaves of plants at the 4 to 6 leaf stage were rubbed with virus particles diluted in 20 mM phosphate buffer (pH 7) and 5% (w/v) carborundum (600 mesh). The infectivity of the diluted particles was adjusted to induce 30 to 60 local lesions when using a total volume of 10 µl on Vigna unguiculata. This dose of inoculum infected 100% of the N. benthamiana and N. tabacum plants. For each plant-vector-insert combination 8 selection marker resistant plants of four to six independent lines were tested in the F1 generation with one repetition. Symptom expression was checked visually 20 and 35 days post infection (dpi) and virus presence/absence was verified by tissue print immunoblot of transverse sections of noninoculated leaves using coat protein-specific polyclonal antiserum AS-0475 (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany) as described [30].
The visual screening of symptoms identified four phenotype classes: immune (no symptoms, negative tissue print), tolerant (no, mild or delayed mild symptoms, positive tissue print), recovery (early symptoms but in late infection no symptoms, positive/negative tissue print) and susceptible (symptoms, positive tissue print). Resistance was defined as the sum of immune, tolerant and recovered plants. Absence of virus was confirmed by RTPCR as described in [8] using primers CMV-CPfor (5’-atg gac aaa tct gra tcw mcc-3’) and CMVrev (5’-ctg gat gga caa ccc gtt c-3’). As an internal control using primers Nadsense (5’-gatgcttcttggggcttcttgtt-3’) and Nadantisense (5’- ctccagtcaccaacattggcataa-3’) a plant specific fragment was amplified from total RNA simultaneously to the virus specific fragment as described in [31].
2.3. Gene Constructs in pLH6000 and pBIN19 Binary Vectors and Plant Transformation
The constructs are based on the coat protein gene (CP) or the 2b gene silencing suppressor gene including the overlapping region with the 2a of the isolate CMVAN. Genes were used either in a translatable and/or a nontranslatable form as well as in an inverted repeat form. In addition a chimeric construct containing a translatable GFP upstream of the inverted 2b repeat was prepared. The constructs are listed in Table 1 and the details of their generation are given in the supporting information. The constructs were transformed using Agrobacterium tumefaciens LBA4404 (pBIN19) or GV3101 (pLH6000) by electroporation.
Leaf discs of Nicotiana benthamiana and Nicotiana tabacum cv. Samsun nn were transformed with Agrobacterium tumefaciens LBA4404 according to [32] using for pLH6000 constructs hygromycin (20 mg/l) and for pBIN19 constructs kanamycin (50 mg/l) as a selection marker, respectively. Integration of the transgene and absence of Agrobacterium was verified by PCR. Transgene F1 were identified by germinating seeds on kanamycin (150 mg/l) and hygromycin (100 mg/l) containing MS-agar [33] before transferring seedlings to soil.
3. Results
3.1. Description of the Constructs
Virus specific inserts were derived from subgroup Ib isolate CMVAN. The inserts of the single gene constructs ΔCP, Δ2a2b and Δ2aΔ2b had a length of 773 bp (ΔCP),
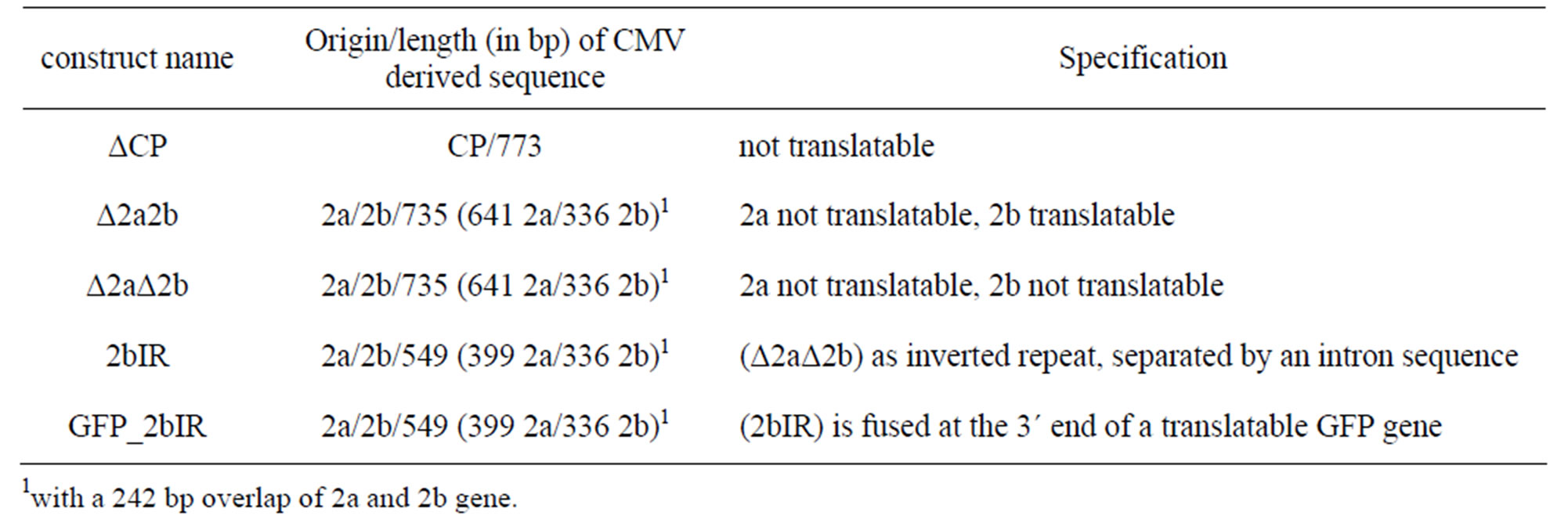
Table 1. Names, origin and length of viral sequences used in pLH6000 and pBIN19 binary vectors.
739 bp (Δ2a2b) and 738 bp (Δ2aΔ2b). In construct ΔCP the start codon ATG was changed to GGT and for Δ2aΔ2b construct the adenine from the start codon of the 2b silencing suppressor was removed resulting in nontranslatable CPand Δ2aΔ2b-genes, respectively. The inverted repeat constructs 2bIR and GFP_2bIR were based on the Δ2aΔ2b construct containing no translatable ORF. The only translatable gene of all constructs was the 2b silencing suppressor in the construct Δ2a2b. The ΔCP is based on RNA 3 while all 2b and Δ2b containing constructs are based on sequences of RNA 2. A more detailed description of the cloning procedure is given in the supporting information. Due to antibiotic resistance conflicts constructs based on pLH6000 were transformed with Agrobacterium strain GV3101, while the pBIN19 constructs were introduced into plants with Agrobacterium strain LBA4404.
3.2. Sequence Comparison of Virus Specific Inserts with the Respective Regions of Heterologous Isolates Used for Resistance Screening
All subgroup Ib isolates (KS44, P3613) used for heterologous resistance screening showed a nucleic acid identity of 90% - 93% for the Δ2a2b region when compared with the respective region of CMVAN, while for subgroup Ia isolate CMVRT52 the identity was 86% and for subgroup II isolate CMVPV0420 69% when compared with CMVAN. For the 2b region the identity of the Ib isolates ranged between 88% and 92%, subgroup Ia isolate CMVRT52 showed 84% and subgroup II isolate CMVPV0420 65% identity. Roughly the same gradation in the identity is present on the CP genes, 93% - 96% for the Ib isolates, 91% for IA and 76% identical nucleotides for the subgroups II isolate, respectively. The identities are summarized in Table 2.
3.3. Analysis of Transgenic Lines
In total, 249 lines were selected from independent calli transformed with five constructs harboring viral or chi-
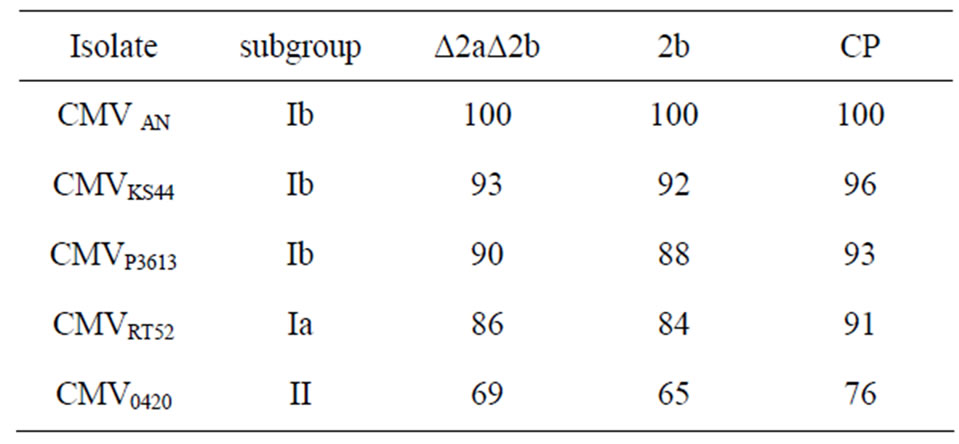
Table 2. Nucleic acid identity (%) of genes Δ2aΔ2b (641 bp 2a and 336 bp 2b with a 241 bp overlap), 2b and CP from subgroups Ia, Ib and II.
meric sequences either in the vector pLH6000 or pBIN19. As controls served plants transformed with either the empty vector or the vector with GFP as insert. All of the control lines showed signs of infection and became infected comparable to the non-transformed plants.
Stable integration was verified by PCR and by segregation patterns after self pollination of the parental generation and subsequent seed germination on selective medium. Seedlings from lines following a segregation pattern of 3:1 (X2 confidence value of P0.05 ≤ 3.84) were transferred into soil in the greenhouse for resistance testing. Each line was tested with 4 individual plants and the testing was repeated.
3.4. Influence of Plant Species and Vector on the Resistance When Inoculated with the Homologous Isolate CMVAN
The overall resistance from N. benthamiana (31% immune plants, Table 3(a) line 11) and N. tabacum cv. Samsun (20% immune plants, Table 3(b), line 11) differed. In N. benthamiana all single gene constructs ΔCP, Δ2a2b and Δ2aΔ2b transformed with the pLH6000 vector resulted in 34% to 50% immune plants derived from 14 lines (Table 3(a), lines 1 - 3) whereas no immune plants were obtained from the 14 lines harboring the same inserts but transformed with the pBIN19 vector. Also no immune plants were obtained when transformed with pLH6000 and pBIN19 in N. tabacum (Table 3(b),
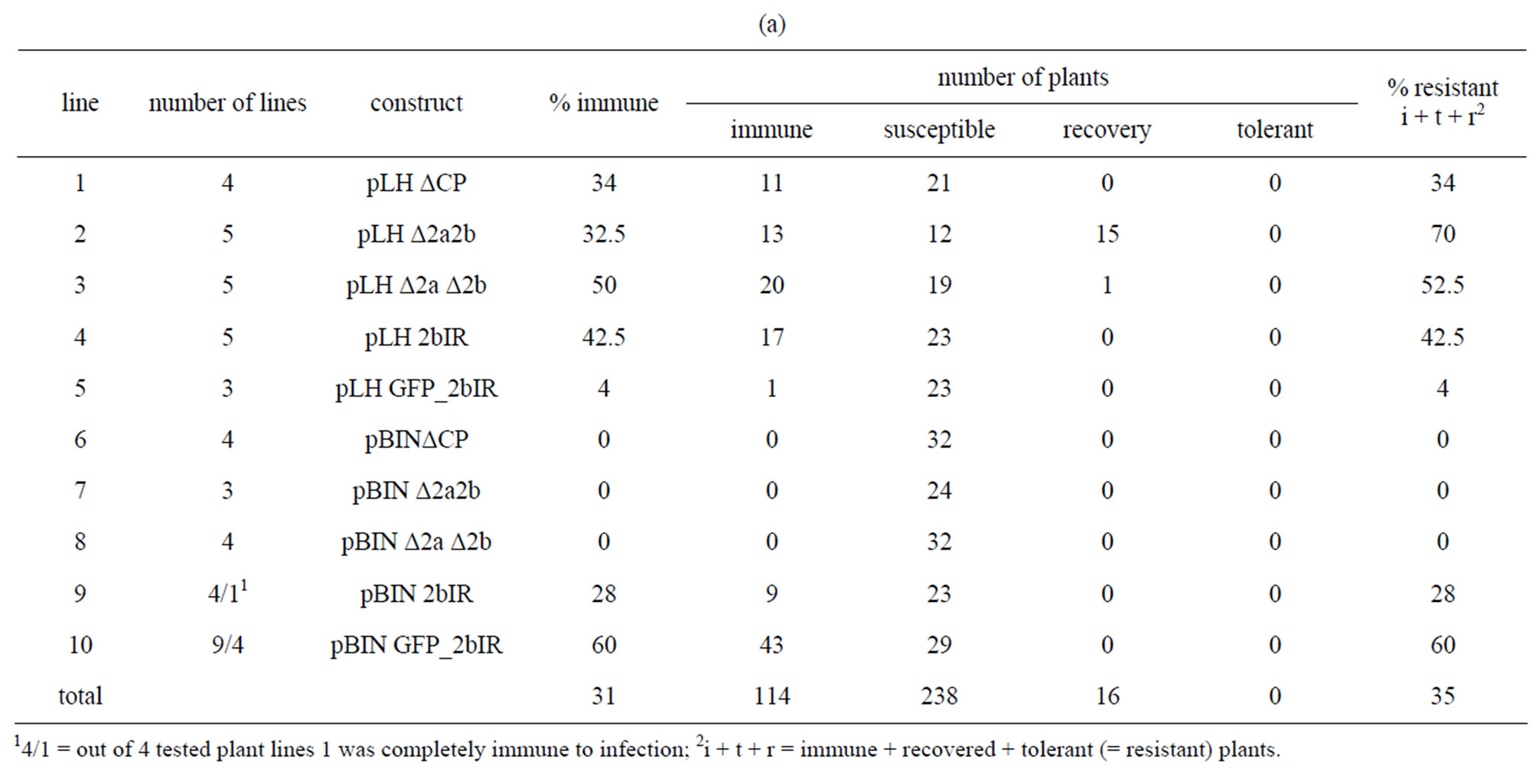
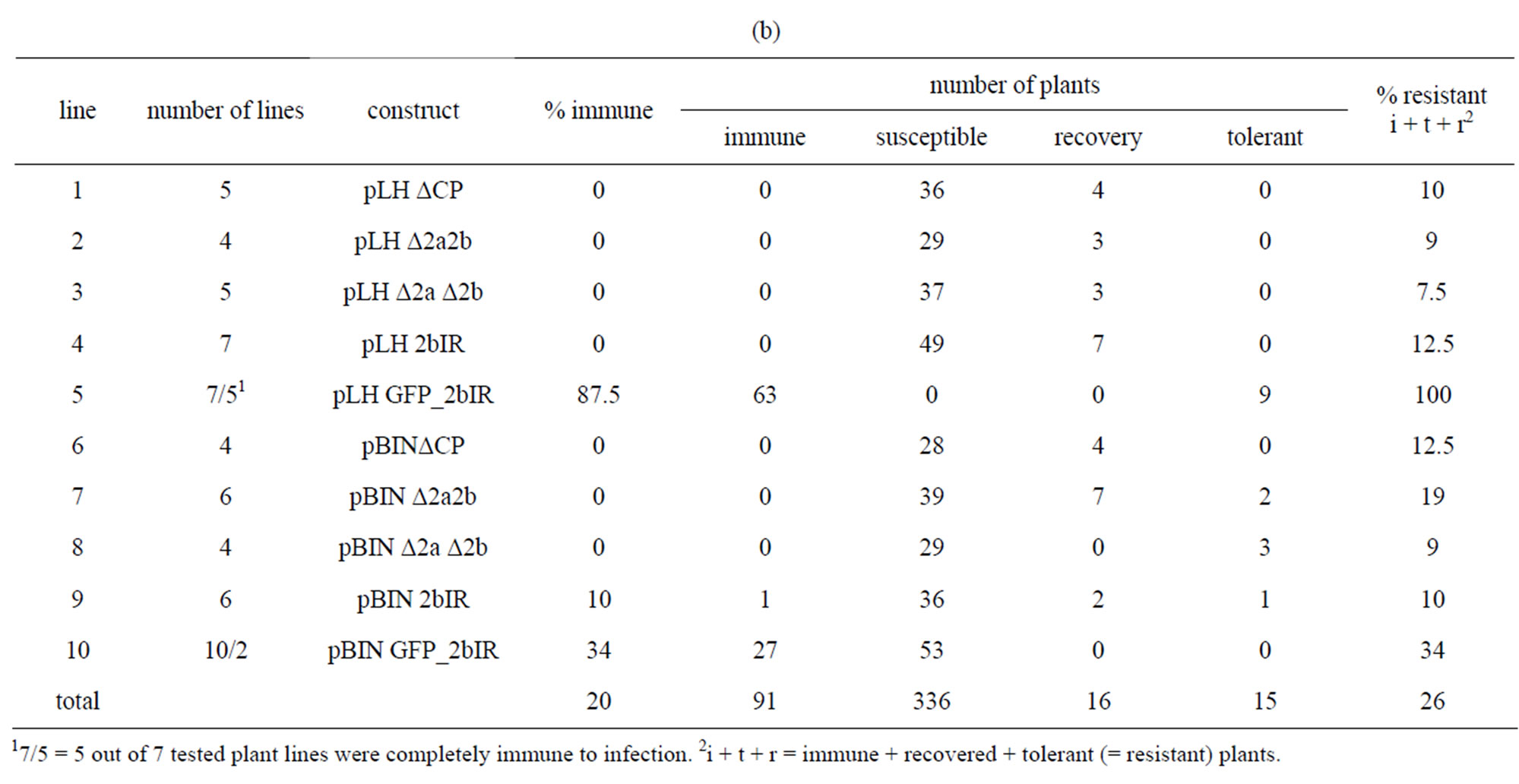
Table 3. (a) Resistance of N. benthamiana transformed with different constructs; (b) Resistance of N. tabacum cv. Samsun transformed with different constructs.
lines 1 - 3 and 6 - 8).
The 2bIR construct cloned in pLH6000 resulted in 42.5% immune N. benthamiana plants (Table 3(a), line 4), while all seven N. tabacum lines were susceptible to CMV infections when transformed with the pLH6000 vector (Table 3(b), line 4). The only observed effect was recovery of seven plants. Similar results were obtained using the pBIN19 vector (Table 3(a), line 9 and Table 3(b), line 9). From N. benthamiana plants transformed with pBIN19 28% immune plants were obtained. From these lines one line was 100% immune to infection.
In summary, N. benthamiana lines showed more immune plants when transformed with single geneor 2bIR-constructs than comparable N. tabacum lines. Transformation of N. benthamiana plants with single gene constructs led to resistance when using the pLH6000 vector but not with the pBIN19 vector. When comparing plants transformed with the inverted repeat construct (2bIR) both plant species, N. benthamiana and N. tabacum, showed immune plants, however in the combination of N. benthamiana plants and pLH6000 vector we obtained with 42.5% the highest score of immune plants.
In contrast to the high number of immune N. benthamiana plants, when transformed with single gene or inverted repeat constructs (2bIR) using the pLH6000 vector, only one out of 24 plants from three lines was not infected harboring the 2bIR construct flanked upstream by the GFP gene (GFP_2bIR, Table 3(a), line 5). All other host species/vector combinations resulted in a high number of resistant plants ranging between 34% and 87% (Table 3(a), line 10 and Table 3(b), lines 5 and 10). With the exception of the N. benthamiana/pLH6000 combination with all other host/vector combinations between one and four lines resulted in 100 % immune plants.
3.5. Testing Immune Lines with Heterologous Isolates from Subgroups Ib, Ia and II
Only plants of lines which were observed to be 100% immune to CMVAN were challenged with heterologous isolates from the subgroups 1b (CMVP3613 and CMVKS44), 1a (CMVRT52) and II (CMVPV0420).
With the exception of one plant which became infected (Table 4B, line 3) the three other N. benthamiana and N. tabacum plant lines transformed with the GFP_2bIR in pBIN19 and pLH6000, respectively, were immune to infection when challenged with the 1b isolates CMVP3613 and CMVKS44 (Table 4A-D, lines 2 and 3). A lower percentage of immune N. tabacum plants, ranging from 12.5% to 100%, was obtained from lines transformed with the pBIN19 vector (Table 4E, F, lines 2 and 3).
When plants of lines transformed with GFP_2bIR in pBIN19 were challenged with the subgroup 1A (CMVRT52) and subgroup II (CMVPV0420) isolates N. benthamiana plants showed 87.5% to 100% immune plants (Table 4A and B, lines 4 and 5) while N. tabacum plants showed not more than 37.5% immune plants (Table 4C-F, lines 4 and 5). N. tabacum plants transformed with GFP_ 2bIR in pBIN19 became completely infected with the serogroup II isolate PV0420 (Table 4E and F, line 5).
In summary, N. benthamiana plants transformed with GFP_2bIR in pBIN19 showed the highest number of immune plants when challenged with isolates from all subgroups.
4. Discussions
Defence against virus infection based on gene silencing may be the result of different strategies. As reviewed [34] the cytoplasmatic pathway is important in virus infection. The endogenous messenger RNA pathway negatively regulates viral gene expression and the suppression of viral transcription may be a result of siRNA guided DNA methylation, which might be activated via the introducetion of a transgene. Since different plant species and even different cultivars have a different genetic background, they might respond to the same virus in different ways although being transformed with the same antiviral construct.
This host dependence became clearly evident in our study when using the single gene constructs ΔCP, Δ2a2b and Δ2aΔ2b and inverted repeat construct (2bIR). Immune plants were only obtained when N. benthamiana plants were transformed, regardless of the translatability of the transgene and origin of the genome segment. N. benthamiana plants express a non functional salicylic acid (SA) inducible RdRP. Yang et al. [35] discussed that due to the missing RdRP the SDE 1-like RdRP produces increasing amounts of siRNAs from the pool of aberrant RNAs in N. benthamiana. This might lead to hyperactive gene silencing and might explain the superiority of N. benthamiana transgenic lines compared to the N. tabacum ones.
Because of antibiotic resistance conflicts the constructs based on pLH6000 were transformed with Agrobacterium strain GV3101, while the pBIN19 constructs were transformed with strain LBA4404. This implies that the influence on resistance of the vector and the Agrobacterium strain have to be discussed together.
Waterhouse et al. [36] summarized that only in plants with multiple, methylated copies of the transgene a co-suppression and virus resistance can be observed. From unpublished data [37] was stated that the copy number in single loci is correlated with Agrobacterium strains, which have different efficiencies to make single or multicopy T-DNA insertions. This is supported by our observation that only for pLH6000 constructs segregation patterns of 15:1 were observed, suggesting multicopy inserts at two different loci (data not shown).
According to [37] it might be possible, that the strain GV3101 did produce plant cells with more than one copy in a single locus arranged in a tandem array. As a consequence gene silencing and resistance is enhanced when comparing the single gene constructs ΔCP, Δ2a2b and Δ2aΔ2b. T-DNA delivery might also be a consequence of stability and replication efficiency in the Agrobacterium due to the origin of replication of the vector, which is pVS1 from Pseudomonas for pLH6000 [38] and RK2 for pBIN 19 [39]. The pVS1 origin of replication ensures good plasmid persistency in Agrobacterium sp. [38]. However, since we did not check for multicopy insertion, this explanation for significantly higher resistance remains highly speculative.
It is, however, reassuring that neither the translatability nor the genome segment origin (ΔCP from RNA 3 and all 2b containing constructs from RNA 2) of the transformed constructs seem to influence virus infection.
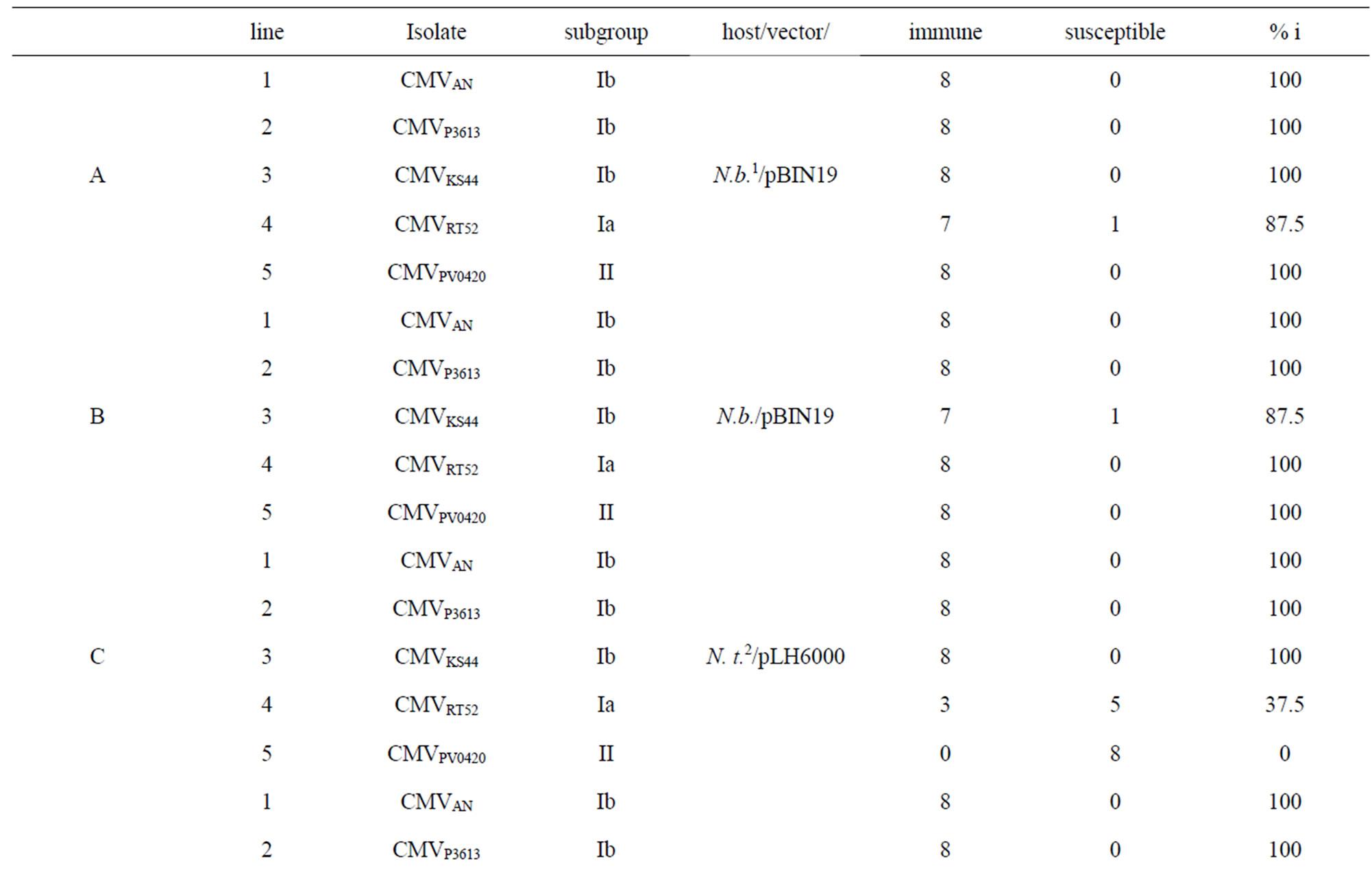
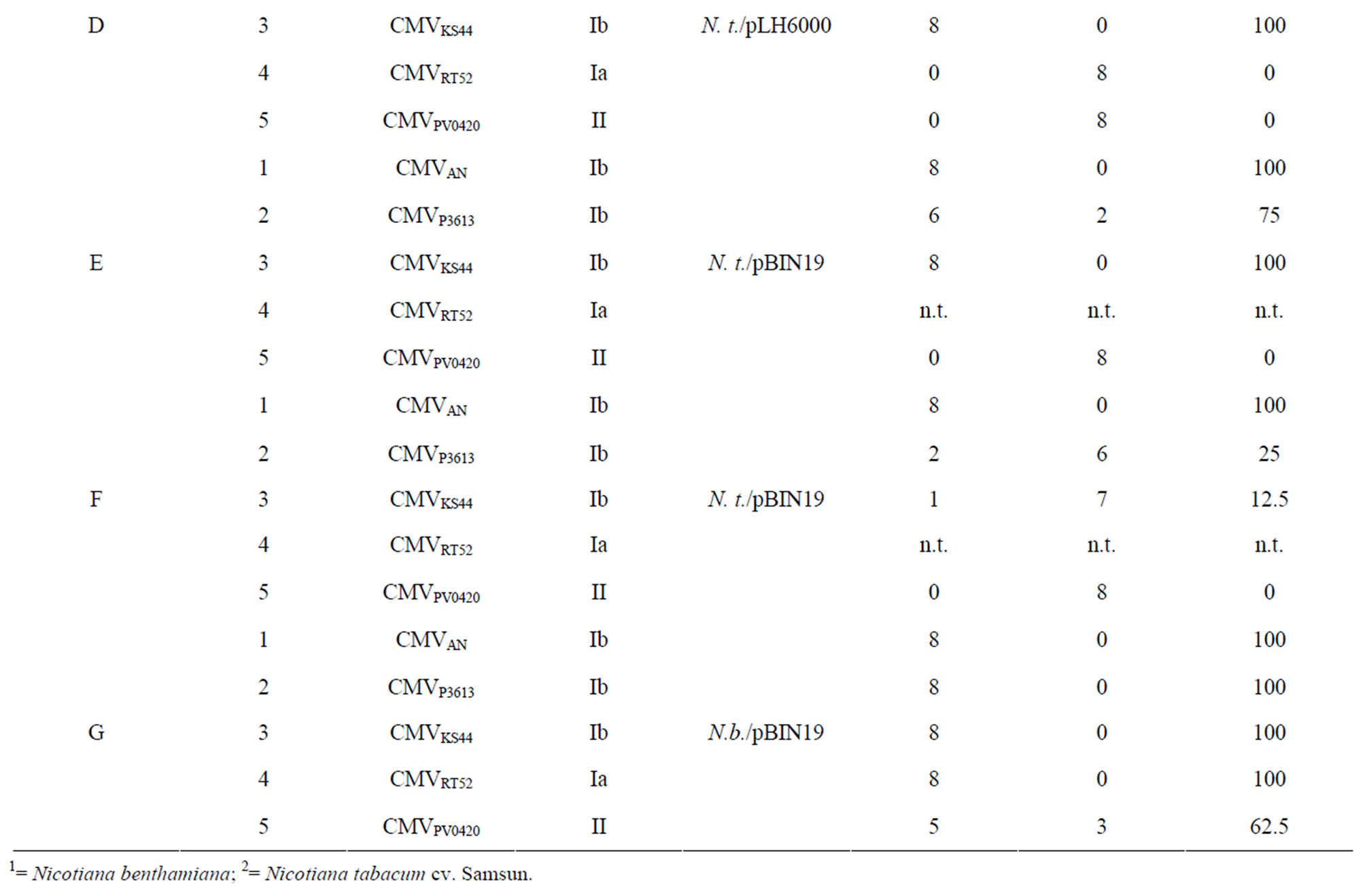
Table 4. Resistance against heterologous isolates of plant lines transformed with GFP_2bIR (A-F) and 2bIR (G), which were 100 % immune against the homologous isolate.
On the other hand the influence of the Agrobacterium strain and/or the vector is not evident when comparing plants transformed with the 2bIR inverted repeat construct. Obviously, the impact of multicopy inserts might be reduced, since the construct itself provides dsRNA. This is in good accordance with findings from [40] who proved the superiority of dsRNA constructs to obtain resistance against CMV. Their construct, based on the CP sequence and transformed into N. tabacum, resulted in 25% to 35% resistant plants, depending on the promoter for transgene expression. Chen et al. [41] extended their studies to several constructs based on the CP as well as the 2a2b sequence in long or short variants and compared the resistance achieved in their transgenic plants transformed with the respective viral regions as single gene constructs. The differences between their and our transgenic plants transformed with the 2a2b single gene constructs in the combination with pBIN19 as the vector and N. benthamiana are the length of the viral part of their constructs and the CMV isolate used for transformation and challenging. The difference in the viral sequence length might explain that we did not get any resistant plants while [41] obtained between 11% and 21% resistant plants in their experiments. Using the CP sequence for transformation, neither [41] nor we got any resistant N. benthamiana plant when using the pBIN19 vector system.
Chen et al. [41] used a CMV RNA2-based inverted repeat construct containing the 3’ part of 2a gene and the 2b gene. The resistance variation of their transgenic N. benthamiana plants against CMV revealed that the differences might be due to the length of the two sequences they used. Their long inverted repeat (LIR) covers 1534 bp and induced resistance in 75% of the plants, while their small inverted repeat (SIR) covered 490 bp and induced only 30% resistant plants. The resistance of the SIR is in good accordance with the data of our 2bIR construct with a viral sequence length of 549 bp. In our study, the resistance frequency ranged between 30% to 40% in N. benthamiana plants derived from pLH6000- 2bIR and pBIN19-2bIR.
When comparing the resistance induced by 2bIR with that of GFP_2bIR, an enhancement for the latter was clearly observed in both tobacco species. The enhancement is consistent with the results obtained by others, who fused GFP with a single fragment or a peptide of the N gene from TSWV and reported enhanced resistance in tobacco plants [42,43]. While all other constructs transformed with the vector pLH6000 in N. benthamiana resulted in highest numbers of immune plants, the performance of this plant/vector combination using GFP_ 2bIR as the antiviral insert resulted in a low number of immune plants. This observation cannot be explained by us.
According to the threshold model of gene silencing [44] it appears possible, that the GFP gene stabilizes the mRNA of 2bIR and prevents rapid degradation, or increases the transport of mRNA from nucleus to cytoplasm to trigger defense response by dsRNA-mediated resistance more efficiently. Probably the effect is not due to the GFP sequence itself, since [45] demonstrated a similar effect caused by a fusion of the NIb gene of Potato virus Y (PVY) with a blue fluorescence protein gene and [30] fusing a nonsense sequence to the N protein gene of Tomato spotted wilt virus (TSWV). However, the nonsense sequence used by [30] was translatable and it cannot be excluded that the effect was protein rather than RNAi mediated. The inoculum dose-dependence for breaking the resistance is supported by our observation of 100% infection when inoculating the GFP_2bIR lines 1 and 6 with two different methods. An immunity of 100% challenged plants was obtained by mechanical inoculation, whereas inoculation by grafting broke resistance (data not shown). A similar effect has been shown by [46] in Solanum lycopersicum L.
In respect to the statement of [47] that for RNA mediated resistance a minimum of 90% identical nucleotides is necessary, our results are unusual. None of the heterologous isolates of subgroup Ia and II did fullfill this condition. However, a common stretch of 23 conserved nucleotides (data not shown) might have led to variable degrees of RNAi mediated immunity. None of the lines with 100% immunity to a challenge with the homologous virus isolate CMVAN used for transformation, revealed a similar level of resistance when challenged with nonhomologous isolates. Only N. benthamiana plants transformed with the pBIN19-GFP_2bIR construct showed 87.5% to 100% immune plants when challenged with isolates from subgroups Ib, Ia and II. All other lines that were immune to the homologous isolate showed a lower degree of resistance when challenged with non-homologous isolates. Possibly, the degree of sequence identity has a similar effect as the dose of inoculum for the resistance effect. A possible explanation might be the different efficiency of the gene silencing due to lower sequence identities between the transgene and the challenging viruses, all applied as inoculum with comparable specific infectivity.
From our comparisons we can conclude that the kind of insert has highest influence for the generation of immune plants. However, also the vector/Agrobacterium system seemed to be highly important—at least when the challenging virus was not the homologous one.
5. Acknowledgements
The skillful technical assistance of Mrs. Mehrmann for laboratory and Mrs. George for the greenhouse work is acknowledged. We thank the German Collection of Microorganisms and Cell Cultures (DSMZ) for providing polyclonal and monoclonal CMV antiserum. The work was part of the project 2001.7860.8-001.00/contract No. 81051899 funded by the Deutsche Gesellschaft für Technische Zusammenarbeit GmbH (GTZ, Germany). P. W. acknowledges the financial support from the Hamburger Senat für Arbeit und Soziales.
REFERENCES
- J. F. Hadden and L. L. Black, “Anthracnose of Pepper Caused by Colletotrichum spp.,” International Symposium on Integrated Management Practices for Tomato and Pepper Production in the Tropics, Taiwan, 21-26 March 1988, Taiwan Asian Vegetable Research and Development Centre, pp. 189-199.
- B. Villalon, “Breeding Peppers to Resist Virus Diseases,” Plant Disease, Vol. 65, No.7, 1981, pp. 557-561. doi:10.1094/PD-65-557
- S. K. Green and J. S. Kim, “Characterization and Control of Pepper Viruses: A Literature Review,” AVRDC Technical Bulletin, Vol. 18, 1991, pp. 1-60.
- P. Palukaitis, and F. Garcia Arenal, “Cucumoviruses,” Advances in Virus Research, Vol. 62, 2003, pp. 241-323. doi:10.1016/S0065-3527(03)62005-1
- P. Palukaitis, M. J. Roossinck, R. G. Dietzgen and R. J. Francki, “Cucumber mosaic virus,” Advances in Virus Research, Vol. 41, 1992, pp. 281-348. doi:10.1016/S0065-3527(08)60039-1
- S. S. Lin, R. Henriques, H. W. Wu, Q. W. Niu, S. D. Yeh, N. H. Chua, P. Palukaitis and F. GarciaArenal, “Cucumoviruses,” Advances in Virus Research, Vol. 62, 2003, pp. 241-323. doi:10.1016/S0065-3527(03)62005-1
- M. J. Roossinck, L. Zhang and K. H. Hellwald, “Rearrangement in the 5′ Nontranslated Region and Phylogenetic Analyses of Cucumber mosaic virus RNA3 Indicate Radial Evolution of Three Subgroups,” Journal of Virology, Vol. 73, No. 9, 1999, pp. 6752-6758.
- D. Zhang, P. Willingmann, C. Heinze, G. Adam, M. Pfunder, B. Frey and J. E. Frey, “Differentiation of Cucumber mosaic virus Isolates by Hybridization to Oligonucleotides in a Microarray Format,” Journal of Virological Methods, Vol. 123, No. 1, 2005, pp. 101-108. doi:10.1016/j.jviromet.2004.09.021
- J. L. Troutman and R. W. Fulton, “Resistance in Tobacco to Cucumber mosaic virus,” Virology, Vol. 6, No. 2, 1958, pp. 303-316. doi:10.1016/0042-6822(58)90084-9
- C. Caranta, A. Palloix, V. Lefebvre and A. M. Daubèze, “QTLs for a Component of Partial Resistance to Cucumber mosaic virus in Pepper: Restriction of Virus Installation in Hostcells,” Theoretical and Applied Genetics, Vol. 94, No. 3-4, 1997, pp. 431-438. doi:10.1007/s001220050433
- C. Caranta, S. Pflieger, V. Lefebvre, A. M. Daubèze, A. Thabuis and A. Palloix, “QTLs Involved in Restriction of Cucumber mosaic virus (CMV) Longdistance Movement in Pepper,” Theoretical and Applied Genetics, Vol. 104, No. 2, 2002, pp. 586-591.
- R. C. Grube, E. R. Radwansky and M. Jahn, “Comparative Genetics of Disease Resistance within the Solanaceae,” Genetics, Vol. 155, No. 2, 2000, pp. 873-887.
- K. Suzuki, T. Kuroda, Y. Miura and J. Muria, “Screening and Weld Traits of Virus Resistant Source in Capsicum spp,” Plant Disease, Vol. 87, No. 7, 2003, pp. 779-783. doi:10.1094/PDIS.2003.87.7.779
- W.H. Kang, N. H. Hoang, H. B. Yang, J. K. Kwon, S.H. Jo, J. K. Seo, K.H. Kim, D. Choi and B. C. Kang, “Molecular Mapping and Characterization of a Single Dominant Gene Controlling CMV Resistance in Peppers (Capsicum annuum L.),” Theoretical and Applied Genetics, Vol. 120, No. 8, 2009, pp. 1587-1596. doi:10.1007/s00122-010-1278-9
- M. Y. Lee, J. H. Lee, H. I. Ahn, M. J. Kim, N. H. Her, J. K. Choi, C. H. Harn and K. H. Ryu, “Identification and Sequence Analysis of RNA3 of a Resistance Breaking Isolate of Cucumber mosaic virus from Capsicum annuum,” Plant Pathology Journal, Vol. 22, No. 3, 2006, pp. 265-270. doi:10.5423/PPJ.2006.22.3.265
- D. Zhang, X. Tan, P. Willingmann, G. Adam and C. Heinze, “Problems Encountered with the Selection of Cucumber mosaic virus (CMV) Isolates for Resistance Breeding Programs,” Journal of Phytopathology, Vol. 159, No. 9, 2011, pp. 621-629. doi:10.1111/j.1439-0434.2011.01816.x
- M. Morroni, J. R. Thompson and M. Tepfer, “Twenty Years of Transgenic Plants Resistant to Cucumber mosaic virus,” Molecular Plant Microbe Interactions, Vol. 21, No. 6, 2008, pp. 675-684. doi:10.1094/MPMI-21-6-0675
- A.Srivastava and S. K. Raj, “Coat Protein-Mediated Resistance against an Indian Isolate of the Cucumber mosaic virus Subgroup IB in Nicotiana benthamiana,” Journal of Biosciences, Vol. 33, No. 3, 2008, pp. 249-257. doi:10.1007/s12038-008-0042-7
- W. M. Wintermantel and M. Zaitlin, “Transgene Translatability Increases Effectiveness of Replicase-Mediated Resistance to Cucumber mosaic virus,” Journal of General Virology, Vol. 81, No. 3, 2000, pp. 587-595.
- R. Shin, J. H. Han, G. J. Lee and K. H. Peak, “The Potential Use of a Viral Coat Protein Gene as a Transgene Screening Marker and Multiple Virus Resistance of Pepper Plants Coexpressing Coat Proteins of Cucumber mosaic virus and Tomato mosaic virus,” Transgenic Research, Vol. 11, No. 2, 2002, pp. 215-219. doi:10.1023/A:1015200622716
- W.Q. Cai, R.X. Fang, H. S. Shang, X. Wang, F. L. Zhang, Y. R. Li, J. C. Zhang, X. Y. Cheng, G. L. Wang and K. Q. Mang, “Development of CMV and TMV Resistant Chili Pepper: Field Performance and Biosafety Assessment,” Molecular Breeding, Vol. 11, No. 1, 2003, pp. 25-35. doi:10.1023/A:1022655204552
- Y. X. Zhu, W. J. OuYang, Y. F. Zhang and Z. L. Chen, “Transgenic Sweet Pepper Plants from Agrobacterium Mediated Transformation,” Plant Cell Reports, Vol. 16, No. 1-2, 1966, pp. 71-75. doi:10.1007/BF01275453
- Y. H. Lee, M. Jung, S. H. Shin, J. H. Lee, S. H. Choi, N. H. Her, J. H. Lee, K. H. Ryu, K. Y. Paek and C. H. Harn, “Transgenic Peppers That Are Highly Tolerant to a New CMV Pathotype,” Plant Cell Reports, Vol. 28, No. 2, 2009, pp. 223-232. doi:10.1007/BF01275453
- T. Christopher and M. V. Rajam, “Effect of Genotype, Explants and Medium on in Vitro Regeneration of Red Pepper,” Plant Cell and Tissue Organ Culture, Vol. 46, No. 3, 1996, pp. 245-250. doi:10.1007/BF02307101
- Y. H. Lee, H. S. Kim, J.Y. Kim, M. Jung, Y. S. Park, J. S. Lee, S. H. Choi, N. H. Her, J. H. Lee, N. L. Hyung, C. H. Lee, S. G. Yang and C. H. Harn, “A New Selection Method for Pepper Transformation: Callus Mediated Shoot Formation,” Plant Cell Reports, Vol. 23, No. 1-2, 2004, pp. 50-58. doi:10.1007/s00299-004-0791-1
- D. Li, K. Zhao, B. Xie, B. Zhang and K. Luo, “Establishment of a Highly Efficient Transformation System for Pepper (Capsicum annuum L.),” Plant Cell Reports, Vol. 21, No. 8, 2003, pp. 785-788.
- M. Manoharan, C. S. Sree Vidya and G. Lakshmi Sita, “Agrobacterium Mediated Genetic Transformation in Hot Chilli (Capsicum annuum L. var. Pusa jwala),” Plant Science, Vol. 131, No. 1, 1998, pp. 77-83. doi:10.1016/S0168-9452(97)00231-8
- V. Mihalka, M. Fari, A. Szasz, E. Balazs and I. Nagy, “Optimised Protocols for Efficient Plant Regeneration and Gene Transfer in Pepper (Capsicum annuum L.),” Journal of Plant Biotechnology, Vol. 2, No. 2, 2000, pp. 143-149.
- H. Lot, J. Marrou, J. B. Quiot and C. Esvan, “Contribution a l’Etude du Virus de la Mosaique du Concombre (CMV),” Annals of Phytopathology, Vol. 4, No. 1, 1972, pp. 25-38.
- F. Schwach, G. Adam and C. Heinze, “Expression of a Modified Nucleocapsidprotein of Tomato spotted wilt virus (TSWV) Confers Resistance against TSWV and Groundnut ringspot virus (GRSV) by Blocking Systemic Spread,” Molecular Plant Pathology, Vol. 5, No. 4, 2004, pp. 309-316. doi:10.1111/j.1364-3703.2004.00229.x
- W. Menzel, W. Jelkmann and E. Maiss, “Detection of Four Apple Viruses by Multiplex RTPCR Assays with Coamplification of Plant mRNA as Internal Control,” Journal of Virological Methods, Vol. 99, No. 1-2, 2002, pp. 81-92. doi:10.1016/S0166-0934(01)00381-0
- R. B. Horsch, J. E. Fry, N. L. Hoffmann, D. Eichholtz, S. G. Rogers and R. T. Fraley, “A Simple and General Method for Transferring Genes into Plants,” Science, Vol. 111, No. 4691, 1985, pp. 1229-1231.
- T. Murashige and F. K. Skoog, “A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Cultures,” Physiologia Plantarum, Vol. 15, No. 3, 1962, pp. 473-497. doi:10.1111/j.1399-3054.1962.tb08052.x
- D. Baulcombe, “RNA Silencing in Plants,” Nature, Vol. 431, No. 7006, 2004, pp. 356-363. doi:10.1038/nature02874
- S. J. Yang, S. A. Carter, A. B. Cole,N. H. Cheng and R. S. Nelson, “A Natural Variant of a Host RNA Dependent RNA Polymerase Is Associated with Increased Susceptibility to Viruses by Nicotiana benthamiana,” Proceedings of the National Academy of Sciences USA, Vol. 101, No. 16, 2004, pp. 6297-6302. doi:10.1073/pnas.0304346101
- P. M. Waterhouse, M. B. Wang and T. Lough, “Gene Silencing as an Adaptive Defence against Virus,” Nature, Vol. 411, No. 6839, 2001, pp. 834-842. doi:10.1038/35081168
- S. B. Gelvin, “Agrobacterium and Plant Transformation: The Biology behind the ‘Gene Jockeying’ Tool,” Microbiology and Molecular Biology Reviews, Vol. 67, No. 1, 2003, pp. 16-37. doi:10.1128/MMBR.67.1.16-37.2003
- Y. Itoh, J. M. Watson, D. Haas and T. Leisinger, “Genetic and Molecular Characterization of the Pseudomonas Plasmid pVS1,” Plasmid, Vol. 11, No. 3, 1984, pp. 206- 220. doi:10.1016/0147-619X(84)90027-1
- M. Bevan, “Binary Agrobacterium Vectors for Plant Transformation,” Nucleic Acids Research, Vol. 12, No. 22, 1984, pp. 8711-8721. doi:10.1093/nar/12.22.8711
- K. Kalanditis, S. Psarsdakis, M. Tabler and M. Tsagris, “The Occurrence of CMV Specific Short RNAs in Transgenic Tobacco Expressing Virus Derived Doublestranded RNA Is Indicative of Resistance to the Virus,” Molecular Plant Microbe Interaction, Vol. 15, No. 8, 2002, pp. 826-833. doi:10.1094/MPMI.2002.15.8.826
- Y. K. Chen, D. Lohuis, R. Goldbach and M. Prins, “High Frequency Induction of RNA Mediated Resistance against Cucumber mosaic virus Using Inverted Repeat Constructs,” Molecular Breeding, Vol. 14, No. 3, 2004, pp. 216-225. doi:10.1023/B:MOLB.0000047769.82881.f5
- S. Z. Pang, F. J. Jan and D. Gonsalves, “Nontarget DNA Sequences Reduce the Transgene Length Necessary for RNA Mediated Tospovirus Resistance in Transgenic Plants,” Proceedings of the National Academy of Sciences USA, Vol. 94, No. 94, 1997, pp. 8261-8266. doi:10.1073/pnas.94.15.8261
- C. Rudolph, P. H. Schreier and J. F. Uhrig, “PeptideMediated Broad-Spectrum Plant Resistance to Tospoviruses,” Proceedings of the National Academy of Sciences USA, Vol. 100, No. 8, 2003, pp. 4429-4434. doi:10.1073/pnas.0730832100
- W. G. Dougherty and T. D. Parks, “Transgenes and Gene Suppression: Telling Us Something New?” Current Opinion in Cell Biology, Vol. 7, No. 3, 1995, pp. 399-405. doi:10.1016/0955-0674(95)80096-4
- J. Schubert, J. Matoušek and D. Mattern, “Pathogenderived Resistance in Potato to Potato virus Y: Aspects of Stability and Biosafety under Field Conditions,” Virus Research, Vol. 100, No. 1, 2004, pp. 41-50. doi:10.1016/j.virusres.2003.12.013
- J. L. Abad, G. Anastasio, M. A. Fraile and F. García Arenal, “A Search for Resistance to Cucumber mosaic virus in the Genus Lycopersicon,” Journal of Plant Pathology, Vol. 82, No. 1, 2000, pp. 39-48.
- C. Ritzenthaler, “Resistance to Plants Viruses: Old Issue, New Answers?” Current Opinion in Biotechnology, Vol. 16, No. 2, 2005, pp. 118-122. doi:10.1016/j.copbio.2005.02.009
- C. Reichel, J. Mathur, P. Eckes, K. Langenkemper, C. Koncz, J. Schell, B. Reiss and C. Maas, “Enhanced Green Fluorescence by the Expression of an Aequorea Victoria Green Fluorescent Protein Mutant in Mono and Dicotyledonous Plant Cells,” Proceedings of the National Academy of Sciences USA, Vol. 93, No. 12, 1996, pp. 5888-5893. doi:10.1073/pnas.93.12.5888
- R. Higuchi, B. Krummel and R. K. Saiki, “A General Method of in Vitro Preparation and Specific Mutagenesis of DNA Fragements: Study of Protein and DNA Interactions,” Nucleic Acids Research, Vol. 16, No. 15, 1998, pp. 7351-7367. doi:10.1093/nar/16.15.7351
Supporting Information
Construction of pLH6000-GFP
The BamHI-restriction recognition site of the plasmid pBlueScript II SK-(Stratagene) was removed by digestion and filling in SK-/ΔBamHI. A fragment of 1867 bp, containing the GFP-gene driven by a double 35S promoter and with a Nos terminator 2x35S/GFP/Nos, was released by HindIII digestion from the plasmid pCKGFPS65C [48], ligated into a HindIII linearized SK-/ΔBamHI vector and transformed in E.coli . In a correct orientation the SpeI site from SK-/ΔBamHI is located upstream of the 35S promoter. The resulting clone was named SK- ΔBamHI-(2x35S/GFP/Nos). The cassette of ΔBamHI- (2x35S/GFP/Nos) from this clone was isolated by SpeI and KpnI digestion and cloned into the SpeI/KpnI site of pLH6000. The resulting plasmid was named pLH6000- GFP.
Construction of pLH6000-ΔCP
The CP gene of CMVAN was amplified by RT-PCT using primers CMV-CP ΔNcoI (5’-ctagagccatggtggacaaatctgg at-3’) and CMV-CP BamHI (5’-gacgtcggatccctggatggac aaccc-3’). To join the ΔCP fragment with the 2x35S promoter and NOS terminator, the GFP gene from the clone SK-ΔBamHI-(2x35S/GFP/Nos) was removed by NcoI/BamHI digestion and the remaining vector was used SK-ΔBamHI-(2x35S/ΔCP/Nos). From this clone the 2x35S/ΔCP/Nos cassette was isolated and ligated into the SpeI/KpnI site of pLH6000 to obtain pLH6000-ΔCP.
Construction of pLH6000-Δ2a2b in Which 2b Is Translatable
A 735 bp fragment containing 641bp of the 3’ part of 2a and the complete 336 bp of the 2b gene from CMVAN with a 242 bp overlap of 2a and 2b, located between nucleotide position 2130 and 2864 on the CMVAN RNA 2 was amplified by RT-PCR with primers 5’-RNA2 (5’-gat gaattcytgtttgctcac-3’) and 3’-RNA2 (5’-ggatggacaacccgt tc-3’) and subcloned (SK-Δ2a + 2b). Two new restriction sites for further subcloning, NcoI and BamHI, were introduced by PCR with the primers CMV-2b-NcoI (5’-cta gagccatggtgaattcttgtttgc-3’) and CMV-2b-BamHI (gacgtc ggatccggatggacaacccgt-3’) using plasmid SK-Δ2a + 2b) as template. The plasmid SK-Δ2a + 2b was digested with NcoI/BamHI and the Δ2a + 2b was isolated and ligated into the NcoI/BamHI linearized plasmid (SK-ΔBamHI- (2x35S/ΔCP/Nos) and transformed into E. coli (SK- ΔBamHI-(2x35S/Δ2a + 2b /Nos). The cassette ΔBamHI- (2x35S/Δ2a + 2b/Nos was isolated by SpeI/KpnI digestion and ligated into the SpeI/KpnI site of pLH6000 (pLH6000-Δ2a2b).
Construction of pLH6000-Δ2a + Δ2b in Which 2b Is Not Translatable
The 2b gene was modified into a non-translatable construct called Δ2a + Δ2b by removing the adenine of the start codon. Using the plasmid SK-Δ2a + 2b as template, the start codon of 2b gene was removed by site-directed mutagenesis according to [49] with the two primer pairs 2b-MS-FOR (5’-gaagaaagtggaattgaacgaaggcgc-3’)/CMV- 2b-BamHI (5’-gacgtcggatccggatggacaacccgt-3’) and CMV-2b-NcoI (5’-ctagagccatggtgaattcttgtttgc-3’)/2bMS-REV (5’-cgttcaattccactttcttctttcgctgc-3’) to generate the Δ2a + Δ2b fragment. The Δ2a + Δ2b fragment was reamplified by primers CMV-2b-NcoI and CMV-2bBamHI with the Δ2a + Δ2b fragment as template and subcloned (SK-Δ2a + Δ2b). The fragment Δ2a + Δ2b from plasmid SK-Δ2a + Δ2b was digested by NcoI/ BamHI, isolated and subcloned into the NcoI/BamHI linearized vector (SK-ΔBamHI-(2x35S/GFP/Nos) to obtain SK-ΔBamHI-(2x35S/Δ2a + Δ2b/Nos). The insert of SK-ΔBamHI-(2x35S/Δ2a + Δ2b/Nos) was digested by HindIII and cloned into pLH6000. The resulting clone was named pLH6000-Δ2aΔ2b.
Construction of 2b with an Inverted Repeat pLH6000-2bIR
For construction of the pLH6000-2bIR, all functional elements were generated separately while introducing restriction sites and subcloned consecutively.
The 198 bp intron from plasmid p1353dsCMVIR was amplified by primers Intron PstI (5’-tatacgatctgcaggcg ctcgcc-3’) and Intron_XbaI (5’-ccctctagataagtttctgc-3’) and ligated into the PstI/XbaI site of SK- (SK-Intron). Antisense and sense strand of the 2b gene, a fragment of 549 bp (containing 335 bp upstream from 2b gene and 455bp downstream from 3’ part of 2a gene but with a 242 bp overlapping region) from position 2253 nt to 2802 nt of CMVAN RNA 2, were amplified with primers 2bAN_ PstI (5’-aatactgcagactcagccc-3’) and 2bAN_BamHI_ XhoI (5’-tacaggatcccaggactcgaggctg-3’) for antisense, 2b_AN_SacI_NcoI (5’-atacagagctccatgggccgaggctgc-3’) and 2b_AN_XbaI (5’-gacagtctagagcaatactgcc-3’) for sense using plasmid SK-Δ2a + Δ2b as template. The two fragments were subcloned to obtain SK-anti2b) and SKsense2b. The anti2b fragment from plasmid SK-anti2b was isolated by PstI/XhoI digestion and ligated with in a PstI and XhoI linearized (SK-Intron) to obtain SK-Intronanti2b. The sense2b fragment was isolated from plasmid SK-sense2b by XbaI and ligated in a XbaI linearized SK-Intron-anti2b vector (SK-ds2bIR). The orientation of the recombinant was identified by BamHI digestion.
The DNA fragments of 2x35S promoter and Nos terminator from plasmid SK-ΔBamHI-(2x35S/Δ2a + Δ2b/ Nos) were isolated by SpeI/NcoI (35S promoter) and BamHI/KpnI (Nos terminator) digestion, respectively. The 2x35S promoter fragment was ligated with the SpeI/ NcoI linearized plasmid SK-ds2bIR to obtain SK-2x35S/ 2bIR. The Nos terminator was ligated in the BamHI/KpnI linearized plasmid SK-2x35S/ds2bIR to obtain SK-2x35S/ 2bIR/Nos. Then the cassette 2x35S/2bIR/Nos from plasmid SK-2x35S/2bIR/Nos was isolated by HindIII digestion and ligated with HindIII linearized pLH6000 (pLH6000-2bIR).
Construction of the Chimeric Construct pLH6000-GFP_2bIR
To join 2bIR with GFP from pCKGFPS65C, a fragment of 2bIR from plasmids of SK-2bIR was isolated with BamHI and ligated in a BamHI linearized SK-ΔBamHI- (2x35S/GFP/Nos). The correct orientation of the recombinant plasmid, GFP: sense2b: intron: antisense2b, was determined by NcoI digestion (SK-ΔBamHI-(2x35S/ GFP_2bIR/Nos). The fragment GFP_2bIR from plasmid SK-ΔBamHI-(2x35S/GFP + 2bIR/Nos) was isolated by SpeI/KpnI digestion and cloned in a SpeI/KpnI linearized pLH6000 (pLH6000-GFP_2bIR). In this construct GFP is translatable.
Transfer of Cassettes into pBIN19
The ΔCP and GFP_2bIR cassettes were cloned in the SpeI/KpnI and the Δ2a2b, Δ2aΔ2b and 2bIR cassettes in the HindIII site of pBIN19.
NOTES
*Corresponding author.

