Advances in Microbiology
Vol.05 No.13(2015), Article ID:62025,13 pages
10.4236/aim.2015.513090
Probiotic Screening of Lactobacilli Isolates from Uttapam Batter Fermented Supplementing with Piper betle L. Leaves
Ashwini Kumar Dubey, Kadirvelu Jeevaratnam*
Department of Biochemistry and Molecular Biology, Pondicherry University, Puducherry, India

Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 16 November 2015; accepted 15 December 2015; published 18 December 2015
ABSTRACT
The in vitro screening of lactobacilli isolates from uttapam batter, fermented supplementing with Piper betle L. leaves, was performed in order to select potent isolates for probiotic use. Their resistance to simulated gastric and intestinal juices as well as their adhesion to epithelial intestinal HCT-15 and vaginal HeLa cell lines were assessed and also evaluated for their immunomodulatory ability in ex vivo condition. Isolates were able to adhere as well as reduce pathogen adhesion to monolayer cell lines’ surfaces and reduced production of pro-inflammatory cytokine TNF-α in LPS treated PBMCs, while enhanced that of anti-inflammatory cytokine IL-10. The isolates exhibited properties of auto-aggregation, co-aggregation, hydrophobicity, bile salt hydrolase activity and strong antimicrobial activity against pathogenic bacteria. Thus, the different lactobacilli isolates displayed potent probiotic and immunomodulatory properties among which AJ7 and AJ82 had a great potential and may have applications in fermented foods as immunomodulatory probiotic additives.
Keywords:
Acid Tolerance, Adhesion, Immune Modulation, Lactobacilli, Probiotic

1. Introduction
The multidrug resistance of common pathogens to several traditional antibiotics has been a serious worldwide problem which has directed to the discovery of new substitutes and consequently there is an increasing concentration towards the usage of probiotic bacteria for the management of various bacterial and fungal infections [1] [2] of digestive tract in humans and animals [3] . Probiotics are live microorganisms that diminish the progression of harmful microbes and encourage well balance of microbiota in the gastrointestinal system [4] . Lactic acid bacteria (LAB) are thought to be dominant in the small intestine [5] and are known to produce lactic acid, hydrogen peroxide, carbon dioxide, diacetyl, bacteriocins [6] [7] which contribute to destroying harmful pathogens in situ. LAB are able to interact with the immune system [8] [9] and could act as bio-therapeutic agent in inflam- matory bowel disease [9] , irritable bowel syndrome [10] , travelers’ diarrhoea [11] , antibiotic associated diarrhoea [12] , enteric diseases [13] , bacterial infections in oral cavity and vaginosis [14] . In recent years, probiotics gained significance because of their roles in gastrointestinal and immunological aids as well as there has been an increased arrival of probiotic products in the world market [15] . A probiotic microorganism must resist gastric and intestinal juices, adhere mucosal surfaces of intestine, and, additionally, suitable traits could be the production of antimicrobial substances, cholesterol reducing property, and the production of β-galactosidase [16] [17] .
During our study on the effect of Piper betle L. leaves on fermentation of Uttapam (traditional south Indian fermented food) batter, lactobacilli with potent antimicrobial activity profile were isolated, characterized and identified [18] . The objective of this study was to evaluate these lactobacilli isolates from uttapam batter fermented supplementing with Piper betle L. leaves for probiotic properties in vitro and immunomodulatory activities ex vivo as per WHO/FAO and ICMR/DBT guidelines [16] [17] .
2. Materials and Methods
2.1. Microorganisms and Chemicals
The following Lactobacillus pentosus and Weissella confusa isolates used in this study to evaluate their probiotic potential were already characterized using 16S rRNA gene sequence analysis and their sequences were submitted to GenBank [18] : Lb. pentosus AJ7 (JX683265), Lb. pentosus AJ9 (JX683266), Lb. pentosus AJ15 (JX683267), Lb. pentosus AJ82 (JX683270), Lb. pentosus AJ94 (JX683271), Lb. pentosus AJ99 (JX683272), Lb. pentosus AJ112 (JX683273), Lb. pentosus AJ116 (JX683274), Lb. pentosus AJ125 (KC533686), W. confusa AJ53 (KC533685), W. confusa AJ67 (JX683268) W. confusa AJ79 (JX683269). The indicator strains Listeria monocytogenes MTCC657, Staphylococcus aureus subsp. aureus MTCC737, Lactobacillus rhamnosus MTCC1408, Lactobacillus plantarum 6160 were procured from MTCC (Microbial Type Culture Collections), Institute of Microbial Technology, Chandigarh, India. The microbial culture media and analytical grade chemicals were procured from HiMedia, (Mumbai, India) and Animal cell culture medium and Histopaque from Sigma-Aldrich, (USA).
2.2. Effect of pH and Bile on Growth of Lactobacilli
The effect of pH and bile on the growth of lactobacilli was studied by growing the isolates in the de Mann Rogosa & Sharpe (MRS) broth adjusted to different pH (3.0, 3.5 and 7.0) using 2 mol∙L−1 HCl or NaOH and MRS broth supplemented with 0.05%, 0.1%, 0.3% (w/v) of bile and MRS broth without bile at pH 6.5 with initial inoculum of overnight culture of isolates to obtain the final optical density (OD) 0.2 at 600 nm, incubated at 37˚C. The OD readings were taken every hour for eight hour then at twenty-fourth hour in a sterile flat bottom ninety- six well microtitre plate using 200 microliters of culture at 600 nm using ELISA reader (Molecular Devices) [19] .
2.3. Resistance to Simulated Gastric Fluid and Intestinal Fluid
The effect of gastric juice on the survival of the isolates was checked at 0, 2 and 4 h because the time from entrance to release from the stomach is 3 - 4 h and similarly, for the intestinal juice also the survivability of isolates was checked at 0, 2 and 4 h [20] . The isolates were propagated overnight in MRS broth, cells were harvested by centrifugation and 109 - 1010 CFU mL−1 were suspended in artificial gastric fluid (NaCl, 0.72 g∙L−1; KCl, 0.05 g∙L−1; NaHCO3 g∙L−1; pepsin, 0.3 g∙L−1) adjusted to pH 2.0, 2.5 and 3.0 and incubated for 0, 2 and 4 h. The intestinal juice resistance was studied by exposing the isolates (109 - 1010 CFU mL−1) to artificial intestinal fluid (0.1% w/v pancreatin and 0.3% w/v bile salts, pH 8.0) for 0, 2 and 4 h [20] [21] . Samples from all trials plated in MRS agar to count colony forming units. The sterile saline (0.85% w/v NaCl) adjusted to pH 7.0 was used as control.
2.4. Bile Salt Hydrolase
The bile salt hydrolase (BSH) activity of LAB isolates was checked by streaking the fresh culture of LAB on BSH medium consisting of MRS Agar supplemented with 0.5% (w/v) sodium salt of TDCA (taurodeoxycholic acid) and 0.37 g∙L−1 CaCl2. The plates were incubated in an anaerobic jar at 37˚C for 36 - 48 h. Precipitation zones around colonies were considered as positive test for the BSH activity [22] .
2.5. Antibiotic Susceptibility Test
The antibiotic susceptibility of the lactobacilli isolates was checked using antibiotic octadiscs (HiMedia, Mumbai, India) towards various antibiotics such as aminoglycosides, penicillins, tetracyclines, cephalosporins, inhibitors of urinary tract antiseptics, nucleic acid synthesis and cytoplasmic membrane functions [23] . The antibiotic discs were placed on the MRS agar media seeded with the LAB isolates and incubated anaerobically at 37˚C for overnight. The susceptibility was recorded by measuring diameter of inhibition zones around the discs.
2.6. Auto-Aggregation
For the aggregation between same cells (auto-aggregation), LAB isolates were grown in MRS broth for overnight at 37˚C followed by harvesting (centrifugation, 10,000 g), washing and re-suspension of pellet in sterile PBS (pH 7.0) and adjusted to 1OD at 600 nm. The cell suspension left for 1 h at room temperature followed by centrifugation at 300 g for 2 min at 20˚C and its OD was read at 600 nm. Auto-aggregation was determined using the following equation.
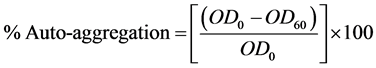
OD0 and OD60 refers to the initial OD and OD of the supernatant determined after 60 min, respectively [19] .
2.7. Co-Aggregation
The co-aggregation (aggregation between genetically different microorganisms) of LAB isolates was performed with the indicator strains Lactobacillus plantarum MTCC 6160 and Lactobacillus rhamnosus MTCC 1408 which were grown in 10 mL of MRS broth and Listeria monocytogenes MTCC 657 and Staphylococcusaureus subsp. aureus MTCC 737 grown in 10 mL of Tryptic soya broth at 37˚C [19] . Briefly, the cells were harvested by centrifugation (10,000 g) after overnight incubation, washed and resuspended in sterile PBS and adjusted to 1 OD at 600 nm. One mL of each cell suspension (i.e., indicator strain and LAB strain to be evaluated for probiotic potential) was transferred to a tube and OD recorded at 600 nm. The mixed cell suspension left for 1h at room temperature followed by centrifugation at 300 g for 2 min at 20˚C and its OD was read at 600 nm. The co-ag- gregation was determined using the following equation.
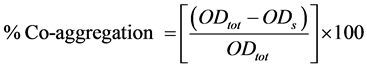
ODtot and ODS refers to the initial OD taken immediately after the relevant strains mixed and OD of the supernatant after 60 min respectively.
2.8. Hydrophobicity
The hydrophobicity assay is an index to measure the ability of isolates to adhere the mucosal surfaces of epithelial lining [23] . The LAB isolates were grown in MRS broth for overnight at 37˚C followed by harvesting, washing and re-suspension of pellet cells in sterile PBS (pH 7.0) and adjusted to OD of 1 at 600 nm. A sample of 1.5 ml LAB suspension was mixed to 1.5 ml of n-hexadecane by vortex and incubated 30 min at room temperature for the separation of aqueous and organic phases. The OD of aqueous phase was read at 600 nm [22] . The hydrophobicity percentage was calculated as follows
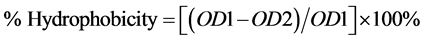
where OD1 refers to the initial OD of the cell suspension and OD2 refers to the OD of the cell suspension in aqueous phase after incubation of 30 min.
2.9. LAB Adhesion and Inhibition of Pathogen Adhesion to Cell Lines
A probiotic strain must adhere the intestinal mucosal surfaces to act in situ and provide gut health benefits. To evaluate the adhesion ability of isolates to intestinal mucosa, epithelial intestinal HCT-15 cell line was chosen [24] . The adhesion of LAB isolates and inhibition of pathogen adhesion to HCT-15was evaluated. Additionally, it is recognized that the application of probiotics can go beyond gut health and it can be used as suppository for vaginal health and thereby may reduce the risk of urinary tract infections, bacterial vaginosis and yeast vaginosis by balancing the microflora [25] - [27] . To study the adhesion ability of isolates at vaginal mucosa, vaginal HeLa cell line was chosen [26] [27] . Both the cell lines were maintained in the DMEM medium supplemented with 10% of fetal bovine serum (FBS), 2 5 mM HEPES, 50 μg∙mL−1 penicillin, 50 μg∙mL−1gentamicin, 50 μg∙mL−1 streptomycin and 1.25 μg∙mL−1 amphotericin-B. The cell lines were used for assay when reached to 90% to 95% confluences. For the LAB adhesion and inhibition of pathogen adhesion to the cell lines, monolayers of cell lines were maintained in the six well tissue culture plates (Tarson, India) and the LAB isolates and pathogen were suspended in serum and antibiotic free DMEM medium. The monolayers of cell lines were washed with the PBS thrice to remove antibiotics followed by the addition of LAB suspensions (1010 - 1011 CFU mL−1) and kept for 90 min at 37˚C [20] . After incubation, wells were washed gently with PBS thrice and the non-adherent recovered LAB cells were plated over MRS agar. The adhesion of LAB isolates were calculated as the amount of adhered bacterial cells, compared to the initial amount of LAB cells added to the cell lines with the following formula,
Amount of adherent LAB cells = log (A) − log (B)
Where A is the No. of viable LAB cells added to the well for adhesion and B is the No. of viable LAB cells non-adhered to the cell lines in the well.
The culture wells, subsequent to incubation with LAB isolates and removing the non-adhered bacterial cells with PBS wash, were fixed with 2% formaldehyde followed by the staining with 0.1% acridine orange dye and observed under fluorescent microscope. For the pathogen (Listeria monocytogenes MTCC 657) exclusion assay, the monolayers were first treated with the LAB isolates (5 ´ 1010 - 1011 CFU mL−1) for 90 min, washed with PBS to remove non-adhered LAB followed by addition of Listeria cells (2 ´ 1010 CFU mL−1) suspended in serum and antibiotic free medium and incubated for 60 min [20] . The excluded Listeria cells were recovered by washing with the PBS and were plated on Listeria selective agar [28] . The amount of excluded Listeria cells were calculated as the amount of Listeria cells recovered after incubation compared to initial amount added to the cell lines with the following formula,
Amount of excluded Listeria cells = log (X) − log (Y)
where X is the no. of viable Listeria cells added to the well for adhesion and Y is the no. of viable Listeria cells adhered to the cell lines in the well.
2.10. Isolation of Human Peripheral Blood Mononuclear Cells (hPBMCs) and Induction of Cytokine Release
The hPBMCs were isolated from the blood of healthy human donor. Briefly, 3.0 mL of Histopaque 1077 (Sigma Aldrich, USA) was placed in a 15 mL centrifuge tube and 3.0 mL of heparinized whole blood was layered on the top followed by centrifugation (400 g, 30 min at room temperature). The upper plasma layer was removed and discarded. The cells separated at the interface was taken up by aspiration and washed with PBS. The cell viability test was determined by trypan blue exclusion and the cell density was adjusted to 107 cells mL−1 in RPMI-1640 medium supplemented with 10% autologous serum, 50 μg∙mL−1 penicillin, 50 μg∙mL−1 gentamicin, 50 μg∙mL−1 streptomycin and 1.25 μg∙mL−1 amphotericin-B and incubated at 37˚C for overnight in a suspension flask. The hPBMCs were incubated with the LPS (1.0 μg∙mL−1) for 30 min before exposed to LAB isolates. Fresh bacterial suspensions of LAB isolates (108 CFU mL−1) were added to hPBMCs (107 cells mL−1) resulting in bacteria-to-cell ratio 10:1 and co-incubated for 24 h in antibiotic and serum free RPMI-1640 medium at 37˚C in a 12-well tissue culture plates [29] . After incubation the supernatants were collected and used immediately for estimation of cytokines. Because, Lipopolysaccharide (LPS) provokes host cells to produce pro-inflammatory mediators including TNF-α, IL-1α, IL-6 by activating several types of transcription factors, LPS stimulated hPBMCs culture was used to study the anti-inflammatory potential of probiotic strains for reduction in TNF-α. The anti-inflammatory cytokine IL-10 was also measured in the supernatant of hPBMCs. The assays for both the pro-inflammatory (TNF-α) and anti-inflammatory (IL-10) cytokines were performed twice independently in duplicates [29] . TNF-α and IL-10 cytokines were measured using anti-human TNF-α and anti-human IL-10 monoclonal antibodies respectively by sandwich ELISA kit (Diasource) as per manufacturer’s recommendations.
2.11. Inhibition of Listeria by Lactobacilli (LAB) Isolates in Situ
In situ inhibition of Listeria by LAB isolates was studied by co-culturing both LAB and Listeria for different time intervals and subsequently their CFU were calculated. The LAB isolates were grown in nutrient broth till the OD reached 0.8 at 600 nm. Overnight culture of Listeria monocytogenes MTCC657 grown in nutrient broth was added to the LAB culture and incubated at 30˚C under shaking condition (180 rpm) in a shaker incubator. The CFU in the co-culture were calculated for LAB and Listeria by selectively plating over MRS agar and Listeria selective agar respectively at 0, 3, 6, and 9 h of intervals. A control LAB and control Listeria culture were also processed in similar way.
3. Results
3.1. Resistance to Simulated Gastric Fluid and Intestinal Fluid
The Lb. pentosus isolates were growing in the MRS broth containing 0.3% of bile (normal concentration in the intestine of healthy man) [4] [30] whereas no growth was observed in case of W. confusa isolates AJ53, AJ67 and AJ79 (Table 1) and were excluded for the further studies. Their resistance to simulated gastric and intestinal juices is shown in Table 2. Based on the growth profile in acidic conditions and presence of bile salts (Table 1) and survivability in the simulated gastric and intestinal juices (Table 2), six isolates AJ7, AJ9, AJ15, AJ82, AJ99 and AJ112 were chosen for the cell line adhesion studies.
3.2. Bile Salt Hydrolase Activity
The isolates AJ9, AJ99 and AJ116 were BSH negative, while others were positive (Table 3).
Table 1. Effect of pH and bile on growth of lactobacilli isolates.
Legends: −, no growth; +, very weak growth, ++, weak growth, +++, good growth, ++++, luxurious growth.
Table 2. Survivability of lactobacilli isolates in simulated gastric and intestinal juices.
Table 3. Bile salt hydrolase, hydrophobicity, auto-aggregation and co-aggregation and cell line adhesion assays.
The values are mean of two independent experiment performed in duplicates, Legends: +, positive; −, negative; ND, not determined, *calculated by formula: amount of adhered LAB cells = log (A) ? log (B), where A is the number of viable LAB cells added to the well and B is the number of viable non-adhered LAB cells, **calculated by formula: amount of excluded Listeria by LAB cells = log (X) ? log (Y), where X is the number of viable Listeria cells added to the well and Y is the number of viable adhered Listeria cells.
3.3. Antibiotic Susceptibility
The LAB isolates were found to be susceptible to most of the antibiotics screened (Table S1). All lactobacilli isolates were sensitive to penicillin, ampicillin, erythromycin, ciprofloxacin, gentamicin, lincomycin, chloramphenicol, streptomycin, amikacin, cephaloridine and tetracycline. However, all the isolates were resistant to colistin.
3.4. Auto-Aggregation, Co-Aggregation and Hydrophobicity
The auto-aggregation between lactobacilli isolates varied from 34% to 63%, while co-aggregation between LAB and non-LAB strains varied from lowest 24% to highest 57% (Table 3). The hydrophobicity of LAB isolates were ranged from least 19% (AJ15) to maximum 90% (AJ82) (Table 3).
3.5. LAB Adhesion and Inhibition of Pathogen Adhesion to Epithelial Cell Lines
The ability of six selected isolates to adhere to epithelial cell lines and pathogen exclusion is shown in Table 3. The isolateAJ15 neither adhered to intestinal HCT-15 cell line and nor inhibited the adhesion of Listeria pathogen, however, strains AJ7, AJ9, AJ82, AJ99 and AJ112 were able to adhere to both the epithelial cell lines HCT-15 and HeLa (Figure 1(A) & Figure 1(B)) considerably better or similar percentage than other lactobacilli strains evaluated in literature [26] as well as inhibited the adhesion of Listeria cells greatly (Table 3).
3.6. Immunomodulation
The production of cytokines TNF-α and IL-10 by four selected LAB isolates (AJ7, AJ82, AJ99 and AJ112) was used as an index of immunomodulation (Table 4). The TNF-α induction was higher in control sample of LPS treated hPBMCs than the hPBMCs incubated with LAB strains. However, there was weak induction of TNF-α in the case of hPBMCs incubated with both the LPS and LAB together (Table 4). It indicates that LAB isolates AJ7, AJ82, AJ99 and AJ112 were able to reduce induction of pro-inflammatory cytokine TNF-α in LPS treated hPBMCs. The stimulation of anti-inflammatory cytokine IL-10 was observed in case of hPBMCs incubated with both the LPS and LAB strains independently and together. The induction of IL-10 in hPBMCs incubated with the LPS was weak in comparison to the hPBMCs incubated with LAB and hPBMCs incubated with LPS and LAB strains both (Table 4). The results show that these isolates AJ7, AJ82, AJ99 and AJ112 induce low TNF-α and high IL-10.
3.7. In Situ Inhibition of Listeria
The two potent isolates AJ7 and AJ82 displayed a very good antagonistic activity against Listeria monocytogenes MTCC657 in nutrient broth when co-cultured (Figure 2(A) & Figure 2(B)). The isolates AJ7 and AJ82 competitively inhibited Listeria. The individual control cultures of both the LAB and Listeria had grown exponentially whereas the number of viable Listeria cells when co-cultured with LAB isolates were dramatically decreased to zero at 9 h of co-incubation probably owing to the effects of various antimicrobial substances released by LAB [6] . On the other hand, the LAB isolates co-cultured with Listeria grew similar to control LAB culture.
4. Discussions
One of the essential properties of a probiotic is inhibition of the growth of pathogenic microorganisms. In the present study, the LAB isolates screened for probiotic had inhibited the growth of different pathogens owing to the production of various antimicrobial substances [6] [18] . In the earlier study these LAB isolates were found to
Table 4. Induction of cytokines TNF-α and IL-10.
Figure 1. (A) Adhesion of LAB isolates on HCT-15 cell line: the control is only HCT-15 with no LAB isolate (a) Control, (b) Lb. pentosus AJ7, (c) Lb. pentosus AJ15, (d) Lb. pentosus AJ82, (e) Lb. pentosus AJ99, (f) Lb. pentosus AJ112; (B) adhesion of LAB isolates on HeLa cell line: the control is HeLa cell line alone without LAB isolate (a) Control, (B) Lb. pentosus AJ7, (c) Lb. pentosus AJ15, (d) Lb. pentosus AJ82, (e) Lb. pentosus AJ99, (f) Lb. pentosus AJ112.
be bacteriocinogenic [18] . However, the LAB isolates did not pose much threat to other LAB strains Lactobacillus rhamnosus MTCC1408 and Enterococcus faecalis MTCC439.
The probiotic strain intended for oral delivery has to travel through the GI tract of the host and therefore it must resist the physiological conditions of the gut such as acidic pH of gastric fluid and presence of bile salts in order to arrive at the action site. The LAB strains in the present study were resistant to acidic pH of artificial gastric fluid and bile concentration of artificial intestinal fluid. The LAB isolates were able to grow at the concentration of intestinal bile in human GIT (0.3% w/v) as well. WHO/FAO recommends a probiotic strain with an ability to hydrolyze bile salts as the BSH positive microorganisms are known to remove cholesterol [31] . The BSH positive isolates of this study may offer a promising use as anti-cholesterolaemic agents for functional foods. Another requirement for a probiotic is that it should not carry transmissible genes for antibiotic resistance

Figure 2. In situ Inhibition of Listeria by LAB isolates (A) Lb. pentosus AJ7 and (B) AJ82.
[29] otherwise undesirable microbes in the gut may acquire resistance via horizontal gene transfer. Since the screened LAB isolates were susceptible to most of the antibiotics resistance transfer cannot happen, however, this necessitates frequent administration of these organisms incorporated foods or the organisms per se in large quantity.
All the LAB isolates had shown high auto-aggregation and co-aggregation characteristics. The aggregation between the same LAB isolates aids in colonization thereby preventing pathogen adhesion in the intestinal mucosa [20] and the co-aggregation of LAB and pathogens enables the LAB to find the pathogen in a close proximity and eliminate pathogen by releasing antimicrobial substances [20] . The antimicrobial substances released by aLAB isolate should inhibit the co-aggregating pathogenic microbe partner otherwise both may co-exist and form mixed culture biofilm. Conversely, a LAB strain co-aggregating with other LAB strain or other normal flora should not antagonistic to each other. In this study, all the LAB isolates had shown high level of hydrophobicity (except AJ9, AJ15 & AJ125). An isolate with higher percentage of hydrophobicity would have strong adhesion characteristics. The adhesion ability of a probiotic strain to the epithelial mucosal surfaces is pivotal for its colonization and in situ antagonism against pathogens subsequently [23] . The LAB isolates adhered to cellline effectively which appears to be a sine quo non for desirable probiotic benefit. It is reported, cell wall composition profile may influence the adhesion of LAB cells to the epithelial cells [4] and the variability observed in the adhesion profile of LAB isolates may be attributed to their variable cell wall composition profile [32] . The adhered LAB isolates to the epithelial cells serve as physical barrier to non-desirable agents. Therefore, the isolates with good adhesion and pathogen exclusion profile (AJ7, AJ82, AJ99, and AJ112) for both the cell lines HCT-15 and HeLa can be applicable to both the anatomic locations, GIT as well as vagina for potential biotheraputic effects subsequent to their in vivo evaluation.
Probiotics may also provide health benefits via immune modulations. Microorganisms including∙Lactobacilli are able to cross the intestinal mucous layer and can penetrate the gut wall either through epithelial layer or Peyer’s patches. Subsequently, the microbial components interact with the immune cells and induce the production of cytokines, chemokines and other innate effectors [33] . The four isolates screened (Table 4) were able to reduce the induction of pro-inflammatory cytokine (TNF-α) and enhanced that of anti-inflammatory cytokine (IL-10).
The immunomodulation ability of a microorganism is believed to be strain specific [34] which varies from one strain to another. The induction of cytokines TNF-α and IL-10 by LAB strains was more or less similar to the other reports [29] [34] . Thus these isolates can be used as anti-inflammatory strains. A probiotic has to act in situ to inhibit the undesirable microorganisms. The two selected isolates AJ7 and AJ82 had inhibited Listeria strain in situ when co-cultured in nutrient broth owing to the effects of antimicrobial substances [18] .
5. Conclusion
The uttapam batter fermented supplementing with the Piper betle L. leaves may offer a promising source for probiotic isolation. The different isolates were resistant to acidic pH, able to adhere human epithelial cell lines, antagonistic to different food borne pathogens and immunomodulatory. Our results had shown that the two Lactobacillus pentosus isolates AJ7 and AJ82, with anti-inflammatory effects, possessed the properties to be used as potential probiotic additives to foods subsequent to their in vivo screening.
Acknowledgements
The authors acknowledge DST-FIST and UGC-SAP for financial assistance to the Department. The author, Ashwini Kumar Dubey, is also thankful to Indian Council of Medical Research (ICMR), New Delhi for Research Fellowships (JRF and SRF).
Cite this paper
Ashwini KumarDubey,KadirveluJeevaratnam, (2015) Probiotic Screening of Lactobacilli Isolates from Uttapam Batter Fermented Supplementing with Piper betle L. Leaves. Advances in Microbiology,05,858-870. doi: 10.4236/aim.2015.513090
References
- 1. Hancock, R.E. and Chapple, D.S. (1999) Peptide Antibiotics. Antimicrobial Agents and Chemotherapy, 43, 1317-1323.
- 2. Vizioli, J. and Salzet, M. (2002) Antimicrobial Peptides versus Parasitic Infections? Trends in parasitology, 18, 475-476. http://dx.doi.org/10.1016/S1471-4922(02)02428-5
- 3. Chou, L.S. and Weimer, B. (1999) Isolation and Characterization of Acid and Bile-Tolerant Isolates from Strains of Lactobacillus acidophilus. Journal of Dairy Science, 82, 23-31.
http://dx.doi.org/10.3168/jds.S0022-0302(99)75204-5 - 4. Kim, P.I., Jung, M.Y., Chan, Y.H., Kim, S., Kim, S.J. and Park, Y.H. (2007) Probiotic Properties of Lactobacillus and Bifidobacterium Strains Isolated from Porcine Gastrointestinal Tract. Applied Microbiology and Biotechnology, 74, 1103-1111. http://dx.doi.org/10.1007/s00253-006-0741-7
- 5. Marco, M.L., Pavan, S. and Kleerebezem, M. (2006) Towards Understanding Molecular Modes of Probiotic Action. Current Opinion in Biotechnology, 17, 204-210.
http://dx.doi.org/10.1016/j.copbio.2006.02.005 - 6. Bhattacharya, S. and Das, A. (2010) Study of Physical and Cultural Parameters on Bacteriocins Produced by Lactic Acid Bacteria Isolated from Traditional Indian Fermented Foods. American Journal of Food Technology, 5, 111-120. http://dx.doi.org/10.3923/ajft.2010.111.120
- 7. Mobaarez, A.M. and Doust, R.H., Sattari, M. and Manthegi, N. (2008) Antimicrobial Effects of Bacteriocins like Substances Produced by L. acidophilus from Traditional Yoghurt on P. aeroginosa and S. aureus. Journal of Biological sciences, 8, 221-224. http://dx.doi.org/10.3923/jbs.2008.221.224
- 8. Ménard, S., Candalh, C., Bambou, J.C., Terpend, K., Cerf-Bensussan, N. and Heyman, M. (2004) Lactic Acid Bacteria Secrete Metabolites Retaining Anti-Inflammatory Properties after Intestinal Transport. Gut, 53, 821-828. http://dx.doi.org/10.1136/gut.2003.026252
- 9. Martín, R., Miquel, S., Ulmer, J., Kechaou, N., Langella, P. and Bermúdez-Humarán, L.G. (2013) Role of Commensal and Probiotic Bacteria in Human Health: A Focus on Inflammatory Bowel Disease. Microbial Cell Factories, 12, 71. http://dx.doi.org/10.1186/1475-2859-12-71
- 10. Clarke, G., Cryan, J.F., Dinan, T.G. and Quigley, E.M. (2012) Probiotics for the Treatment of Irritable Bowel Syndrome—Focus on Lactic Acid Bacteria. Alimentary Pharmacology and Therapeutics, 35, 403-413.
http://dx.doi.org/10.1111/j.1365-2036.2011.04965.x - 11. Paredes-Paredes, M., Flores-Figueroa, J. and DuPont, H.L. (2011) Advances in the Treatment of Travelers’ Diarrhea. Current Gastroenterology Reports, 13, 402-407. http://dx.doi.org/10.1007/s11894-011-0208-6
- 12. Szajewska, H., Ruszczynski, M. and Radzikowski, A. (2006) Probiotics in the Prevention of Antibiotic-Associated Diarrhea in Children: A Meta-Analysis of Randomized Controlled Trials. The Journal of Pediatrics, 149, 367-372. http://dx.doi.org/10.1016/j.jpeds.2006.04.053
- 13. Casadei, G., Grilli, E. and Piva, A. (2009) Pediocin A Modulates Intestinal Microflora Metabolism in Swine in Vitro Intestinal Fermentations. Journal of Animal Science, 87, 2020-2028.
http://dx.doi.org/10.2527/jas.2008-1438 - 14. Dover, S.E., Aroutcheva, A.A., Faro, S. and Chikindas, M.L. (2008) Natural Antimicrobials and Their Role in Vaginal Health: A Short Review. International Journal of Probiotics and Prebiotics, 3, 219-230.
- 15. Vieira, A.V., Teixeira, M.M. and Martins, F.S. (2013) The Role of Probiotics and Prebiotics in Inducing Gut Immunity. Frontiers in Immunology, 4, 445. http://dx.doi.org/10.3389/fimmu.2013.00445
- 16. Indian Council of Medical Research Task Force; Co-ordinating Unit ICMR; Co-ordinating Unit DBT (2011) ICMR-DBT guidelines for evaluation of probiotics in food. Indian Journal of Medical Research, 134, 22-25.
- 17. FAO/WHO (2002) Guidelines for the Evaluation of Probiotics in Food. Ontario, Canada, 1-11.
- 18. Dubey, A.K. and Jeevaratnam, K. (2015) Identification of Lactobacillus pentosus and Weissella confusa Isolated from Uttapam Batter Fermented with Piper betle Leaves. Malaya Journal of Biosciences, 2, 13-25.
- 19. Todorov, S.D., Furtado, D.N., Saad, S.M.I., Tome, E. and Franco, B.D.G.M. (2011) Potential Beneficial Properties of Bacteriocin-Producing Lactic Acid Bacteria Isolated from Smoked Salmon. Journal of Applied Microbiology, 110, 971-986. http://dx.doi.org/10.1111/j.1365-2672.2011.04950.x
- 20. Vidhyasagar, V. and Jeevaratnam, K. (2013) Evaluation of Pediococcus pentosaceus Strains Isolated from Idly Batter for Probiotic Properties in Vitro. Journal of Functional Foods, 5, 235-243. http://dx.doi.org/10.1016/j.jff.2012.10.012
- 21. Ripamonti, B., Agazzi, A., Bersani, C., De Dea, P., Pecorini, C., Pirani, S., Rebucci, R., Savoini, G., Stella, S., Stenico, A., Tirloni, E. and Domeneghini, C. (2011) Screening of Species-Specific Lactic Acid Bacteria for Veal Calves Multi-Strain Probiotic Adjuncts. Anaerobe, 17, 97-105.
http://dx.doi.org/10.1016/j.anaerobe.2011.05.001 - 22. Lee, H., Yoon, H., Ji, Y., Kim, H., Park, H., Lee, J., Shin, H. and Holzapfel, W. (2011) Functional Properties of Lactobacillus Strains Isolated from Kimchi. International Journal of Food Microbiology, 145, 155-161.
http://dx.doi.org/10.1016/j.ijfoodmicro.2010.12.003 - 23. Agaliya, P.J. and Jeevaratnam, K. (2012) Screening of Lactobacillus plantarum Isolated from Fermented Idli Batter for Probiotic Properties. African Journal of Biotechnology, 11, 12856-12864.
- 24. Varma, P., Kavitha, R.D., Menon, K.K. and Biswas, R. (2010) Lactobacillus fermentum Isolated from Human Colonic Mucosal Biopsy Inhibits the Growth and Adhesion of Enteric and Foodborne Pathogens. Journal of Food Science, 75, M546-M551. http://dx.doi.org/10.1111/j.1750-3841.2010.01818.x
- 25. Hsieh, P.S., An, Y., Tsai, Y.C., Chen, Y.C., Chuang, C.J., Zeng, C.T., Wang, C.T. and King, V.A.E. (2013) Potential of Probitoic Strains to Modulate the Inflammatory Responses of Epithelial and Immune Cells in Vitro. New Microbiologica, 36, 167-179.
- 26. Ayeni, F.A., Sánchez, B., Adeniyi, B.A., de Los Reyes-Gavilán, C.G., Margolles, A. and Ruas-Madiedo, P. (2011) Evaluation of the Functional Potential of Weissella and Lactobacillus Isolates Obtained from Nigerian Traditional Fermented Foods and Cow’s Intestine. International Journal of Food Microbiology, 147, 97-104. http://dx.doi.org/10.1016/j.ijfoodmicro.2011.03.014
- 27. Coman, M.M., Verdenelli, M.C., Cecchini, C., Silvi, S., Orpianesi, C., Caspani, M., Mondello, F. and Cresci, A. (2015) In Vitro Evaluation on HeLa Cells of Protective Mechanisms of Probiotic Lactobacilli against Candida Clinical Isolates. Journal of Applied Microbiology, 119, 1383-1390. http://dx.doi.org/10.1111/jam.12947
- 28. Osmanagaoglu, O., Kiran, F. and Ataoglu, H. (2010) Evaluation of in Vitro Probiotic Potential of Pediococcus pentosaceus OZF Isolated from Human Breast Milk. Probiotics and Antimicrobial Proteins, 2, 162-174. http://dx.doi.org/10.1007/s12602-010-9050-7
- 29. Ben Salah, R., Trabelsi, I., Ben Mansour, R., Lassoued, S., Chouayekh, H. and Bejar, S. (2012) A New Lactobacillus plantarum Strain, TN8, from the Gastro Intestinal Tract of Poultry Induces High Cytokine Production. Anaerobe, 18, 436-444. http://dx.doi.org/10.1016/j.anaerobe.2012.05.001
- 30. Goldin, B.R., Gorbach, S.L., Saxelin, M., Barakat, S., Gualtieri, L. and Salminen, S. (1992) Survival of Lactobacillus Species (Strain GG) in Human Gastrointestinal Tract. Digestive Diseases and Sciences, 37, 121-128. http://dx.doi.org/10.1007/BF01308354
- 31. Patel, A.K., Singhania, R.R., Pandey, A. and Chincholkar, S.B. (2010) Probiotic Bile Salt Hydrolase: Current Developments and Perspectives. Applied Biochemistry and Biotechnology, 162, 166-190.
http://dx.doi.org/10.1007/s12010-009-8738-1 - 32. Dimitrov, Z., Gotova, I. and Chorbadjiyska, E. (2014) In Vitro Characterization of the Adhesive Factors of Selected Probiotics to Caco-2 Epithelium Cell Line. Biotechnology & Biotechnological Equipment, 28, 1079-1083. http://dx.doi.org/10.1080/13102818.2014.969948
- 33. Quinto, E.J., Jiménez, P., Caro, I., Tejero, J., Mateo, J. and Girbés, T. (2014) Probiotic Lactic Acid Bacteria: A Review. Food and Nutrition Sciences, 5, 1765-1775.
http://dx.doi.org/10.4236/fns.2014.518190 - 34. Helwig, U., Lammers, K.M., Rizzello, F., Brigidi, P., Rohleder, V., Caramelli, E., Gionchetti, P., Schrezenmeir, J., Foelsch, U.R., Schreiber, S. and Campieri, M. (2006) Lactobacilli, Bifidobacteria and E. coli Nissle Induce Pro- and Anti-Inflammatory Cytokines in Peripheral Blood Mononuclear Cells. World Journal of Gastroenterology, 12, 5978-5986.
Supplementary Table
Table S1. Antibiotic susceptibility of the lactobacilli isolates.
Legends: R, resistant; S, sensitive. All the isolates were sensitive to penicillin-G, tetracycline, lincomycin, gentamicin erythromycin, ciprofloxacin, chloramphenicol, streptomycin, amikacin, cephaloridine and ampicillin while resistant to colistin.
NOTES
*Corresponding author.






