Open Journal of Veterinary Medicine
Vol.4 No.5(2014), Article ID:46270,11 pages DOI:10.4236/ojvm.2014.45012
Identification of ompL1 and lipL32 Genes to Diagnosis of Pathogenic Leptospira spp. Isolated from Cattle
Hernández-Rodríguez Patricia1,2,3*, Gomez Ramirez Arlen2,4, Baquero Mónica4, Quintero Gladys1,3
1Molecular Biology and Immunogenetics Research Group—BIOMIGEN, Bogotá, Colombia
2Health Research Center—CISVI, Bogotá, Colombia
3Department of Basic Sciences, Universidad de la Salle, Bogotá, Colombia
4Faculty of Agricultural Sciences, Veterinary Medicine Program, Universidad de la Salle, Bogotá, Colombia
Email: *phernandez@unisalle.edu.co
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 13 March 2014; revised 10 April 2014; accepted 18 April 2014
ABSTRACT
Diagnosis of leptospirosis in Colombia is based on clinical history and serological testing. However, disease symptoms are nonspecific and there is no uniform criteria regarding the qualifications considered positive. Therefore, it is important to identify and characterize genes associated with pathogenicity in native strains for the development of new diagnostic tests and vaccine production. The aim of this study was to identify the ompL1 and lipL32 genes in Leptospira strains isolated from urine samples of cattle. Sixteen strains were obtained from urine samples and, DNA was isolated to perform two Polymerase Chain Reaction (PCR) tests which identified lipL32 and ompL1 genes. As positive control, a reference strain of L. interrogans was used. L. biflexa and Escherichia coli strains were used as a negative control. In 100% of the samples were identified amplicons of 960 bp and 423 bp corresponding to ompL1 and lipL32 genes respectively. Thus, the pathogenic property and conservation of genes in the isolated strains were confirmed. This study is presented as a contribution to the diagnosis of leptospirosis to use these genes as molecular markers of infection. The results of this study might provide clues for future clinical, epidemiological and molecular research leading to implement new diagnostic strategies and to expand knowledge of the pathophysiology of a disease of public health impact on human and animal.
Keywords:Leptospirosis, PCR, ompL1, lipL32

1. Introduction
Leptospirosis is a worldwide distributed disease, affecting most of mammals (cattle, goats, sheep, pigs, dogs, horses, and rodents) in which it causes several signs including subclinical symptoms, abortions, stillbirths and mummification. These changes affect production cycles causing multiorgan complications that can lead to death [1] -[3] . The etiologic agent of the disease belongs to the genus Leptospira, which includes saprophytic and pathogenic species. Wild and domesticated mammals are the main sources of leptospires, which harbor in kidneys and excrete with urine [4] . Direct contact with contaminated urine or indirect exposures through contaminated water with urine is one of the ways of infection to humans [5] .
Leptospirosis is considered as an important emerging global public health problem because of increasing incidence in both developing and developed countries, being recognized as a disease with epidemic proportions [6] . However, the impact of this disease in some regions and countries is unknown due to few epidemiological reports. Even so, the reported data infer a high incidence in human and livestock industry worldwide. Serovar Hardjo of Leptospirainterrogans is the most frequently isolated agent in infections with Leptospira in cattle [7] -[9] and it is one of responsible for abortions in bovines [8] . Bovine leptospirosis has an impact in livestock industry due to economic losses for reproductive and non-reproductive causes such as abortions and septicemia respectively [1] .
There are three recognized stages of the disease: acute, subacute and chronic. Acute disease is the less common stage, it is generated by accidental serovars and it is usually the most severe and mortal in calves with clinical signs as pyrexia, hemoglobinuria, hematuria, anemia, pulmonary congestion, and meningitis. On the other hand, cows present agalactia and abortions due to pyrexia [10] -[12] . Subacute stage is common and the less severe; cows present pyrexia and agalactia. Milk has calostrum appearance and the udder is soft [12] -[14] . Chronic leptospirosis is the most common stage; pregnant cows present abortions, weak calves and placental retention. This stage is associated with serovar Hardjo [12] [15] [16] . Molecular classification indicates that within serovar Hardjo there are two important species in bovine leptospirosis: Leptospirainterrogans serovar Hardjo (hardjoprajitno) and Leptospiraborgpetersenii serovar Hardjo (hardjobovis). Epidemiological studies have demonstrated that Leptospiraborgpetersenii serovar Hardjo (hardjobovis) is the most common cause of bovine leptospirosis worldwide. Leptospirainterrogans serovar Hardjo (hardjoprajitno) has been isolated mostly in cattle from United Kingdom [2] . However, Pomona and Grippotyphosa serovars are also associated with reproductive losses [17] [18] .
Several studies have investigated about the pathogenetic mechanisms of leptospirosis, particularly host-parasite interactions and identification of virulence genes in different serovars of pathogenic and non-pathogenic leptospires [19] [20] . In this way, antigenic proteins expressed during infection have been identified. These have an important implication for the development of new techniques for diagnosis and vaccine production.
Spirochetes, including bacteria of the genus Leptospira, have a cytoplasmic membrane and outer membrane [21] . Identification and characterization of components of the outer membrane of Leptospira species are complex. Nevertheless, several transmembrane proteins, lipoproteins and peripheral membrane proteins have been determined [22] . By its strategic location, these outer membrane proteins (OMP’s) are important in research because are responsible for the interaction between pathogen and host [23] . This fact is more important if it is assumed that the OMP’s participate in mechanisms of evasion of the immune response as well as the persistence of spirochetes in the host [24] . Among isolated proteins from L. interrogans serovar Copenhageni are found: LipL32, LipL21, LipL45, LipL31, OmpL1, Flagelin/FlaB1, LipL36, LipL41, LipL71, and LigA [25] . Among these, LipL32 and OmpL1 proteins are candidates for designing new diagnostic techniques and producing immunogens.
First studies for identification of Leptospira were performed using techniques such as direct observation of the microorganism by dark field microscopy. Samples of blood, urine and tissues (kidney or liver) can be obtained for isolation of these bacteria by using biopsy or immediately after death in semisolid media such as fletechers medium or EMJH. Microscopic agglutination test (MAT) is used as gold standard test. However, this serological test shows a low sensitivity and specificity. In addition, immunoenzymatic methods as enzymeLinked Immuno Sorbent Assay (ELISA) and immunochromatography have been used. In the 80’s, it was implemented the use of restriction enzymes for the taxonomic knowledge of some serovars of Leptospira [26] [27] . Polymerase Chain Reaction (PCR) has been used for diagnosis of leptospirosis in human. This technique offers: 1) Sensitivity, because this microorganism is detected in a sample with very small amounts of genetic material; 2) Specificity, because through strict conditions, the microorganism to be detected is capable of being amplified. 3) Speed, because it allows fast processing compared with other techniques to detect bacteria, which require more complex procedures. And 4) It can be mentioned that this test offers versatility because it is allowed to the diagnosis of various microorganisms [28] -[31] .
Diagnosis of leptospirosis in Colombia is based on clinical history and serological testing. However, disease symptoms are nonspecific and there is no uniform criteria regarding the qualifications considered positive. Therefore, the aim of this study is to identify lipL32 and ompL1 genes associated with pathogenicity and antigenicity in native strains as molecular markers of infection with Leptospira.
2. Materials and Methods
2.1. Bacterial Strains
Sample size from 70 isolated strains of bovine urine characterized as pathogenic by PCR in a previous study was calculated by using the program Win Episcope 2.0. [39] . It was used a confidence level of 95% from which it was obtained a minimum sample size of 16. An ATCC reference strain of L. interrogans (23581) and a strain of L. interrogans donated by the Colombian Agricultural Institute (ICA—Instituto Colombiano Agropecuario) were used as positive controls. Escherichia coli and an ATCC reference of L. biflexa strain (23582) were used as negative controls.
2.2. DNA Extraction
DNA was isolated from 16 samples and controls. DNA was extracted from 1 mL of each culture of isolated strains and controls by using QIAamp® DNA Mini Kit (QIAGEN Inc., USA). The total concentration and purity of the DNA were quantified by spectrophotometry using absorption of light at 260 and 280 nm (A260/280). DNA was considered good quality with range = ~1.8 - 2.0.
2.3. Bioinformatic Analysis
Two pairs of primers: forward primer lipL32 (5’-CGC TGA AAT GGG AGT TCG TAT GAT T-3’) combined with the reverse primer lipL32 (5’-CCA ACA GAT GCA ACG AAA GAT CCT TT-3’) and forward primer ompL1 (5’-TTG ATT GAA TTC TTA GAG TTC GTG TTT ATA-3’) combined with the reverse primer ompL1 (5’-AAG GAG AAG CTT ATG ATC CGT AAC ATA AGT-3’) [32] -[34] were used to amplify DNA products of 423 bp and 960 bp of the L. interrogans lipL32 and ompL1 genes, respectively. Resources and database of the National Center for Biotechnology Information (NCBI) were used to confirm sequence, size, position genes and binding site of used primers in PCR.
2.4. PCR Amplification
Two PCR were performed to identify lipL32 and ompL1 genes associated with pathogenicity by using a modification of a procedure previously reported [35] . Two microliters of DNA extract were used for amplification in a total reaction mixture of 48 µL containing final concentrations of 16.25 µM Tris-HCl, 0.812 µM MgCl2, 108.33 µM of each deoxynucleoside triphosphate (dNTP), 0.677 U of Taq polymerase and 2 pmol/µL of each of primers. The PCR was carried out as follows: 94˚C for 5 min, forty cycles of 94˚C for 15 s, 56˚C for 35 s, 72˚C for 40 s. The PCR products were separated on 2% agarose gel, stained with ethidium bromide and examined with ultraviolet light.
3. Results
3.1. Bioinformatic Analysis
The results obtained by using Nucleotide resource of NCBI indicate that the lipL32 gene sequence has been reported only in pathogenic strains of Leptospira sp. The gene has 820 bp and it is located on chromosome 1 at position 5’-1’938, 935-1’939, 755 bp-3’. We identified 118 reports of this gene in pathogenic strains of Leptospira sp. Leptospira species in which the gene has been reported are: L. interrogans, L. borgpetersenii, L. santarosai, L. kirschneri, L. noguchi and L. wali. ompL1 gene has been reported in the following pathogenic strains: L. interrogans, L. borgpetersenii, L. santarosai, L. kirschneri, L. noguchi and L. weilii. This gene has 960 bp and it is located on chromosome at position 5’-686.212 - 687.174 bp-3’. The sequences of the lipL32 primers were aligned with the genome of L. interrogans, L. borgpetersenii, L. santarosai, L. kirschneri, L. noguchii and L. weilii by using Blast program; while the sequences of the ompL1 primers were aligned with the genome of L. interrogans, L. borgpetersenii and L. noguchii. This confirmed the alignment site of primers for ompL1 and lipL32 genes and that the selected primers were associated exclusively with pathogenic leptospires.
3.2. PCR Amplification
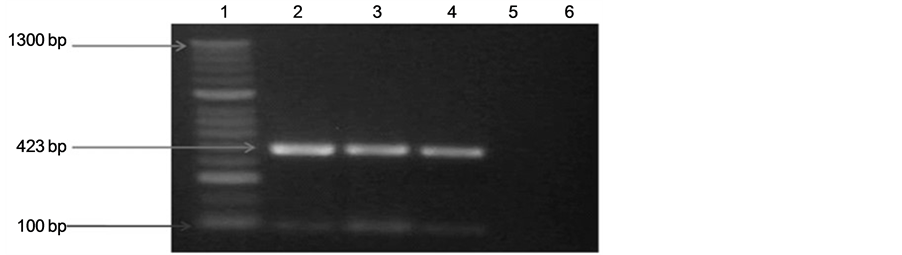
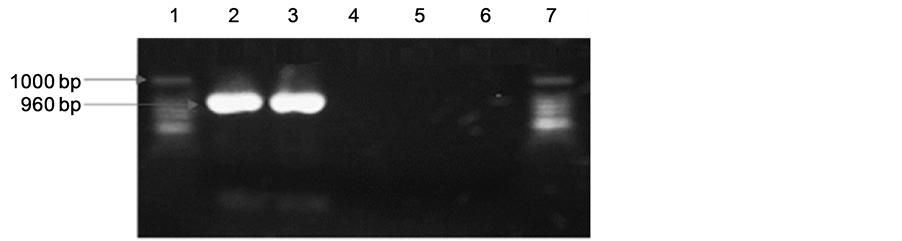
We identified the amplified products of 423 and 960 bp, corresponding to lipL32 and ompL1 genes respectively from 100% of the samples. The amplified product of 423 bp corresponding to lipL32 gene is showed in Figure 1. Figure 2 shows the amplified product of 960 bp of ompL1 gene presents in pathogenic strains isolated from cattle. Positive and negative controls are showed in both figures.
4. Discussion
To contribute to a better understanding of molecular pathophysiology of leptospirosis, the identification of two OMP’s genes was performed in strains of pathogenic leptospires. OMP’s and lipoproteins are the main components of leptospiral surface [36] . These proteins play a role in signal transduction, nutrition uptake, and immunogenicity [37] . In this study, we identified lipL32 and ompL1 genes in strains of L. interrogans isolated from bovine urine. The bioinformatic analysis indicated that the lipL32 gene is exclusively in pathogenic strains of
 (a)
(a)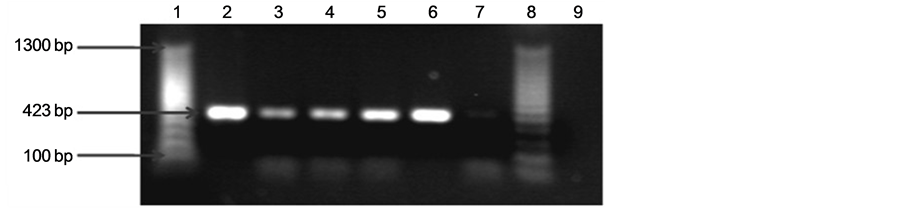 (b)
(b)
Figure 1. Agarose gel electrophoresis of polymerase chain reaction (PCR) analysis products for lipL32 (423 bp) in pathogenic Leptospira. A 100-bp molecular weight marker was used. The PCR products were separated on 2.0% agarose gel, stained with ethidium bromide, examined with UV light, and visualized with a transilluminator (Bio-Rad). Analysis of amplified products, (a) Line 1: Molecular weight marker. Line 2: Leptospirainterogans (ATCC). Lines 3 and 4: Positive samples amplified from bovine urine (pathogenic leptospireslipL32, 423 bp). Line 5: Negative control of PCR. Line 6: Leptospirabiflexa (ATCC); (b) Line 1: Molecular weight marker. Line 2: Leptospira interogans (ATCC). Lines 3-6: Positive samples amplified from bovine urine (pathogenic leptospireslipL32, 423 bp). Line 7: Leptospirabiflexa (ATCC). Line 8: Molecular weight marker. Line 9: Negative control of PCR.
 (a)
(a) (b)
(b)
Figure 2. Representative polymerase chain reaction (PCR) for ompL1 DNA in urine samples from bovine. The PCR products were separated on 1.5% agarose gel, stained with ethidium bromide, examined with UV light, and visualized with a transilluminator (Bio-Rad). Analysis of amplified products, (a) Line 1: Pathogenic Leptospirainterrogans (ATCC). Line 2: Positive samples amplified from bovine urine. Line 3: Negative control of PCR. Line 4: E coli. Line 5: Saprophytic Leptospirabiflexa (ATCC). Line 6: Negative control PCR. Line 7: Molecular weight marker; (b) Line 1: Pathogenic Leptospirainterrogans (ATCC). Line 2-10: Positive samples amplified from bovine urine. Line 11: Negative control of PCR. Line 12: E coli. Line 13: Saprophytic Leptospirabiflexa (ATCC). Line 14: Molecular weight marker.
Leptospira sp., which agrees with reports by various authors [22] [38] [39] . Similar to lipL32 gene, ompL1 gene has been reported only in pathogenic strains of Leptospira sp. [40] [41] . Both genes were identified in existing strains with public health impact. The primers used were obtained from reports in which it was confirmed the size of the amplified products [30] [31] [41] -[43] . The use of theseprimers for the identification of lipL32 gen by real time or conventional PCR has been postulated to the development of diagnosis tests for leptospirosis in samples from patients who are in antibiotic therapy or in early stages of infection [30] [44] . Therefore, bioinformatic analyses are postulated as other useful tool for the study of Leptospira, which have become a significant method for researchers. During more than 20 years of taxonomic and phylogenetic studies, an important number of Leptospira sequences have been generated. All this information has been deposited in large databases such as GenBank (www.ncbi.nlm.nih.gov/Genbank/index.html) [45] .
The determination of ompL1 and lipL32 genes in reference strains of L. interrogans and L. biflexa used in this study agreed with reports related to the presence of these genes in pathogenic strains and their absence in saprophytic strains [19] [37] [41] [46] [47] . The identification of ompL1 and lipL32 genes specific bands of 960 bp [30] [44] [48] and 423 bp [31] [49] [50] respectively, indicates presence of these genes in native pathogenic strains isolated from bovine urine. This result suggests that OMP’s are conserved proteins among pathogenic leptospires, which may have the potential of inducing comprehensive immunity and play an important role in virulence [5] .
It is known that OMP’s act as adhesions [23] [51] -[53] , antibodies fixation points [54] [55] , porins [56] -[58] , receptors for soluble proteins such as siderophores [59] , and complement proteins [60] . It is also possible that they are responsible for the interaction of pathogens with hosts due to their strategic localization [23] . It is presumed that OMP’s participate in evasion mechanisms of immune response and the persistence of leptospiras in the host [24] .
lipL32 is a highly conserved gen in pathogenic leptospires during infection in mammals [61] and it has been identified as an extracellular matrix-binding protein specifically to laminin, collagen I, and fibronectin [62] . It increases the cell permeability and accelerates the apoptosis process [63] . OmpL1 is a porin expressed in pathogenic Leptospira strains [33] [46] that allows the diffusion of hydrophilic solutes through the external membrane to the periplasm [22] .
However, the function of the majority of OMP’s and molecular pathogenesis remain almost unknown [5] [64] , despite the above mentioned and the fact that several major OMP’s express during infection have been potentially employed in the development of subunit vaccines and serologic test for leptospirosis diagnosis [31] [41] [65] -[67] . It is known that OMP’s have an important role in the pathogenesis because they facilitate the adaptation and interaction between bacteria and host [68] .
Leptospirosis is considered one of the most common zoonotic diseases worldwide [1] . However, it is neglected due to the fact that the endemicity of the disease is related to socioeconomic and climatic conditions of developing countries affecting impoverished populations [69] [70] . Leptospirosis has emerged as an important problem, especially where some risk factors are present such as inadequate sanitation that allows rat-borne transmission [3] [70] [71] . Actually there are over 500,000 reports of severe leptospirosis every year with fatality rates over 10% [72] . On the other hand, leptospirosis is a disease of livestock with an important economic impact due to reproductive losses. Abortions, stillbirths, embryo mortality and infertility are some of the factors involved in the presentation of leptospirosis in cattle [73] -[76] .
According to several studies, bovine leptospirosis is an important disease that affects production systems in Colombia. Since 1991 researchers from different Colombian regions have been studying the seropositivity of the disease [16] [77] -[81] . Only one research established the presence of Leptospira spp. in bovine using PCR, and they determined a prevalence of 37% for Sabana de Bogotá [82] . It is evident the presence of Leptospira spp. in bovine production systems and the need of using new detection techniques. Due to the epidemiologic and economic relevance it is important that leptospirosis must be considered a disease with impact which should be studied in order to improve methods for diagnosis, prevention and treatment.
5. Conclusions
This study is presented as a contribution for the diagnosis of leptospirosis through the use of these genes as molecular markers of infection. The results of this study might provide clues for future clinical, epidemiological and molecular research leading to implement new diagnostic strategies and to expand knowledge of the pathophysiology of a disease of public health impact.
Both, ompL1 and lipL32 are considered key genes to be studied in order to develop new methods of diagnosis and prevention. For instance, molecular diagnosis based on the detection of these genes has been identified [30] [42] [83] [84] . Several OMPs have been evaluated as potential vaccine candidates [33] [85] [86] . Vaccines based in LipL32 include vectors such as Mycobacterium bovis [66] and adenovirus [34] expressing the LipL32 antigen. It has been shown that OmpL1 and LipL41 have immunoprotective effects on the hamster model [33] . The function and location of many proteins are not yet known in Leptospira. A recent study reevaluated the position of LipL32 in subsurface [87] .
Further studies involving the identification of genes associated with the adaptation of leptospires to different hosts [88] and their expression profiles under several conditions should be under this way in an effort for a better understanding of molecular pathogenesis of this disease as well as for the development of strategies for control and prevention of leptospirosis as an emerging pathology with public health impact.
Acknowledgements
This study was sponsored by Universidad de La Salle in Bogotá D.C., Colombia. Authors thank the Basic Science Department and the Veterinary Medicine Program in Universidad de La Salle for their support, the Colombian Agricultural Institute (ICA—Instituto Colombiano Agropecuario) for the donation of Leptospirainterrogans and Rosario Santos for their guidance in microbiologic procedures.
Conflict of Interest Statement
The findings and conclusions in this report are those of the authors and do not necessarily represent the view of the funding agencies. The use of trade names and commercial sources are only for identification and it does not imply endorsement by Universidad de la Salle in Bogotá D.C.-Colombia.
References
- Levett, P.N. (2001) Leptospirosis. Clinical Microbiology Reviews, 14, 296-326. http://dx.doi.org/10.1128/CMR.14.2.296-326.2001
- Grooms, D. (2006) Reproductive Looses Caused by Bovine Viral Diarrhea Virus and Leptospirosis. Theriogenology, 66, 624-628. http://dx.doi.org/10.1016/j.theriogenology.2006.04.016
- Romero-Vivas, C.M., Cuello-Pérez, M., Agudelo-Flórez, P., Thiry, D., Levett, P.N. and Falconar, A.K. (2013) CrossSectional Study of Leptospira Seroprevalence in Humans, Rats, Mice, and Dogs in a Main Tropical Sea-Port City. The American Journal of Tropical Medicine and Hygiene, 88, 178-183. http://dx.doi.org/10.4269/ajtmh.2012.12-0232
- Jorge, S., Hartleben, C.P., Seixas, F.K., Coimbra, M.A., Stark, C.B., Larrondo, A.G., Amaral, M.G., Albano, A.P., Minello, L.F., Dellagostin, O.A. and Brod, C.S. (2012) Leptospira borgpetersenii from Free-Living White-Eared Opossum (Didelphis albiventris): First Isolation in Brazil. Acta Tropica, 24, 147-151. http://dx.doi.org/10.1016/j.actatropica.2012.07.009
- Cinco, M. (2010) New Insights into the Pathogenicity of Leptospires: Evasion of Host Defences. New Microbiologica, 33, 283-292.
- Maites, E., Jay, M.T., Deresinski, S., Shieh, W.J., Zaki, S.R., Tompkins, L. and Smith, D.S. (2004) Reemerging Leptospirosis, California. Emerging Infectious Disease, 10, 406-412. http://dx.doi.org/10.3201/eid1003.030431
- Srivastava, S.K. (2006) Prospects of Developing Leptospiral Vaccines for Animals. Indian Journal of Medical Microbiology, 24, 331-336. http://dx.doi.org/10.4103/0255-0857.29412
- Escamilla, H.P., Martínez, M.J., Medina, C.M. and Morales, S.E. (2007) Frequency and Causes of Infectious Abortion in a Dairy Herd in Queretaro, Mexico. Canadian Journal of VeterinaryResearch, 71, 314-317.
- Hernández, P., Quintero, G., Díaz, C. and Dalmau, E. (2008) Comparación del Cultivo Microbiológico y Visualización por Campo Oscuro Para el Diagnóstico de Leptospirosis en Bovinos de la Sabana de Bogotá. Revista de Investigación, Universidad de La Salle, 8, 9-15.
- Guijarro, R. and Calvo, E. (1999) Tratamiento y Control de Leptospirosis Bovina. Producción Animal, 1, 27-36.
- Perdomo, E. and Garin, A. (2002) Leptospirosis Animal. Guía de Control y Manejo de Leptospirosis, OPS/HCP/HCV/ URU.ZOO, 224-226.
- Prescott, J.F. (2007) Leptospirosis. In: Jubb, K.V., Ed., Pathology of Domestic Animals, Elsevier, Edinburgh, 481-490.
- Ellis, W.A. (1983) Recent Developments in Bovine Leptospirosis. Veterinary Annual, 91-95.
- Vanasco, N.,Sequeiro, G., Fontana, M., Fusco, S. and Sequeiro, M. (2000) Descripción de un Brote de Leptospirosis en la Ciudad de Santa Fé, Argentina,Marzo-Abril 1998. Revista Panamericana de Salud Pública, 7, 27-32. http://dx.doi.org/10.1590/S1020-49892000000100006
- Daniel-Givens, M. and Marley, S.D. (2008) Infectious Causes of Embryonic and Fetal Mortality. Theriogenology, 70, 270-285. http://dx.doi.org/10.1016/j.theriogenology.2008.04.018
- Díaz, C.A., Dalmau, E.A., Laiceca, V. and García, J.G. (2007) Correlación de las Alteraciones Producidas por la Leptospirosis a Nivel Hepato-renal con VariablesProductivas y Reproductivas en Bovinos de la Sabana de Bogotá. Revista Medicina Veterinaria, 13, 7-17.
- Kingscote, B.F. and Wilson, D. (1986) Leptospira pomona Abortion Storm in Cattle Herdin Saskatchewan. The Canadian Veterinary Journal, 27, 440-442.
- Barr, B.C. and Anderson, B.L. (1993) Infectious Diseases Causing Bovine Abortion and Fetal Loss. Veterinary Clinics of North America: Food Animal Practice, 9, 343-368.
- Cullen, P.A., Cordwell, S.J., Bulach, D.M., Haake, D.A. and Adler, B. (2002) Global Analysis of Outer Membrane Proteins from Leptospira interrogans Serovar Lai. Infection and Immunity, 70, 2311-2318. http://dx.doi.org/10.1128/IAI.70.5.2311-2318.2002
- Thongboonkerd, V., Chiangjong, W., Saetun, P., Sinchaikul, S., Chen, S.T. and Kositanont, U. (2009) Analysis of Differential Proteomes in Pathogenic and Non-Pathogenic Leptospira: Potential Pathogenic and Virulence Factors. Proteomics, 9, 3522-3534. http://dx.doi.org/10.1002/pmic.200700855
- Holt, S. (1978) Anatomy and Chemistry of Spirochetes. Microbiology, 42, 114-160.
- Haake, D.A. and Matsunaga, J. (2002) Characterization of the Leptospiral Outer Membrane and Description of Three Novel Leptospiral Membrane Proteins. Infection and Immunity, 70, 4936-4945. http://dx.doi.org/10.1128/IAI.70.9.4936-4945.2002
- Sansonetti, P.J. (1991) Genetic and Molecular Basis of Epithelial Cell Invasion by Shigella Species. Reviews of Infectious Diseases, 13, 285-292. http://dx.doi.org/10.1093/clinids/13.Supplement_4.S285
- Blanco, D.R., Walker, E.M., Haake, D.A., Champion, C.I., Miller, J.N. and Lovett, M.A. (1990) A Complement Activation Limits the Rate of in Vitro Treponemicidal Activity and Correlates with Antibody-Mediated Aggregation of Treponema pallidum Rare Outer Membrane Protein “TROMP”. Journal of Immunology, 144, 1914-1921.
- Nally, J.E., Whitelegge, J.P., Bassilian, S., Blanco, D.R. and Lovett, M.A. (2007) Characterization of the Outer Membrane Proteome of Leptospira interrogans Expressed during Acute Lethal Infection. Infection and Immunity, 75, 766- 773. http://dx.doi.org/10.1128/IAI.00741-06
- Tomonori, K., Uchida, Y., Fujishima, N., Yasunaga, A., Fujitomi, Y., Shibata, O., Hadama, T., Shirabe, J., Yokoyama, S. and Tsuji, K. (1988) The Pathogenesis of Adenocarcinoma of the Stomach after Successful Radiotherapy of Squamous Cell Carcinoma of the Lower Intrathoracic Esophagus—Case Report. Gan No Rinsho. Japan Journal of Cancer Clinics, 34, 1185-1189.
- Tsuyoshi, Y. (1988) Cloning and Expression of Antigen Genes of Leptospira interrogans Serovar Canicola in Escherichia coli, and Restriction Endonuclease DNA Analysis of Antigenic Variants of Leptospires Selected by Monoclonal Antibodies. Japanese Journal of Veterinary Research, 36, 180-189.
- Barnett, J., Barnett, D., Bolin, C.A., Summers, T.A., Wagar, E.A., Cheville, N.F., Hartskeerl, R.A. and Haake, D.A. (1999) Expression and Distribution of Leptospiral Outer Membrane Components during Renal Infection of Hamsters. Infection and Immunity, 67, 853-861.
- Laras, K., Cao, B.V., Bounlu, K., Nguyen, T.K., Olson, J.G., Thongchanh, S. Tran, N.V., Hoang, K.L., Punjabi, N., Ha, B.K., Ung, S.A., Insisiengmay, S., Watts, D.M., Beecham, H.J. and Corwin, A.L. (2002) The Importance of Leptospirosis in Southeast Asia. The American Journal of Tropical Medicine and Hygiene, 67, 278-286.
- Levett, P.N., Morey, R.E., Galloway, R.L., Turner, D.E., Steigerwalt, A.G. and Mayer, L.W. (2005) Detection of Pathogenic Leptospires by Real-Time Quantitative PCR. Journal of Medical Microbiology, 54, 45-49. http://dx.doi.org/10.1099/jmm.0.45860-0
- Natarajaseenivasan, K., Vijayachari, P., Sharma, S., Sugunan, A.P. and Sehgal, S.C. (2005) Phenotypic & Genotypic Conservation of ompL1 & lipL41 among Leptospiral Isolates of Andaman Islands. Indian Journal of Medical Research, 122, 343-347.
- Woo, T.H., Patel, B.K., Smythe, L.D., Norris, M.A., Symonds, M.L. and Dohnt, M.F. (1998) Identification of Pathogenic Leptospira by Taq Man Probe in a Light Cycler. Analytical Biochemistry, 256, 132-134. http://dx.doi.org/10.1006/abio.1997.2503
- Haake, D.A., Mazel, M.K., Mccoy, A.M., Milward, F., Chao, G., Matsunaga, J. and Wagar, E.A. (1999) Leptospiral Outer Membrane Proteins OmpL1 and LipL41 Exhibit Synergistic Immuno Protection. Infection and Immunity, 67, 6572-6582.
- Branger, C., Sonrier, C., Chatrenet, B., Klonjkowski, B., Ruvoen-Clouet, N., Aubert, A., André-Fontaine, G. and Eloit, M. (2001) Identification of the Hemolysis-Associated Protein 1 as a Cross-Protective Immunogen of Leptospira interrogans by Adenovirus-Mediated Vaccination. Infection and Immunity, 69, 6831-6838. http://dx.doi.org/10.1128/IAI.69.11.6831-6838.2001
- Léon, A., Pronost, S., Tapprest, J., Foucher, N., Blanchard, B., André-Fontaine, G., Laugier, C., Fortier, G. and Leclercq, R. (2006) Identification of Pathogenic Leptospira Strains in Tissues of a Premature Foal by Use of Polymerase Chain Reaction Analysis. Journal of Veterinary Diagnostic Investigation, 18, 218-221. http://dx.doi.org/10.1177/104063870601800216
- Cullen, P.A., Haake, D.A. and Adler, B. (2004) Outer Membrane Proteins of Pathogenic Spirochetes. FEMS Microbiology Reviews, 28, 291-318. http://dx.doi.org/10.1016/j.femsre.2003.10.004
- Cullen, P.A., Xu, X., Matsunaga, J., Sanchez, Y., Ko, A., Haake, D.A. and Adler, B. (2005) Surfaceome of Leptospira spp. Infection and Immunity, 73, 4853-4863. http://dx.doi.org/10.1128/IAI.73.8.4853-4863.2005
- Guerreiro, H., Croda, J., Flannery, B., Mazel, M., Matsunaga, J., Reis, M.G., Levett, P.N., Ko, A.I. and Haake, D.A. (2001) Leptospiral Proteins Recognized during the Humoral Immune Response to Leptospirosis in Humans. Infection and Immunity, 69, 4958-4968. http://dx.doi.org/10.1128/IAI.69.8.4958-4968.2001
- Zhang X., Yu, Y., He, P., Zhang, Y.X., Hu, B.Y., Yang, Y., Nie, Y.X., Jiang, X.G., Zhao, G.P. and Guo, X.K. (2005) Expression and Comparative Analysis of Genes Encondig Outer Membrane Proteins LipL21, LipL32 and OmpL1 in Epidemic Leptospires. Acta Biochimica et Biophysica Sinica, 37, 649-656. http://dx.doi.org/10.1111/j.1745-7270.2005.00094.x
- Shang, E.S., Exner, M.E., Summers, T.A., Martinich, C., Champion, C.I., Hankcock, R.E. and Haake, D.A. (1995) The Rare Outer Membrane Protein, OmpL1, of Pathogenic Leptospira Species Is a Heat-Modifiable Porin. Infection and Immunity, 63, 3174-3181.
- Dong, H., Hu, Y., Xue, F., Sun, D., Ojcius, D., Mao, Y. and Yan, J. (2008) Characterization of the ompL1 Gene of Pathogenic Leptospira Species in China and Cross-Immunogenicity of the OmpL1 Protein. BMC Microbiology, 8, 223- 235. http://dx.doi.org/10.1186/1471-2180-8-223
- Reitstetter, R.E. (2006) Development of Species-Specific PCR Primer Sets for the Detection of Leptospira. FEMS Microbiology Letters, 264, 31-39. http://dx.doi.org/10.1111/j.1574-6968.2006.00431.x
- Guerra, B., Schneider, T., Luge, E., Draeger, A., Moos, V., Loddenkemper, C., Jansen, A. and Nöckler, K. (2008) Detection and Characterization of Leptospira interrogans Isolates from Pet Rats Belonging to a Human Immunodeficiency Virus-Positive Patient with Leptospirosis. Journal of Medical Microbiology, 57, 133-135. http://dx.doi.org/10.1099/jmm.0.47452-0
- Agudelo-Flórez, P., Londoño, A.F., Quiroz, V.H., Ángel, J.C., Moreno, N., Loaiza, E.T., Muñoz, L.F. and Rodas, J.D. (2009) Prevalence of Leptospira spp. in Urban Rodents from a Groceries Trade Center of Medellín, Colombia. The American Journal of Tropical Medicine and Hygiene, 81, 906-910. http://dx.doi.org/10.4269/ajtmh.2009.09-0195
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J. and Sayers, E.W. (2010) GenBank. Nucleic Acids Research, 38, D46-D51. http://dx.doi.org/10.1093/nar/gkp1024
- Haake, D.A., Champion, C.L., Martinich, C., Shang, E.S., Blanco, D.R., Miller, J.N. and Love, M. (1993) Molecular Cloning and Sequence Analysis of the Gene Encoding OmpL1, a Transmembrane Outer Membrane Protein of Pathogenic Leptospira spp. Journal of Bacteriology, 35, 4225-4234.
- Haake, D.A., Suchard, M.A., Kelley, M.M., Dundoo, M., Alt, D.P. and Zuerner, R.L. (2004) Molecular Evolution and Mosaicism of Leptospiral Outer Membrane Proteins Involves Horizontal DNA Transfer. Journal of Bacteriology, 186, 2818-2828. http://dx.doi.org/10.1128/JB.186.9.2818-2828.2004
- Mayer-Scholl, A., Draeger, A., Luge, E., Ulrich, R. and Nöckler, K. (2011) Comparison of Two PCR Systems for the Rapid Detection of Leptospira spp. from Kidney Tissue. Current Microbiology, 62, 1104-1106. http://dx.doi.org/10.1007/s00284-010-9829-5
- Chen, F., Fan, Q., Qiu, W., Yin, G. and Xing, Y. (2010) Cloning and Analysis of the Sequence of Outer MembranceProtein Gene of a Serovar Leptospira canicola Interrogans. China Animal Husbandry & Veterinary Medicine, 6, 55-59.
- Vedhagiri, K., Natarajaseenivasan, K., Chellapandi, P., Prabhakaran, S.G., Selvin, J., Sharma, S. and Vijayachari, P. (2009) Evolutionary Implication of Outer Membrane Lipoprotein-Encoding Genes ompL1, lipL32 and lipL41 of Pathogenic Leptospira Species. Genomics, Proteomics & Bioinformatics, 7, 96-106. http://dx.doi.org/10.1016/S1672-0229(08)60038-8
- Isberg, R. and Falkow, S. (1985) A Single Genetic Locus Encoded by Yersinia pseudotuberculosis Permits Invasion of Cultured Animal Cells by Escherichia coli K-12. Nature, 317, 262-264. http://dx.doi.org/10.1038/317262a0
- Bessen, D. and Gotschlich, E. (1986) Interactions of Gonococci with HeLa Cells: Attachment, Detachment, Replication, Penetration, and the Role of Protein II. Infection and Immunity, 54, 154-160.
- Miller, V.L. and Falkow, S. (1988) Evidence for Two Genetic Loci in Yersinia enterocolitica That Can Promote Invasion of Epithelial Cells. Infection and Immunity, 56, 1242-1248.
- Saukkonen, K., Abdillahi, H., Poolman, J.T. and Leinonen, M. (1987) Protective Efficacy of Monoclonal Antibodies to Class 1 and Class 3 Outer Membrane Proteins of Neisseria meningitides B:15:P1.16 in Infant Rat Infection Model: New Prospects for Vaccine Development. Microbial Pathogenesis, 3, 261-267. http://dx.doi.org/10.1016/0882-4010(87)90059-3
- Murphy, T.F. and Bartos, L.C. (1988) Human Bactericidal Antibody Response to Outer Membrane Protein P2 of Nontypeable Haemophilus Influenzae. Infection and Immunity, 56, 2673-2679.
- Mcguinness, B., Barlow, A.K., Clarke, I.N., Farley, J.E., Anilionis, A., Poolman, J.T. and Heckels, J.E. (1990) Comparative Sequence Analysis of the Class 1 Protein Gene (porA) from Three Strains of Neisseria meningitidis: Synthetic Peptides Define the Epitopes Responsible for Serosubtype Specificity. The Journal of Experimental Medicine, 171, 1871-1882. http://dx.doi.org/10.1084/jem.171.6.1871
- Jeanteur, D., Lekey, J.H. and Pattus, F. (1991) The Bacterial Porin Superfamily: Sequence Alignment and Structure Prediction. Molecular Microbiology, 5, 2153-2164. http://dx.doi.org/10.1111/j.1365-2958.1991.tb02145.x
- Li, Z.M., Hannah, J.H., Stibitz, S., Nguyen, N.Y., Manclark, C.R. and Brennan, M.J. (1991) Cloning and Sequencing of the Structural Gene for the Porin Protein of Bordetella pertussis. Molecular Microbiology, 5, 1649-1656. http://dx.doi.org/10.1111/j.1365-2958.1991.tb01912.x
- Stoebner, J.A. and Payne, S.M. (1988) Iron-Regulated Hemolysin Production and Utilization of Heme and Hemoglobin by Vibrio cholerae. Infection and Immunity, 56, 2891-2895.
- Hoffman, P.S., Ripley, M. and Weeratna, R. (1992) Cloning and Nucleotide Sequence of a Gene (ompS) Encoding the Major Outer Membrane Protein of Legionella pneumophila. Journal of Bacteriology, 174, 914-920.
- Haake, D.A., Chao, G., Zuerner, R.L., Barnett, J.K., Barnett, D., Mazel, M., Matsunaga, J., Levett, P.N. and Bolin, C.A. (2000) The Leptospiral Major Outer Membrane Protein LipL32 Is a Lipoprotein Expressed during Mammalian Infection. Infection and Immunity, 68, 2276-2285.http://dx.doi.org/10.1128/IAI.68.4.2276-2285.2000
- Hoke, D.E., Egan, S., Cullen, P.A. and Adler, B. (2008) LipL32 Is an Extracellular Matrix-Interacting Protein of Leptospira spp. and Pseudoalteromonas tunicata. Infection and Immunity, 76, 2063-2069. http://dx.doi.org/10.1128/IAI.01643-07
- Sun, Z., Bao, L., Li, D.K., Huang, B. and Wu, B. (2010) Effect of Leptospira interrogans Outer Membrane Proteins LipL32 on HUVEC. Microbial Pathogenesis, 49, 116-121.http://dx.doi.org/10.1016/j.micpath.2010.05.006
- Xue, F., Dong, H., Wu, J., Wu, Z., Hu, W., Sun, A., Troxell, B., Yang, X.F. and Yan, J. (2010) Transcriptional Responses of Leptospira interrogans to Host Innate Immunity: Significant Changes in Metabolism, Oxygen Tolerance, and Outer Membrane. PLoS Neglected Tropical Diseases, 4, e857. http://dx.doi.org/10.1371/journal.pntd.0000857
- Croda, J., Ramos, J.G., Matsunaga, J., Queiroz, A., Homma, A., Riley, L.W., Haake, D.A., Reis, M.G. and Ko, A.I. (2007) Leptospira Immunoglobulin-Like Proteins as a Serodiagnostic Marker for Acute Leptospirosis. Journal of Clinical Microbiology, 45, 1528-1534. http://dx.doi.org/10.1128/JCM.02344-06
- Seixas, F.K., da Silva, É.F., Hartwig, D.D., Cerqueira, G.M., Amaral, M., Fagundes, M.Q., Dossa, R.G. and Dellagostin, O.A. (2007) Recombinant Mycobacterium bovis BCG Expressing the LipL32 Antigen of Leptospira interrogans Protects Hamsters from Challenge. Vaccine, 26, 88-95. http://dx.doi.org/10.1016/j.vaccine.2007.10.052
- Picardeau, M., Bulach, D.M., Bouchier, C., Zuerner, R.L., Zidane, N., Wilson, P.J., Creno, S., Kuczek, E.S., Bommezzadri, S., Davis, J.C., Mcgrath, A., Johnson, A., Boursaux-Eude, C., Seemann, T., Rouy, Z., Coppel, R.L., Rood, J., Lajus, A., Davies, J.K., Médigue, C. and Adler, B. (2008) Genome Sequence of the Saprophyte Leptospira biflexa Provides Insights into the Evolution of Leptospira and the Pathogenesis of Leptospirosis. PLoS ONE, 3, e1607.
- Nguyen, T.X., Alegre, E.R. and Kelley, S.T. (2006) Phylogenetic Analysis of General Bacterial Porins: A Phylogenomic Case Study. Journal of Molecular Microbiology and Biotechnology, 11, 291-301. http://dx.doi.org/10.1159/000095631
- Bharti, A.R., Nally, J.E., Ricaldi, J.N., Matthias, M.A., Diaz, M.M., Lovett, M.A., Levett, P.N., Gilman, R.H., Willig, M.R., Gotuzzo, E. and Vinetz, J.M. (2003) Leptospirosis: A Zoonotic Disease of Global Importance. The Lancet Infectious Diseases, 3, 757-771. http://dx.doi.org/10.1016/S1473-3099(03)00830-2
- Ko, A.I., Goarant, C. and Picardeau, M. (2009) Leptospira: The Dawn of the Molecular Genetics Era for an Emerging Zoonotic Pathogen. Nature Reviews Microbiology, 7, 736-747. http://dx.doi.org/10.1038/nrmicro2208
- Riley, L.W., Ko, A.I., Unger, A. and Reis, M.G. (2007) Slum Health: Diseases of Neglected Populations. BMC International Health and Human Rights, 7, 2-11. http://dx.doi.org/10.1186/1472-698X-7-2
- Mcbride, A.J., Athanazio, D.A., Reis, M.G. and Ko, A.I. (2005) Leptospirosis. Current Opinion in Infectious Disease, 18, 376-386. http://dx.doi.org/10.1097/01.qco.0000178824.05715.2c
- Sanderson, M.W. and Gnad, D.P. (2002) Biosecurity for Reproductive Diseases. Veterinary Clinics of North America: Food Animal Practice, 18, 78-98. http://dx.doi.org/10.1016/S0749-0720(02)00003-8
- Muñoz-Zanzi, C.A., Thurmond, M.C. and Hietala, S.K. (2004) Effect of Bovine Viral Diarrhea Virus Infection on Fertility of Dairy Heifers. Theriogenology, 61, 1085-1099. http://dx.doi.org/10.1016/j.theriogenology.2003.06.003
- Bon Durant, R.H. (2007) Selected Diseases and Conditions Associated with Bovine Conceptus Loss in the First Trimester. Theriogenology, 68, 461-473. http://dx.doi.org/10.1016/j.theriogenology.2007.04.022
- Givens, M.D. and Marley, M.S. (2008) Infectious Causes of Embryonic and Fetal Mortality. Theriogenology, 70, 270- 285. http://dx.doi.org/10.1016/j.theriogenology.2008.04.018
- Zuluaga, A.G. (2009) Risk Factors Associated to Leptospirosis in Bovine Cattle Ranches in Pereira between the Years 2002 and 2005. Investigación Andina, 11, 109-117.
- Aricapa, H.J., Pérez, J.E., Cabrera, I.C. and Rivera, K. (2008) Valoración de la Respuesta de Anticuerpos Tipo IgM e IgG Frente a Leptospira en Bovinos. Biosalud, 7, 29-39.
- Vásquez, L., Mosquera, A., Rivera, N., Campo, V. and Agudelo, P. (2006) Prevalencia Serológica de Leptospirosis en Ganado Bovino en el Matadero del Municipio de Popayán, Cauca. Revista Universidad de Cauca.
- Gallego, J.F. (2001) Leptospirosis in Colombian Dairy Cattle: Microbiological, Serological and Molecular Aspects of the Disease. Unpublished Doctoral Dissertation, The Royal Veterinary College, University of London, London.
- Ochoa, J.E., Sánchez, A. and Ruiz, I. (2000) Epidemiología de la Leptospirosis en una Zona Andina de Producción Pecuaria. Revista Panamericana de Salud Pública, 7, 325-331. http://dx.doi.org/10.1590/S1020-49892000000500006
- Hernández-Rodríguez, P., Díaz, C.A., Dalmau, E.A. and Quintero, G.M. (2011) A Comparison between Polymerase Chain Reaction (PCR) and Traditional Techniques for the Diagnosis of Leptospirosis in Bovines. Journal of Microbiological Methods, 84, 1-7. http://dx.doi.org/10.1016/j.mimet.2010.10.021
- Bomfim, M.R., Barbosa-Stancioli, E.F. and Koury, M.C. (2008) Detection of Pathogenic Leptospires in Urine from Naturally Infected Cattle by Nested PCR. The Veterinary Journal, 178, 251-256. http://dx.doi.org/10.1016/j.tvjl.2007.07.029
- Ahmed, A., Anthony, R.M. and Hartskeerl, R.A. (2010) A Simple and Rapid Molecular Method for Leptospira Species Identification. Infection, Genetics and Evolution, 10, 955-962. http://dx.doi.org/10.1016/j.meegid.2010.06.002
- Branger, C., Chatrenet, B., Gauvrit, A., Aviat, F., Aubert, A., Bach, J.M. and André-Fontaine, G. (2005) Protection against Leptospira interrogans Sensu Lato Challenge by DNA Immunization with the Gene Encoding Hemolysin-Associated Protein. Infection and Immunity, 73, 4062-4069. http://dx.doi.org/10.1128/IAI.73.7.4062-4069.2005
- Faisal, S.M., Yan, W., Chen, C.S., Palaniappan, R.U., Mcdonough, S.P. and Chang, Y.F. (2008) Evaluation of Protective Immunity of Leptospira Immunoglobulin Like Protein A (LigA) DNA Vaccine against Challenge in Hamsters. Vaccine, 26, 277-287. http://dx.doi.org/10.1016/j.vaccine.2007.10.029
- Pinne, M. and Haake, D.A. (2013) LipL32 Is a Subsurface Lipoprotein of Leptospira Interrogans: Presentation of New Data and Reevaluation of Previous Studies. PLoS One, 8, e51025. http://www.ncbi.nlm.nih.gov/pubmed/?term=leptospira+lipl32++subsurface http://dx.doi.org/10.1371/journal.pone.0051025
NOTES

*Corresponding author.

