American Journal of Plant Sciences
Vol.3 No.1(2012), Article ID:16620,4 pages DOI:10.4236/ajps.2012.31005
Molecular Characterization of Pseudomonas spp. Isolated from Root Nodules of Various Leguminous Plants of Shekhawati Region, Rajasthan, India
![]()
1Department of Science, FASC, Mody Institute of Technology and Science, Sikar, India; 2Institute of Genomics and Integrative Biology, Delhi, India.
Email: *gaurrajarshi@hotmail.com
Received April 15th, 2011; revised July 1st, 2011; accepted August 2nd, 2011
Keywords: PGPRs; 16S Sequencing; Pseudomonas; Root-Nodule Bacteria; 16S rDNA
ABSTRACT
Plant growth promotory Pseudomonas strains were isolated from root nodules of five plant species, viz., Trifolium pretense, Cicer arietinum, Amaranthus polygamus, Vigna mungo, and Trigonella foenum; that plants were denizen of Shekhawati region of Rajasthan. A total of 8 bacterial isolates were evaluated for growth promotion using PGP properties. Partial 16S rDNA sequencing data showed that these 8 bacterial isolates belonged to genus Pseudomonas. MEGA 4.0.2, software was used to construct a neighbor joining tree by employing boot strap method. Result exhibited significant diversity among recovered Pseudomonas strains.
1. Introduction
Bacterial diversity is of particular importance in human sustenance since these small creatures comprise the majority of earth’s species diversity. Bacterial diversity is considered as one of the most useful resource with considerable significance in the global form of bioremediation and bio-prospecting [1]. Interaction between bacteria and roots of plants has been reported to be beneficial, detrimental or neutral and this delicate balance is a consequence of both soil and plant type [2]. Bacteria, beneficial to plants may be symbiotic or free living, and are abundant near the roots. Such beneficial free-living bacteria have been termed PGPR or plant growth promoting rhizobacteria [3]. They benefit plants through, 1) production of plant hormones, such as auxins, 2) asymbiotic N2 fixation, 3) antagonism against phytopathogenic microorganisms by production of antibiotics, siderophroes, -(1, 3)-glucanase, chitinase, and cyanide, and 4) solubilization of mineral phosphates and other nutrients [4-8]. A number of PGPR such as Bacillus, Pseudomonas and Arthrobacter have been used for enhancement of plant performance [9,10].
The use of fluorescents pseudomonads (FLPs) as PGPR requires precise understanding of the interactions between plant-bacteria, among bacteria-microbiota, and how biotic and abiotic factors influence these relationships. Over the last few years, modern technologies, such as immunofluorescence microscopy and reporter genes, have improved the study of Pseudomonas inoculants in soil and have markedly enhanced the knowledge about their behaviour in this environment [11].
Nevertheless it is still necessary to better understand the plant response to the presence of the introduced bacteria. An important consideration is the characterization of the rhizosphere populations. Comprehension of the dynamics of the microbial populations could shed light on the process of selecting successful strains that promote plant growth and/or suppressing diseases. Recent advances in the study of the intraand inter-species signaling are providing an important area for scientific research, as well as, relevant application. Understanding quorum sensing systems in antifungal metabolite production and identification of promoters that can be induced or increased in the rhizosphere provides new approaches for the development of new biological control agents [12].
In the present study a molecular approach was used to characterize pseudomonads population recovered from root nodules of five plant species, viz., Trifolium pretense, Cicer arietinum, Amaranthus polygamus, Vigna mungo, and Trigonella foenum; that plants were denizen of Shekhawati region of Rajasthan, India.
2. Materials and Methods
2.1. Isolation of Bacteria
Root nodules of different leguminous plants were collected from different arid areas of Shekhawati region of Rajasthan. For the isolation of bacterial strains, the nodule surface was sterilized with 0.1% HgCl2 for 2 - 3 min and crushed in sterilized distilled water. Bacterial population was enumerated by employing yeast extract mannitol agar (YEMA) [13], and chromeazurol “S”(CAS) agar for siderophore producers [14]. Plates were incubated in triplicates at 28˚C.
2.2. Functional Attributes of the Bacterial Isolates
Isolates were tested for their ability to produce indole acetic acid [15,16], HCN, and siderophore.
2.3. IAA Production
Bacteria were grown overnight in five ml of M-9 minimal medium [15] supplemented with L-tryptophan to achieve a final concentration of 0, 50, 100, 200 and 500 g·ml–1. After incubation for 42 h, bacterial growth was measured spectrophotometrically at 600 nm; cells were removed from culture medium by centrifugation at 7500 rpm for 10 min. A 1 ml aliquot of supernatant was mixed with 4 ml of Salkowski’s reagent (150 ml of concentrated H2SO4, 250 ml of D.W., 7.5 ml of 0.5 M FeCl3·6H2O). Samples were left at room temperature for 25 min and absorbance was read at 535 nm. The concentration of IAA was determined by referring to a standard curve.
2.4. DNA Isolation and PCR
Eight strains (TF-L, CH-L, CP-K, CP-L, MB-K, MB-L, TK-L, and TK-S) were characterized employing 16S rRNA sequencing. For Genomic DNA extraction, the bacteria were grown in Nutrient Broth on an incubator shaker (120-rev·min–1) at 28˚C for 24 hours. The HiPurATM Genomic DNA Miniprep Purification Spin Kit for gram negative bacteria (HIMEDIA, Mumbai, India) was used to isolate DNA as described by manual of manufacturer. For DNA amplification, pure 3 μl DNA sample was used. The fD1 and rD1 primers were used for DNA amplification, which are used for the amplification of 16S rDNA gene [17]. PCR reactions were performed in a final volume of 60 μl with 1.2 μl of each primer, 1.2 μl of MgCl2 (25 mM), 1.5 μl of dNTP mix (10 mM), 1.2 μl of Taq DNA polymerase with 6.0 μl PCR buffer, 3.0 μl DMSO (99.9%) and 41.7 μl Nuclease free water. This reaction mixture was made for 10 bacterial samples. The DNA was amplified over 30 cycles of Denaturation for 1 min at 95˚C, annealing at 59˚C for 1 min and extension at 72˚C for 1 min. The final extension at which DNA extend, was 72˚C for 7 min. The conformation of Amplification was done by Agarose Gel Electrophoresis by running 5 μl of PCR reaction mixture on 1% Agarose gel.
2.5. Sequencing of 16S rDNA
16S PCR product was purified with exo-sap. The sequences were determined by using Big dye terminator Kit (ABI Foster city, CA, USA) on an ABI 310 DNA sequencer. For NJ tree, sequences were aligned using MEGA 4.0.2 software. The resulting sequences were compared with those stored in the gene bank data system using BLAST search [18]. The accession numbers of the 16S rDNA sequences are described: TF-L (HQ890310), CH-L (HQ890311), CP-K (HQ890312), CP-L (HQ890313), MB-K (HQ890314), MB-L (HQ890315), TK-L (HQ890317), and TK-S (HQ890318).
3. Results and Discussion
Molecular tools for the identification of soil bacteria were used and 16S rRNA gene analysis was intensively used to understand the phylogenetic relationships. Bacterial phylogenetic classification is based on sequence analysis of the SSU 16S rRNA molecule or its genes. Over 20,000 SSU RNA gene sequences have now been deposited in specialist r-RNA databases such as the rRNA Database Project (RDP) [19]. The 5-end region of the gene is useful for discrimination of Bacillus species [20], corynebacteria [21], and mycobacteria [22], whereas the distal region was used for identification of various clinical microorganisms [23]. A particularly exciting development is the use of real time DNA sequencing based on pyrosequencing to provide the DNA sequence for database searching [24]. Given the conservation of 16S rRNA gene, at least 99% similarity seems to be a commonly accepted score for identification [25,26].
For further identification at genus level, bacterial isolates were identified through homology search with BLAST and FASTA using partial sequence of 16S rDNA. Sequencing data showed that these eight isolates belonged to genus, Pseudomonas spp. being a dominant species. Only one bacterial isolate, CH-L showed distinct group from others and exhibited similarity with reference strains, P. aeruginosa and P. mendocina. A large group was made by isolates, TF-L, TK-S, MB-L with the reference strains, P. putida, P. gingeri, P. teessidea, P. moorei, P. koreensis, and P. fluorescens. Other isolates exhibited significant discriminatory relatedness with reference strains (Figure 1). All the sequences obtained from eight isolates were aligned with each other to de-
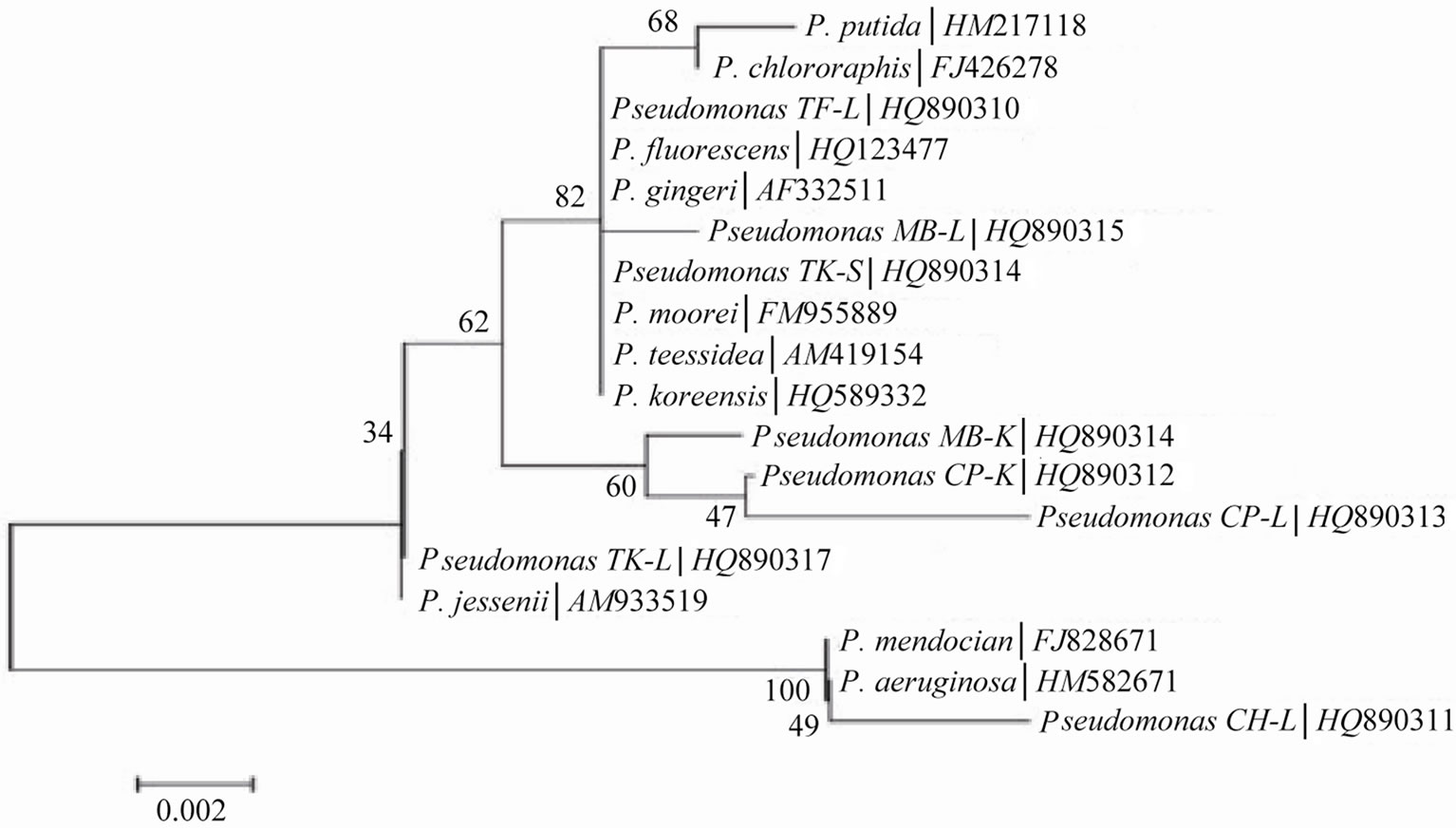
Figure 1. Homology tree constructs by employing bootstrap method.
termine genetic diversity amongst the root-nodule bacteria. A comparison of 16S rDNA sequence with the reference strain, Pseudomonas spp. to which they matched, was performed. A consensus tree was drawn from these aligned sequences using MEGA version 4.0.2. On the basis of 16S rDNA sequence homology four major clusters were formed (Figure 1).
Homology tree based on sequence alignment of 16S rDNA of bacterial isolates permitted rapid phylogenetic analysis. However, strains isolated from different geographic location shared similar DNA homology. Phylogenetic analysis on the basis of 16S rDNA sequences provided better understanding in evaluation of genetic diversity of bacteria isolated from same and different ecological niche; phylogenetic analysis of 500 bp of terminal region of 16S rDNA from cultivated strain has been found to show existence of large bacterial diversity [27].
4. Conclusions
Finally, it is concluded that all bacterial isolates obtained from root nodules of various leguminous plants, were found more similar with Pseudomonas spp. after phylogenetic analysis and they were also showing Plant Growth Promoting activities, which are essential for plant growth and nutrition.
So, these agronomic strategies are relevant for achieving sustainable agriculture as one of the goals for using PGPRs is to make them reliable, assessable product for the farmers. Consequently, continued research is needed to develop new approaches to ameliorate the efficiency of PGPRs and to understand the ecological, genetic and biochemical relationships in their habitat.
5. Acknowledgements
Authors are thankful to Department of Science and Technology, Rajasthan, India for their financial support under student project scheme.
REFERENCES
- M. C. Homer-Devine, K. M. Carney and B. J. M. Bohannan, “An Ecological Perspective on Bacterial Biodiversity,” Proceedings of the Royal Society of London, Vol. 271, 2004, pp. 113-122.
- X. Latour, T. Carberand, G. Lagurre, F. Alland and P. Lemanceu, “The Composition of Fluorescent Pseudomonad Population Associated with Roots Is Influenced by Plant and Soil Type,” Applied and Environmental Microbiology, Vol. 62, No. 7, 1996, pp. 2449-2456.
- J. W. Kloepper, R. Lifshiftz and R. M. Zablotowicz, “Free Living Bacterial Inocula for Enhancing Crop Productivity,” Trends in Biotechnology, Vol. 7, No. 2, 1989, pp. 39-44. doi:10.1016/0167-7799(89)90057-7
- A. Renwick, R. Campbell and S. Coc, “Assessment of in Vivo Screening Systems for Potential Biocontrol Agents of Gaeumannomces graminis,” Plant Pathology, Vol. 40, No. 4, 1991, pp. 524-532. doi:10.1111/j.1365-3059.1991.tb02415.x
- M. A. Flaishman, A. Eyal, C. Zilberstein, C. Voisard and D. Hass, “Suppression of Septoria tritici Blotch and Leaf Rust of Wheat by Recombinant Cyanide Producing Strains of Pseudomonas putida,” Molecular Plant-Microbe Interactions, Vol. 9, No. 7, 1996, pp. 642-645. doi:10.1094/MPMI-9-0642
- F. J. Gutierrez Manero, N. Acero, J. A. Lucas and A. Probanza, “The Influence of Native Rhizobacteria on European Alder [Alnus glutinosa (L.) Gaertan] Growth. II. Characterization of Growth Promoting and Growth Inhibiting Strains,” Plant and Soil, Vol. 182, No. 1, 1996, pp. 67-74.
- J. R. de Freitas, M. R. Banerjee and J. J. Germida, “Phosphate Solubilizing Rhizobacteria Enhance the Growth and Yield But Not Phosphorus Uptake of Canola (Brassica napus),” Biology and Fertility of Soils, Vol. 24, No. 4, 1997, pp. 358-364. doi:10.1007/s003740050258
- L. R. Kennedy, C. Pereg-Gerk, R. Wood, K. Deaker, K. Gilchrist and S. Katupitya, “Biological Nitrogen Fixation in Non Leguminous Field Crops: Facilitating the Evolution of an Effective Association between Azospirillum and Wheat,” Plant and Soil, Vol. 194, No. 1-2, 1997, pp. 65-79. doi:10.1023/A:1004260222528
- F. B. Holl, C. P. Chanway, R. Turkingon and R. Radley, “Growth Response of Crested Wheatgrass (Agropyron cristatum L.), White Clover (Trifolium repens L.) to Inoculation with Bacillus polymixa,” Soil Biology and Biochemistry, Vol. 20, No. 1, 1998, pp. 19-24.
- G. A. O’ Neill, R. A. Radley and C. P. Chanway, “Variable Effects of Emergence—Promoting Rhizobacteria on Conifer Seedling Growth under Nursery Conditions,” Biology and Fertility of Soils, Vol. 13, No. 1, 1992, pp. 45-49. doi:10.1007/BF00337237
- J. Sørensen, L. E. Jensen and O. Nybroe, “Soil and Rhizosphere as Habitats for Pseudomonas Inoculants: New Knowledge on Distribution, Activity and Physiological State Derived from Micro-Scale and Single-Cell Studies,” Plant and Soil, Vol. 232, No. 1-2, 2001, pp. 97- 108.
- N. G. Rumjanek, M. C. C. Fonseca and G. R. Xavier, “Quorum Sensing Emsistemas Agrícolas: Comportamento Multicelular em Procariotos via Comunicação Intercellular,” Biotecnologia ciência e desenvolvimento, Vol. 33, No. 1, 2004, pp. 35-50.
- M. N. Vincet, L. A. Harrison, J. M. Brackin, P. A. Kovacevich, P. Mukherji, D. M. Weller and E. A. Pierson, “Genetic Analysis of the Antifungal Activity of a Soilborne Pseudomonas aureofaciens Strain,” Applied and Environmental Microbiology, Vol. 57, No. 10, 1991, pp. 2928-2934.
- B. Schwyn and J. B. Neilands, “Universal Chemical Assay for Detection and Determination of Siderophores,” Analytical Biochemistry, Vol. 160, No. 2, 1987, pp. 40-47.
- S. A. Gordon and R. P. Weber, “Colorimetric Estimation of Indole Acetic Acid,” Plant Physiology, Vol. 26, No. 1 1951, pp. 192-195. doi:10.1104/pp.26.1.192
- H. John, J. Leaven, E. Steven and S. E. Lindow, “Utilization of Plant Hormones Indole 3 Acetic Acid for Growth by Pseudomonad putida Strain 1290,” Applied and Environmental Microbiology, Vol. 71, No. 5, 2005, pp. 2365- 2371. doi:10.1128/AEM.71.5.2365-2371.2005
- W. G. Weisburg, S. M. Barns, D. A. Pelletier and D. J. Lane, “16S Ribosomal DNA Amplification for Phylogenetic Study,” Journal of Bacteriology, Vol. 173, No. 2, 1991, pp. 697-703.
- S. F. Altschul, T. L. Madden, A. A. Schaffer, Z. Zhang, W. Miller and D. J. Lipman, “Gapped BLAST and PSIBLAST: A New Generation of Protein Data Base,” Nucleic Acid Research, Vol. 25, No. 17, 1997, pp. 389-3402. doi:10.1093/nar/25.17.3389
- B. L. Maidack, J. R. Cole, T. G. Lilburn, C. T. J. Parker, P. R. Saxman, P. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt and J. M. Tiedje, “The RDP-II (Ribosomal Database Project),” Nucleic Acids Research, Vol. 29, No. 1, 2001, pp. 82-85.
- K. Goto, T. Omura, Y. Hara and Y. Sadaie, “Application of the Partial 16S rDNA Sequence as an Index for Rapid Identification of Species in the Genus Bacillus,” Journal of General Applied Microbiology, Vol. 46, No. 1, 2000, pp. 1-8. doi:10.2323/jgam.46.1
- Y. W. Tang, A. Von Graevenitz, M. G. Waddington, M. K. Hopkins, D. H. Smith, H. Li, C. P. Kolbert, S. O. Montbornery and D. H. Persing, “Identification of Coryneform Bacterial Isolates by Ribosomal DNA Sequence Analysis,” Journal of Clinical Microbiology, Vol. 38, No. 4, 2000, pp. 1676-1678.
- J. Baldus-Patel, D. G. B. Leonard, X. Pan, J. M. Musser, R. E. Berman and I. Nachamkin, “Sequence-Based Identification of Mycobacterium Species Using the Microseq 500, 16S rDNA Bacterial Identification System,” Journal of Clinical Microbiology, Vol. 38, No. 1, 2000, pp. 246- 251.
- R. Trotha, T. Hanck, W. König and B. König, “Rapid Rebosequencing—An Effective Diagnostic Tool for Detecting Microbial Infection,” Infection, Vol. 29, No. 1 2001, pp. 12-16. doi:10.1007/s15010-001-0064-7
- H. Unnerstad, H. Ericsson, A. Alderborn, W. Tham, M. L. Danielsson-Tham and J. G. Mattsson, “Pyrosequencing as Method for Grouping of Listeria monocytogenes Strains on the Basis of Single Nucleotide Polymorphisms,” Applied and Environmental Microbiology, Vol. 67, No. 11, 2001, pp. 5339-5342. doi:10.1128/AEM.67.11.5339-5342.2001
- M. Drancourt, C. Bollet, A. Carlioz, R. Martelin, J.P. Gayral and D. Raoult, “16S Ribosomal Sequence Analysis of a Large Collection of Environmental and Clinical Unidentifiable Bacterial Isolates,” Journal of Clinical Microbiology, Vol. 38, No. 10, 2000, pp. 3623-3630.
- K. L. Simpson, B. Petterson and F. G. Priest, “Characterization of Iactobacilli from Scotch Malt-Whisky Distilleries and Description of Lactobacillus ferintoschensis sp. nov., a New Species Isolated from Malt Whisky Fermentations,” Microbiology, Vol. 147, No. 4, 2001, pp. 1007-1016
- J. C. Hunter-Cerva, “The Value of Microbial Diversity,” Current Opinion of Microbiology, Vol. 1, No. 3, 1998, pp. 278-285. doi:10.1016/S1369-5274(98)80030-1
NOTES
*Corresponding author.
#Equally contributed.

