Advances in Microbiology
Vol.4 No.4(2014), Article ID:44191,9 pages DOI:10.4236/aim.2014.44028
Clostridium bifermentans as an Aero-Tolerant Exponent of Strictly Anaerobe Genera
Katarzyna Leja*, Kamila Myszka, Mariola Olkowicz, Wojciech Juzwa, Katarzyna Czaczyk
Department of Biotechnology and Food Microbiology, Poznan University of Life Sciences, Poznan, Poland
Email: *katleja@up.poznan, kmyszka@up.poznan.pl, Olkowicz@up.poznan.pl, wojciech.juzwa@up.poznan.pl, kasiacz@up.poznan.pl
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 14 January 2014; revised 14 February 2014; accepted 21 February 2014
ABSTRACT
Strictly anaerobic bacteria in the evolutionary way formed some mechanisms of oxygen tolerance. These changes enable strictly anaerobic bacteria existing in natural environment in which avoiding oxygen is not possible. Clostridium bifermentans is described as a strictly anaerobe species; however, in the literature, there is some information about its oxygen tolerance. Thus, in this work, the level of C. bifermentans aero-tolerance, its mechanisms, and the ability to metabolite production in presence of oxygen in cultivation medium were investigated. It was found out that C. bifermentans is able to survive in the presence of oxygen. Moreover, they are able to utilize oxygen and product metabolites when the level of oxygen is below 10%. In bacteria cells superoxide dismutase was detected.
Keywords:C. bifermentans; Microaerophilic; Oxygen Tolerance

1. Introduction
The occurrence of oxygen in the Earth’s atmosphere two billion years ago, as a consequence of developing of photosynthetic bacteria, caused evolution changes in an anaerobic bacteria metabolism. These changes provided form mechanisms of protection against toxic by-products of oxygen in bacteria cells and enable strictly anaerobic bacteria to exist in natural environment in which avoiding oxygen is not possible [1] [2] . The strictly anaerobic microorganisms are not able to gain energy from oxygen respiration but first of all they are not able to long-time survive in the presence of oxygen at atmospheric level [3] . However, some strictly anaerobic bacteria species are able to survive short-time oxygenation, and they continue their growth after bringing back anaerobic conditions [1] [4] [5] . Moreover, the ability of utilizing of small amounts of oxygen by several species was developed. Some scientists explain the physiology of oxygen utilization by strictly anaerobic microorganisms by the fact that the main respiration process carried out with slight partial oxygen pressure [6] . However, the main meaning in aero-tolerance mechanisms in microorganisms plays: enzymes (catalase, superoxide dismutase, peroxydase) [3] [7] -[10] occurrence of negative aerotropism, occurrence of intermembrane chemoreceptors, and reduction of oxygen bonds [11] [12] . The very important role against oxygen-stress plays superoxide dismutase. The genes encoding this enzyme which were detected is some anaerobic microorganisms, such as Bacteroides fragilis, Porphyromonas gingivalis [13] , Methanobacterium thermoautotrophicum [14] [15] Desulfovibrio vulgaris [16] and Clostridium perfringens [17] . Despite the fact that Clostridium bifermentans is described as a strictly anaerobe species [18] -[21] , in the literature there is some information about its oxygen tolerance [22] -[24] . Moreover, Hewitt and Morris [2] detected in C. bifermentans cells enzyme superoxide dismutase.
In this work, the level of C. bifermentans aero-tolerance, its mechanisms, and the ability to metabolite production in the presence of oxygen in cultivation medium were investigated.
2. Materials and Methods
2.1. Microorganisms
C. bifermentans strains (KM 371, KM 374, and KM 376) were isolated from samples that were collected from a manure in the Wielkopolska Region (Poland) and it has been identified and characterized in a previous work [25] . Cultures of C. bifermentans are kept at culture collection of Department of Biotechnology and Food Microbiology at Poznan University of Life Sciences.
2.2. Cultivation Medium and Conditions
Bacteria were cultivated in the modified PY medium consisted of (g/l): BactoPeptone 10; yeast extract 10; CaCl2, MgSO4 × 7H2O 0.96; K2HPO4 2; NaHCO3 20; NaCl 4; glycerol 50. The anaerobic conditions in flask were maintained by an anaerobic gas generating kit (Oxoid, UK), microanaerobic conditions by CampyGen Kit (Oxoid, UK).
2.3. Bath Fermentation Procedures
A preculture was carried out in a 500 ml flask containing 300 ml PY medium with glycerol at 37˚C for 24 h. It was inoculated into a five liters bioreactor (Sartorius Stedim, Germany) with three liters PY medium. According to Myszka et al. [25] , a blanket of a high-purity grade gas mixture of 5% O2 and 95% CO2 was maintained during cultivation. Gas flow rate was at up to 1.0 l/min, the stirrer speed varied between 200 and 500 rpm. The fermentation was run at 30˚C for seven days. Samples were taken in 24 h intervals to further analyses.
2.4. Analytical Procedures
The redox potential of bacteria cells was analyzed by flow cytometer BD FACS Aria III (Becton Dickinson). The cells were characterized by two non-fluorescent parameters: forward scatter (FSC) and side scatter (SSC), and two fluorescent parameters: green fluorescence (FL1) from RedoxSensor #8482; Green reagent and red fluorescence (FL2) from propidium iodide (PI) reagent. For excitation of both fluorescent reagents 488 nm blue laser was employed. The flow cytometric analyses were performed by using logarithmic gains and specific detectors settings. The threshold was set on the FSC and FL2 (PI) signals in order to discriminate microbial cells from the background and to put all the cells on scale. Data were acquired in a five-decade logarithmic scale as area signals (FSC-A, SSC-A, FL1-A and FL2-A) and analyzed with FACS DIVA software (Becton Dickinson).
The ability to metabolite production in microaerophilic conditions were delineated with a high liquid performance chromatography (HPLC) technique. The Hewlett Packard system consisted of an auto sampler and a pump, and a refractive index detector was used. The analysis was performed isocratically at flow rate 0.6 ml/min. at 65˚C, on a column Aminex HPX- 87H300 × 7.8 (Bio-Rad, USA). 0.5 M H2SO4 as a mobile phase was also used. The standards were applied to identify peaks in chromatograms, and peak areas were measured to determine the chemical compounds concentration (ChemStation, Agilent, USA).
The protein profile of C. bifermentans cells was tested by mass spectrometry. Analyses were done in LC-MS/MS configuration using nanoACQUITY UPLC (Ultra Performance Liquid Chromatography) coupled with mass spectrometer Orbitrap Velos (Thermo). Chromatography analyses were done on a column RP-C18 (Waters, BEH130 C18, 75 μm × 250 mm). Proteins were identified using MASCOT algorithm.
Oxygen concentration in the cultivation environment was analyzed by gas chromatography (Agilent Technologies, 7890A System) with injector and Thermal Conductivity Detector (TCD). Analyzes were done using TG Bond Msieve 5A (Thermo Scientific) column (30 m × 0.53 mm × 50 μm). The mobile phase was helium (99.99%). The temperature of injector and detector was set up to 230˚C.
3. Results
In the first step of investigation on the aero-tolerance of C. bifermentans strains, the changes of redox potential of its cells was measured by the technic of flow cytometer. The aim of this experiment was to confirm the ability of these bacteria to survive and maintained the metabolic activity in the microaerophilic conditions. Bacteria were cultivated in 500 ml flask with CampyGen kit. As a result it was find out that independently on cultivation conditions bacteria cells have generally comparable metabolic activity level, measured as an intensity of fluorescence (Figure 1). The significant differences in the redox potential were only observed in 24 hour of cultivation in case of KM 374 and KM 376, and in 48 hour in KM 371 strain. However, in 72 cultivation hour and later this potential was closely related in all investigated strains. C. bifermentans produce metabolites in the stationary phase of growth (between 24 and 120 hour of cultivation) (unpublished data) so it indicated the assumption that the presence of oxygen in cultivation environment not influenced on the efficiency of metabolite production by C. bifermentans strains.
Thus, the authors compared the metabolic profile of investigated strains cultivated both in anaerobic and microanaerobic a five liters bioreactors. Bacteria were cultivated in PY medium, with glycerol as a carbon source. The results are presented in Table 1. The main product of glycerol metabolic pathway was 1,3-propanediol (1,3-PD), and the by-products were lactic, fumaric, acetic, succinic acids, and ethanol in trace amounts (excluding KM 371).
The metabolites were synthetized on the same level, independently on the cultivation conditions. It indicated that C. bifermentans, despite the fact that they are recognized as a strictly anaerobic, have same mechanisms of an aero-tolerance.
Thus, the level of oxygen tolerance by C. bifermentans was investigated. Bacteria were cultivated in a five liters bioreactors with automatic microaeration (1%, 5%, and 10% of O2). The ability of 1,3-PD production by investigated strains in these conditions were tested. The obtained results are presented in Figure 2.
It occurred that investigated C. bifermentans strains were able to the main metabolite production in the presence of small amount of oxygen (1% and 5%). The aeration on the level of 10% of oxygen inhibited 1,3-PD synthesis, however bacteria survived in such conditions (unpublished data). Moreover, in cultivations with mi-
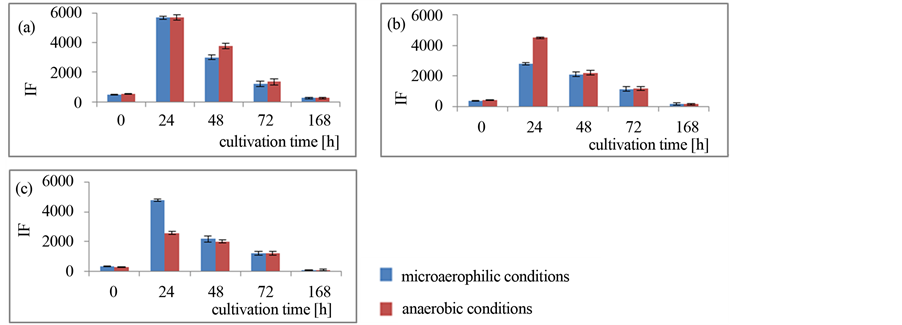
Figure 1. The redox potential of C. bifermentans cells during cultivations: A KM 371; B KM 374; C KM 376; IF – fluorescence intensity.
Table 1. Metabolic profile of investigated strains of C. bifermentans cultivated in anaerobic and microanaerobic conditions.
AC—anaerobic condition; MC—microanaerobic condition; LA—lactic acid; FA—fumaric acid; AA—acetic acid; SA—succinic acid; EtOH—ethanol; nd—not detected.
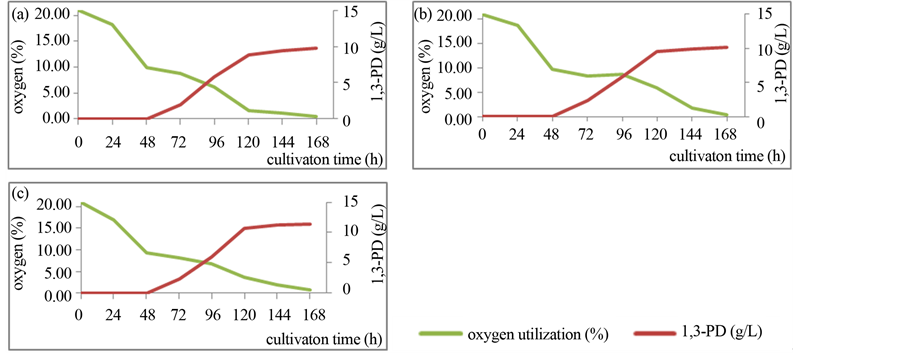
Figure 2. The kinetics of oxygen utilization (green line) and 1,3-PD production (red line) by C. bifermentans strains: (a) KM 371; (b) KM 374; (c) KM 376.
croaeration 1,3-PD was synthetized in comparable amounts such as in strictly anaerobic conditions. These observations conduced the above mentioned assumption about the presence of aero-tolerance mechanisms in C. bifermentans cells.
In the aim to develop the mechanisms of aero-tolerance of C. bifermentans, the ability to oxygen utilization and the protein profile were investigated. The oxygen utilization by investigated strains was tested in 500 ml flask cultivations by the technique of gas chromatography. As a result it was find out that C. bifermentans are able to oxygen utilization. In Figure 2 the kinetics of oxygen utilization by investigated C. bifermentans strains were confront with the kinetics of 1,3-PD production. It was observed that the ability to 1,3-PD synthesis is strictly dependent on the level of oxygen in the fermentation environment. The necessary condition to begin the main metabolite of glycerol pathway production by tested strains is limitation of oxygen below 10%. This observation confirmed the earlier conclusion that bacteria were not able to 1,3-PD synthetize in bioreactor cultivations with aeration on the level of 10% of oxygen (Figure 3).
The important aero-tolerance factors in microorganisms are enzymes, such as peroxidase, superoxide dismutase, and catalase. Thus, the protein profile of C. bifermentans strains was tested by tandem mass spectrometry. Bacteria were cultivated in microaerophilic conditions (CampyGen) in 500 ml flasks. It was find out that despite the presence of oxygen during bacteria cultivation, the typical enzymes taking part in glycerol pathway were detected (Table 2). Also the enzyme responsible for catalyzing the dismutation of superoxide into oxygen and hydrogen peroxide were found (Table 2).
4. Discussion
The natural environment is a rich source of strains with high metabolic potential, which might be used in many branches of industry. Some of them evaluated to adapt to conditions in which they live. An example of such
Table 2. Proteins isolated from C. bifermentans cells.
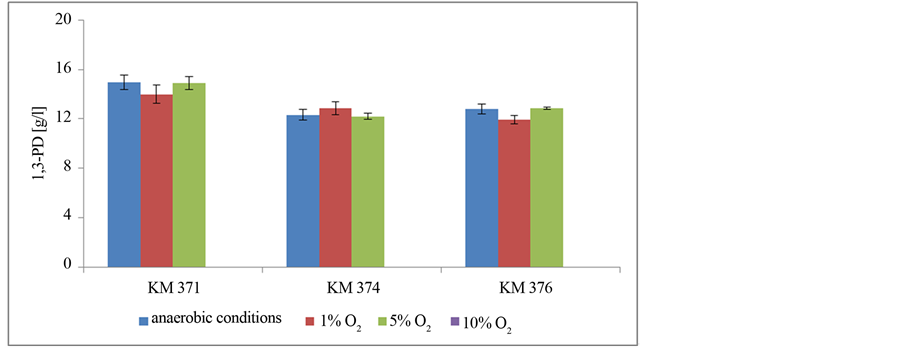
Figure 3. The level of oxygen tolerance of C. bifermentans strains presents as an ability to 1,3-PD production in different conditions.
evolution is an aero-tolerance of strains generally recognized as strictly anaerobe. The aero-tolerance of strains is a very important issue from the industrial point of view. It caused that application of microorganisms in a large production scale is easier and chemical synthesis of varied chemical compounds may be substitute by biotechnological way [26] [27] .
Bacteria from Clostridium genera exist commonly in the natural environment, for example in dust, soil [28] , water, sediments and in the alimentary canal in people and animals [29] . They play a key role in many solvents (among other butyrate, acetate, and iso-propanole) [30] -[32] , organic acids (acetic, fumaric, succinic, lactic) [32] , diols (1,3-PD, 2,3-butanediol) [33] [34] and gases (carbon dioxide, methanol, hydrogen) [35] [36] production.
In the work, the aero-tolerance of C. bifermentans strains isolated from the probes obtained from the natural environment [25] was investigated. The isolation and selection processes of these strains were described in earlier publications [25] [37] . The authors investigated the not-known yet ability of this species to 1,3-PD synthesis. During these researches it was find out that despite the fact that bacteria belong to the Clostridium genera are strictly anaerobic, obtained isolates proved some tolerance to the oxygen.
In the first step of this work, the redox potential of bacteria cells, cultivated both in anaerobic and microaerophilic conditions, was compared. The aim of this step was to confirm assumption that C. bifermentans strains are able to maintain the metabolic activity when oxygen is presents in the fermentation environment. As a result it was found out the oxygen did not significantly influence on the metabolism of bacteria cells and it was the foundation for further experiments. Also other scientists used the redox potential to measure the metabolic activity of bacteria cells [38] [39] . Also the ability of 1,3-PD, the main metabolite of glycerol pathway, production in microaerophilic was tested and the results were compared with the results obtained in anaerobic fermentations. It occurred that independently on cultivation conditions, the concentration of produced metabolite is on the same level. This observation contributed the earlier thesis that investigated bacteria are able to survive and maintain the metabolic activity in the presence of oxygen. The synthesis of 1,3-PD by C. bifermentans in microaerophilic conditions is very important issue, because there is no literature data about the ability of bacteria from the Clostridium genera to 1,3-PD production in microaerophilic conditions. But we can find information about microaerophilic synthesis of 1,3-PD by among other some strains belong to Lactobacillus (eg. Lactobacillus diolivorans) [40] , Klebsiella (eg., Klebsiella pneumoniae, Klebsiella oxytoca) [41] , and Pediococcus (eg., Pediococcus pentosaceus) [40] genera.
Despite the fact that generally bacteria C. bifermentans are regarded as strictly anaerobic [18] -[21] , in the literature there are some data about their tolerance to oxygen [22] [23] . Gibs [23] stated that in the presence of oxygen, the sporulation process of C. bifermentans is more effective (even when the oxygen is on the atmospheric level). Hewitt and Morris [2] detected in C. bifermentans cells the antioxidative enzyme—superoxide dismutase and Kawasaki et al. [24] detected peroxidase NADH/NADPH. Researches carried out by Admassu et al. [22] indicated that C. bifermentans maintained metabolic activity in the presence of oxygen. However, there is no information about the concentration of oxygen which they tested. Thus, the authors decided to check the level of oxygen tolerance. In this aim, bacteria were cultivated in bioreactors with aeration on the level of 1%, 5%, and 10% of oxygen. As a result of conducing maintaining of metabolic activity of bacteria cells, concentration of synthetized 1,3-PD was tested. It was found out that in 1% and 5% of oxygen 1,3-PD was normally produced, but in cultivation with 10% of oxygen synthesis of metabolite was inhibited, because the metabolic enzymes associated with the detected metabolites proteins were inactive. Because the key role in microorganism aero-tolerance plays enzymes, such as superoxide dismutase, catalase, peroxidase [7] [9] , the protein profile of investigated strains were checked. These analyses conducted the thesis presented by Admassu et al. [22] that in C. bifermentans superoxide dismutase, taking part in antioxidation reactions, is present. The very important thing is also the fact that despite the presence of oxygen during bacteria cultivations, the main enzyme of glycerol pathway was detected. Moreover, in the literature data there is no information about protein profile of C. bifermentans. Only two papers demonstrated some information about the presence of cells low-molecular proteins located in inactive spores of C. bifermentans [42] . The common mechanism of aero-tolerance is also the ability to oxygen consumption, thus it was also checked in investigated strains. As a result, it was found out that investigated bacteria are able to consume oxygen. Moreover, the limitation of oxygen below 10% is a necessary condition to beginning synthesis of 1,3-PD.
The mechanisms which protect live cells against toxic by-products generated during uncompleted decomposition of oxygen were formed in evolution processes. They are indispensable for further survive of particular anaerobic bacteria species. Generally, these mechanisms have not been precisely described. C. bifermentans is able to survive in presence of oxygen. It also produces metabolites in microaerophilic conditions. The aero-tolerance mechanisms of this species based mainly on the ability to oxygen utilization and the presence of superoxide dismutase in bacteria cells.
Acknowledgements
The paper was prepared within the framework of project PO IG 01.01.02-00-074/09, co-funded by The European Union from The European Regional Development Fund within the framework of the Innovative Economy Operational Programme 2007-2013.
References
- Brioukhanov, A.L. and Netrusov, A.I. (2007) Aerotolerance of Strictly Anaerobic Microorganisms and Factors of Defense against Oxidative Stress: A Review. Applied Biochemistry and Microbiology, 43, 637-654. http://dx.doi.org/10.1134/S0003683807060014
- Hewitt, J. and Morris, J.G. (1975) Superoxide Dismutase in Some Obligately Anaerobic Bacteria. FEBS Letters, 50, 315-318. http://dx.doi.org/10.1016/0014-5793(75)90058-7
- Brioukhanov, A.L., Thauer, R.K. and Netrusov, A.I. (2002) Catalase and Superoxide Dismutase in the Cells of Strictly Anaerobic Microorganisms. Journal of Medical Microbiology, 71, 330-335.
- Karnholz, A., Kusel, K., Gossner, A., Schramm, A. and Drake, H.L. (2002) Tolerance and Metabolic Response of Acetogenic Bacteria toward Oxygen (O2). Applied and Environmental Microbiology, 2, 1005-1009. http://dx.doi.org/10.1128/AEM.68.2.1005-1009.2002
- Rocha, E.R., Selby, T., Coleman, J.P. and Smith, C.J. (1995) Oxidative Stress Response in an Anaerobe, Bacteroides fragilis: A Role for Catalase in Protection against Hydrogen Peroxide. Journal of Bacteriology, 178, 6895-6903.
- Cypionka, H. (2000) Oxygen Respiration by Desulfovibrio species. Annual Review of Microbiology, 54, 827-848. http://dx.doi.org/10.1146/annurev.micro.54.1.827
- Hillmann, F., Riebe, O., Fischer, R.J., Mot, A., Caranto, J.D., Kurtz, D.M. and Bahl, H. (2009) Reductive Dioxygen Scavenging by Flavo-Diiron Proteins of Clostridium Acetobutylicum. FEBS Letters, 583, 241-245. http://dx.doi.org/10.1016/j.febslet.2008.12.004
- Jędrzejczak-Krzepkowska, M. and Bielecki, S. (2011) Bifidobacteria and Fructane Stimulating Their Growth. Postępy Biochemii, 57, 1-9 (in Polish).
- Lombard, M., Fontecave, M., Touati, D. and Nivière, V. (2002) Reaction of the Desulfoferrodoxin from Desulfoarculus baarsii with Superoxide Anion Evidence for a Superoxide Reductase Activity. Journal of Biological Chemistry, 275, 115-121. http://dx.doi.org/10.1074/jbc.275.1.115
- Rolfe, R.D., Hentges, D.J., Campbell, B.J. and Barrett, J.T. (1978) Factors Related to the Oxygen Tolerance of Anaerobic Bacteria. Applied and Environmental Microbiology, 36, 306-313.
- Kawasaki, S., Watamura, Y., Ono, M., Watanabe, T., Takeda, K. and Niimura, Y. (2005) Adaptive Responses to Oxygen Stress in Obligatory Anaerobes Clostridium acetobutylicum and Clostridium aminovalericum. Applied and Environmental Microbiology, 71, 8442-8450. http://dx.doi.org/10.1074/jbc.275.1.115
- Morris, J.G. (1976) Oxygen and the Obligatory Anaerobe. Journal of Applied Bacteriology, 40, 229-244. http://dx.doi.org/10.1111/j.1365-2672.1976.tb04171.x
- Nakayama, K. (1990) The Superoxide Dismutase-Encoding Gene of the Obligately Anaerobic Bacterium Bacteroides gingivalis. Gene, 96, 149-150. http://dx.doi.org/10.1016/0378-1119(90)90357-W
- Meile, L., Fischer, K. and Leisinger, T. (2005) Characterization of the Superoxide Dismutase Gene and Its Upstream Region from Methanobacterium thermoautotrophicum Marburg. FEMS Microbioogyl Letters, 128, 247-253. http://dx.doi.org/10.1111/j.1574-6968.1995.tb07532.x
- Takao, M., Oikawa, A. and Yasui, A. (1990) Characterization of a Superoxide Dismutase Gene from the Archaebacterium Methanobacterium thermoautotrophicum. Archivum of Biochemistry and Biophysis, 283, 210-216. http://dx.doi.org/10.1016/0003-9861(90)90633-A
- Shenvi, N.V. and Kurtz, D.M. (1977) Desulfovibrio vulgaris Superoxide Dismutase (sod) Gene. Complete eds, EMBL/GenBank Direct Submission. Accession number AF034841,
- Geissmann, T.A., Teuber, M. and Meile, L. (1999) Transcriptional Analysis of the Rubrerythrin and Superoxide Dismutase Genes of Clostridium perfringens. Journal of Bacteriology, 181, 7136-7139.
- Archer, R.H. Maddox, I.S. and Chong, R. (1982) Transformation of Cholic Acid by Clostridium bifermentans. Journal of Applied Microbiology, 52, 49-56.
- Chamkha, M., Patel, B.K.C., Garcia, J.L. and Labata, M. (2001) Isolation of Clostridium bifermentans from Oil Mill Wastewaters Converting Cinnamic Acid to 3-Phenylpropionic Acid and Emendation of the Species. Anaerobe, 7, 189-197. http://dx.doi.org/10.1006/anae.2001.0382
- Verhulst, A., Semjen, G., Meerts, U., Janssen, G., Parmentier, G., Asselberghs, S. and van Hespen, H. (1985) Biohydrogenation of Linoleic Acid by Clostridium sporogenes, Clostridium bifermentans, Clostridium sordellii and Bacteroides sp. FEMS Microbiology Letters, 31, 255-259. http://dx.doi.org/10.1111/j.1574-6968.1985.tb01157.x
- Zhang, S., Kim, T.H., Lee, Y. and Hwang, S.J. (2012) Effects of VFAs Concentration on Bio-Hydrogen Production with Clostridium bifermentans 3AT-ma. Energy Procedia, 14, 518-523. http://dx.doi.org/10.1016/j.egypro.2011.12.968
- Admassu, W., Sethuraman, A.V., Crawford, R. and Korus, R.A. (1998) Growth Kinetics of Clostridium bifermentans and Its Ability to Degrade TNT Using an Inexpensive Alternative Medium. Bioremediation Journal, 2, 17-28. http://dx.doi.org/10.1080/10889869891214187
- Gibbs, P.A. (1964) Factors Affecting the Germination of Spores of Clostridium bifermentans. Microbiology, 37, 41-48. http://dx.doi.org/10.1099/00221287-37-1-41
- Kawasaki, S., Tomoyuki, N., Yoshitaka, N., Yoshimi, B., Tai, U., Kazuo, K., Michio, K. and Youichi, N. (1998) Effect of Oxygen on the Growth of Clostridium butyricum (Type Species of the Genus Clostridium), and the Distribution of Enzymes for Oxygen and for Active Oxygen Species in Clostridia. Journal of Fermentation and Bioengineering, 86, 368-372. http://dx.doi.org/10.1016/S0922-338X(99)89006-0
- Myszka, K., Leja, K., Olejnik-Schmidt, A.K. and Czaczyk, K. (2012) Isolation Process of Industrially Useful Clostridium bifermentans from Natural Samples. Journal of Bioscience and Bioengineering, 113, 631-633. http://dx.doi.org/10.1016/j.jbiosc.2012.01.003
- Kaeberlein, T., Lewis, K. and Epstein, S.S. (2002) Isolating “Uncultivable” Microorganisms in Pure Culture in a Simulated Natural Environment. Science, 296, 1127-1129. http://dx.doi.org/10.1126/science.1070633
- Nicol, R.W., Marchand, K. and Lubitz, W.D. (2012) Bioconversion by Crude Glycerol by Fungi. Applied Microbiology and Biotechnology, 93, 1865-1875. http://dx.doi.org/10.1007/s00253-012-3921-7
- Cremonesi, P., Vanoni, L., Silvetti, T., Morandi, S. and Brasca, M. (2012) Identification of Clostridium beijerinckii, C butyricum, C sporogenes, C tyrobutyricum Isolated from Silage, Raw Milk and Hard Cheese by a Multiplex PCR Assay. Journal of Dairy Research, 79, 318-323. http://dx.doi.org/10.1017/S002202991200026X
- Meier, T.R., Myers, D.D., Ko, E.K.A., Holden, M. and Claire, H.F. (2007) Gangrenous Clostridium perfringens Infection and Subsequent Wound Management in a Rhesus Macaque (Macaca mulatta). Journal of the American Association for Laboratory Animal Science, 46, 68-73.
- Beckers, L., Hiligsmann, S., Lambert, S.D., Heinrichs, B. and Thonart, P. (2013) Improving Effect of Metal and Oxide Nanoparticles Encapsulated in Porous Silica on Fermentative Biohydrogen Production by Clostridium butyricum. Bioresource Technology, 133, 109-117. http://dx.doi.org/10.1016/j.biortech.2012.12.168
- Buckel, W. (2005) Special Clostridial Enzymes and Fermentation Pathways. In: Buckel, W., Ed., Handbook on Clostridia, CRC Press LLC, Boca Raton, 81-83. http://dx.doi.org/10.1201/9780203489819.ch9
- Yang, X., Tu, M., Xie, R., Adhikari, S. and Tong, Z. (2013) A Comparison of Three pH Control Methods for Revealing Effects of Undissociated Butyric Acid on Specific Butanol Production Rate in Batch Fermentation of Clostridium acetobutylicum. AMB Express, 3, 3-6. http://dx.doi.org/10.1186/2191-0855-3-3
- Collas, F., Kuit, W., Clément, B., Marchal, R., López-Contreras, A.M. and Monot, F. (2012) Simultaneous Production of Isopropanol, Butanol, Ethanol and 2,3-Butanediol by Clostridium acetobutylicum ATCC 824 Engineered Strains. AMB Express, 2, 45-47. http://dx.doi.org/10.1186/2191-0855-2-45
- Wilkens, E., Ringel, A.K., Hortig, D., Willke, T. and Vorlop, K.D. (2012) High-Level Production of 1,3-Propanediol from Crude Glycerol by Clostridium butyricum AKR102a. Applied Microbiology and Biotechnology, 93, 1057-1063. http://dx.doi.org/10.1007/s00253-011-3595-6
- Huyen, N.T.T., Yen, D.T., Yen, N.T., Nga, V.T. and Hien, L.T. (2012) Using of Response Surface Methodology for Optimization of Biohydrogen Production by Clostridium spp. Isolated in Vietnam. Journal of Biology, 43, 1-12.
- Kubiak, P., Leja, K., Myszka, K., Celińska, E., Spychała, M., Powałowska-Szymanowska, D., Czaczyk, K. and Grajek, W. (2012) Physiological Predisposition of Various Clostridium Species to Synthetize 1,3-Propanediol from Glycerol. Process Biochemistry, 47, 1308-1319. http://dx.doi.org/10.1016/j.procbio.2012.05.012
- Leja, K., Myszka, K., Kubiak, P., Wojciechowska, J., Olejnik-Schmidt, A.K., Czaczyk, K. and Grajek, W. (2011) Isolation and Identification of Clostridium spp from Natural Samples that Performs Effective Conversion of Glycerol to 1,3-Propanediol. Acta Scientarum Polonorum-Biotechnologia, 10, 25-34.
- Braeckman, B.P., Houthoofd, K., Vreese, A.D. and Vanfleteren, J.R. (2002) Assaying Metabolic Activity in Ageing Caenorhabditis elegans. Mechanisms of Ageing and Development, Genetic Effects on Aging III, 123, 105-119.
- Springer, J.E., Azbill, R.D. and Carlson, S.L. (1998) A Rapid and Sensitive Assay for Measuring Mitochondrial Metabolic Activity in Isolated Neural Tissue. Brain Research Protocols, 2, 259-263. http://dx.doi.org/10.1016/S1385-299X(97)00045-7
- Pflügl, S., Marx, H., Mattanovich, D. and Sauer, M. (2012) 1,3-Propanediol Production from Glycerol with Lactobacillus diolivorans. Bioresource Technology, 119, 133-140. http://dx.doi.org/10.1016/j.biortech.2012.05.121
- Marçal1, D., Rêgo, A.T., Carrondo, M.A. and Enguita, F.J. (2008) 1,3-Propanediol Dehydrogenase from Klebsiella pneumoniae: Decameric Quaternary Structure and Possible Subunit Cooperativity. Journal of Bacteriology, 191, 1143- 1151. http://dx.doi.org/10.1128/JB.01077-08
- Setlow, P. and Waites, W.M. (1976) Identification of Several Unique, Low-Molecular-Weight Basic Proteins in Dormant Spores of Clostridium bifermentans and Their Degradation during Spore Germination. Journal of Bacteriology, 127, 1015-1017.
NOTES

*Corresponding author.


